Comparative Metabolomics to Unravel the Biochemical Mechanism Associated with Rancidity in Pearl Millet (Pennisetum glaucum L.)
Abstract
:1. Introduction
2. Results
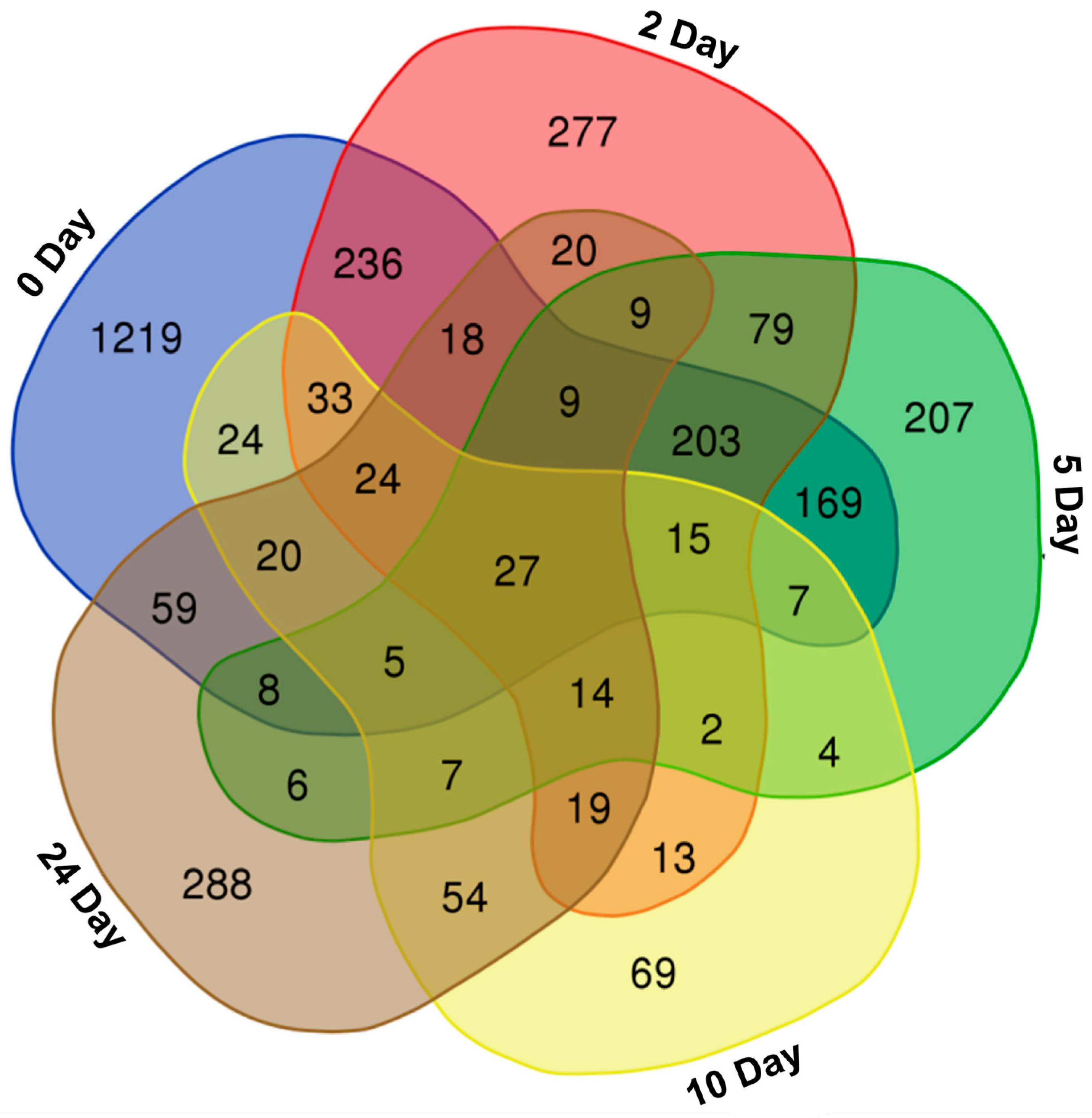
2.1. Differential Accumulation of Metabolites in Flours of Pear Millet Genotypes
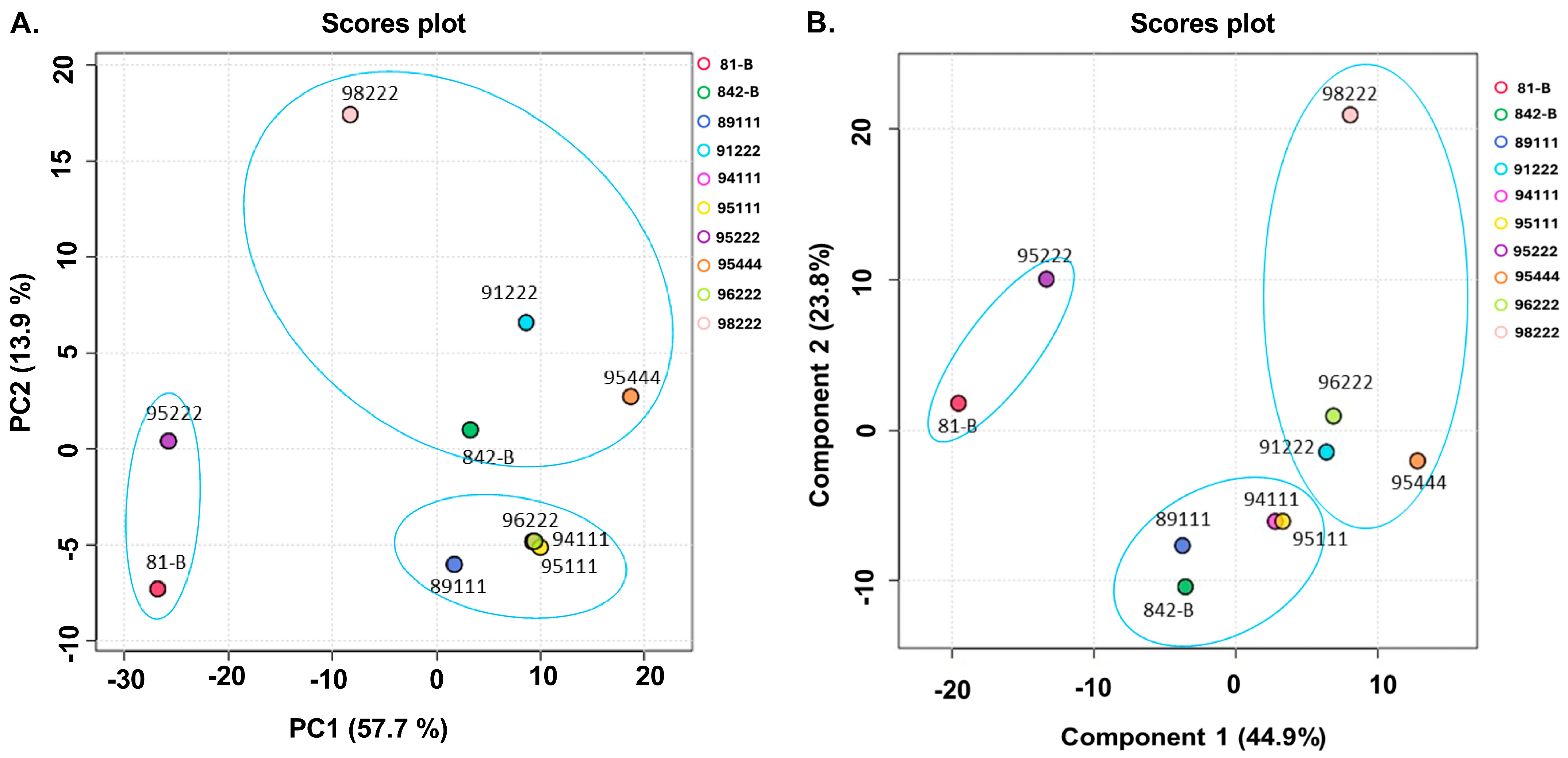
2.2. Classification of Pearl Millet Genotypes Based on Principal Component Analysis (PCA) and Partial Least Squares-Discriminant Analysis (PLS-DA)
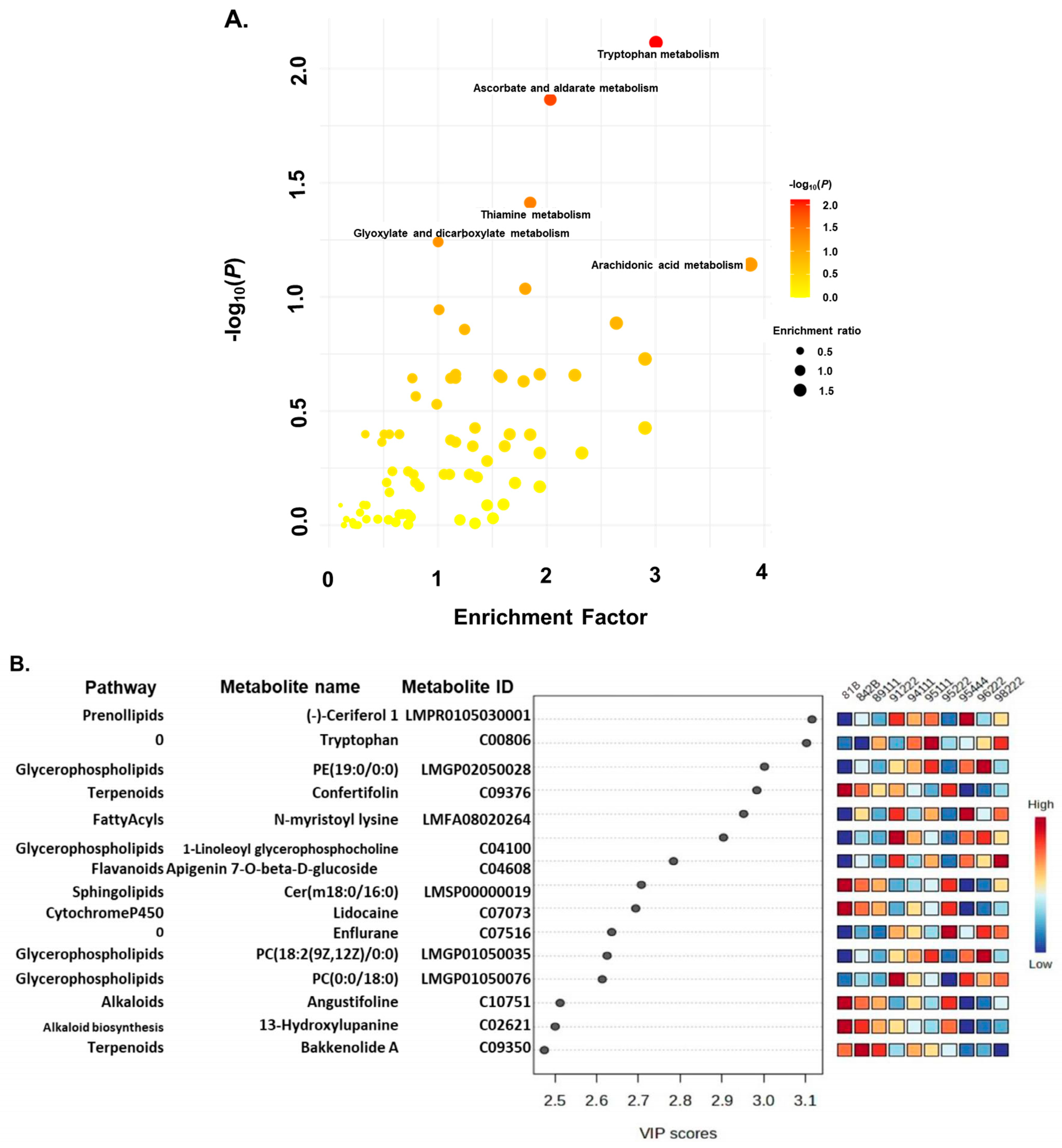
2.3. Pathway Enrichment Analysis
2.4. Identification of Metabolites Associated with Rancidity
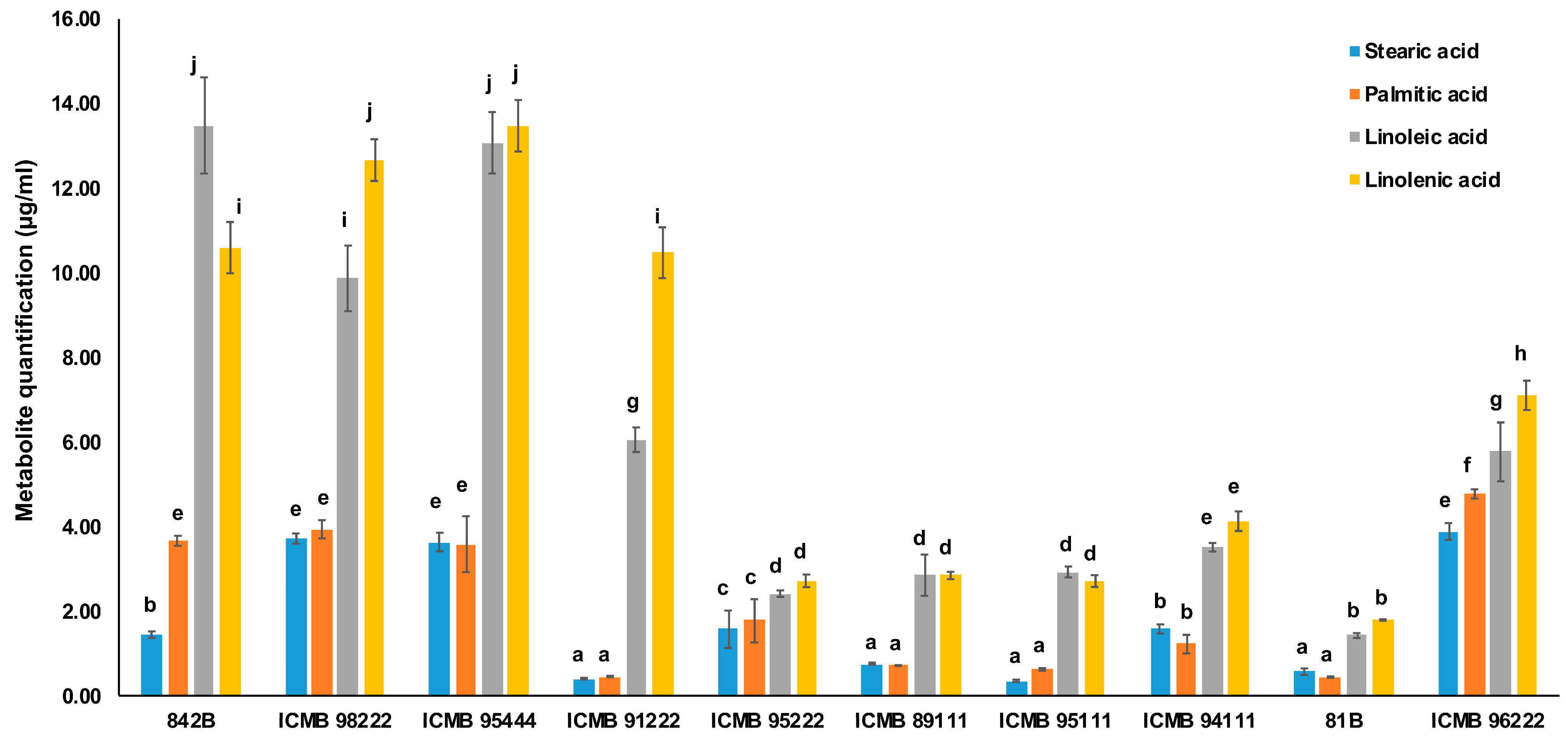
2.5. Validation of Key Metabolites Implicated in the Process of Rancidity
3. Discussion
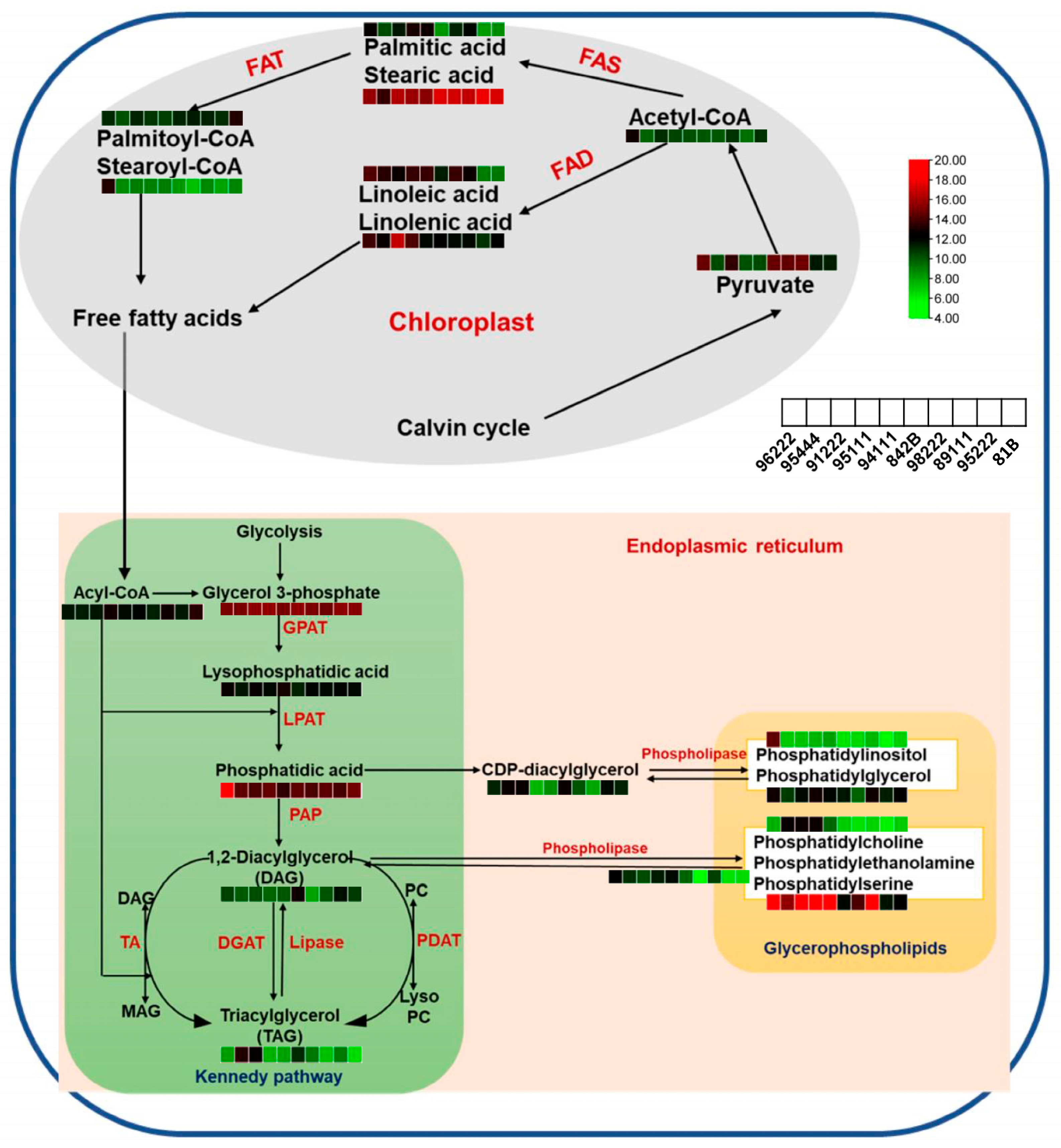
3.1. Rancidity in Pearl Millet Is Associated with the Presence of Metabolites Derived from Fatty Acids
3.2. Pearl Millet Rancidity Is Associated with the Presence of Flavonoid-Related Metabolites
4. Materials and Methods
4.1. Plant Genotypes and Storage Conditions
4.2. Metabolite Extraction and LC-High-Resolution MS (LC-HRMS) Analysis
4.3. Metabolomic Data Analysis
4.4. Estimation of Saturated and Unsaturated Fatty Acids Using LC-MS
4.4.1. Extraction
4.4.2. Quantification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. 2020. Available online: http://www.fao.org/faostat/en/#data (accessed on 24 March 2024).
- Mahendrakar, M.D.; Kumar, S.; Singh, R.B.; Rathore, A.; Potupureddi, G.; Kishor, P.K.; Gupta, R.; Srivastava, R.K. Genetic variability, genotype× environment interaction and correlation analysis for grain iron and zinc contents in recombinant inbred line population of pearl millet [Pennisetum glaucum (L). R. Br.]. Indian J. Genet. Plant Breed. 2019, 79, 545–551. [Google Scholar] [CrossRef]
- Srivastava, R.K.; Satyavathi, C.T.; Mahendrakar, M.D.; Singh, R.B.; Kumar, S.; Govindaraj, M.; Ghazi, I.A. Addressing iron and zinc micronutrient malnutrition through nutrigenomics in pearl millet: Advances and prospects. Front. Genet. 2021, 12, 723472. [Google Scholar] [CrossRef] [PubMed]
- Goswami, S.; Asrani, P.; Ansheef Ali, T.; Kumar, R.D.; Vinutha, T.; Veda, K.; Kumari, S.; Sachdev, A.; Singh, S.P.; Satyavathi, C.T. Rancidity matrix: Development of biochemical indicators for analysing the keeping quality of pearl millet flour. Food Anal. Methods 2020, 13, 2147–2164. [Google Scholar] [CrossRef]
- Aher, R.R.; Reddy, P.S.; Bhunia, R.K.; Flyckt, K.S.; Shankhapal, A.R.; Ojha, R.; Everard, J.D.; Wayne, L.L.; Ruddy, B.M.; Deonovic, B. Loss-of-function of triacylglycerol lipases are associated with low flour rancidity in pearl millet [Pennisetum glaucum (L.) R. Br.]. Front. Plant Sci. 2022, 13, 962667. [Google Scholar] [CrossRef]
- Selvan, S.S.; Mohapatra, D.; Kate, A.; Kar, A.; Modhera, B. Mapping and analysis of volatomes from pearl millet (Pennisetum glaucum L.) grains during different storage conditions with solid-phase microextraction–gas chromatography–mass spectrometry. Cereal Chem. 2023, 100, 1114–1122. [Google Scholar] [CrossRef]
- Goswami, S.; Kumar, R.R.; Praveen, S. Hydrolytic and oxidative decay of lipids: Biochemical markers for rancidity measurement in pearl millet flour. Omics Meet Plant Biochem. Appl. Nutr. Enhanc. One Health Perspect. 2019, 221, 147–149. [Google Scholar]
- Bhargavi, H.; Singh, S.P.; Goswami, S.; Yadav, S.; Aavula, N.; Shashikumara, P.; Singhal, T.; Sankar, S.M.; Danakumara, T.; Hemanth, S. Deciphering the genetic variability for biochemical parameters influencing rancidity of pearl millet (Pennisetum glaucum LR Br.) Flour in a set of highly diverse lines and their categorization using rancidity matrix. J. Food Compos. Anal. 2024, 128, 106035. [Google Scholar] [CrossRef]
- Goyal, P.; Chugh, L.K. Partial purification and characterization of peroxidase from pearl millet (Pennisetum glaucum [L.] R. Br.) grains. J. Food Biochem. 2014, 38, 150–158. [Google Scholar] [CrossRef]
- Boncompagni, E.; Nielsen, E.; Sanogo, M.D.; Coghetto, V.; Temporiti, M.E.; Daglia, M.; Di Lorenzo, A.; Doria, E. Anti-nutritional metabolites in six traditional African cereals. J. Food Nutr. Res. 2019, 58, 115. [Google Scholar]
- Ali, A.; Kumar, R.R.; Bansal, N.; Bollinedi, H.; Singh, S.P.; Satyavathi, C.T.; Praveen, S.; Goswami, S. Characterization of biochemical indicators and metabolites linked with rancidity and browning of pearl millet flour during storage. J Plant Biochem Biotech. 2023, 32, 121–131. [Google Scholar] [CrossRef]
- Deepak, Y.; Chhabra, A.; Laxman, C.; Behl, R. Genotypic variability for rancidity and its association with morphological and biochemical traits in pearl millet [Pennisetum Glaucum (L.) R. Br.]. Ann. Biol. 2010, 26, 9–12. [Google Scholar]
- Grebenteuch, S.; Kanzler, C.; Klaußnitzer, S.; Kroh, L.W.; Rohn, S. The formation of methyl ketones during lipid oxidation at elevated temperatures. Molecules. 2021, 26, 1104. [Google Scholar] [CrossRef] [PubMed]
- Hamany Djande, C.Y.; Pretorius, C.; Tugizimana, F.; Piater, L.A.; Dubery, I.A. Metabolomics: A tool for cultivar phenotyping and investigation of grain crops. Agronomy 2020, 10, 831. [Google Scholar] [CrossRef]
- Mazumdar, S.; Gupta, S.; Banerjee, R.; Gite, S.; Durgalla, P.; Bagade, P. Determination of variability in rancidity profile of select commercial pearl millet varieties/hybrids. In Proceedings of the CGIAR Research Program on Dryland Cereals Review Meeting, Hyderabad, India, 5–6 October 2016. [Google Scholar]
- De Vos, R.C.; Moco, S.; Lommen, A.; Keurentjes, J.J.; Bino, R.J.; Hall, R.D. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2007, 2, 778–791. [Google Scholar] [CrossRef] [PubMed]
- Avuthu, T.; Sanivarapu, H.; Prasad, K.; Sharma, N.; Sudini, H.K.; Yogendra, K. Comparative metabolomics analysis reveals secondary cell wall thickening as a barrier to resist Aspergillus flavus infection in groundnut. Physiol. Plant 2024, 176, e14169. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Takahashi, H.; Suzuki, H.; Suda, K.; Yamazaki, Y.; Takino, A.; Kim, Y.-I.; Goto, T.; Iijima, Y.; Aoki, K.; Shibata, D. Long-chain free fatty acid profiling analysis by liquid chromatography–mass spectrometry in mouse treated with peroxisome proliferator-activated receptor α agonist. Biosci. Biotechnol. Biochem. 2013, 77, 2288–2293. [Google Scholar] [CrossRef]
- Hummel, J.; Segu, S.; Li, Y.; Irgang, S.; Jueppner, J.; Giavalisco, P. Ultra performance liquid chromatography and high resolution mass spectrometry for the analysis of plant lipids. Front. Plant Sci. 2011, 2, 54. [Google Scholar] [CrossRef]
- Padmaja, P.; Kalaisekar, A.; Venkateswarlu, R.; Shwetha, S.; Rao, B.D.; Tonapi, V. Thermal treatment in combination with laminated packaging under modified atmosphere enhances the shelf life of pearl millet flour. Food Chem. Adv. 2023, 2, 100190. [Google Scholar] [CrossRef]
- Satyavathi, C.T.; Praveen, S.; Mazumdar, S.; Chugh, L.; Kawatra, A. Enhancing Demand of Pearl Millet as Super Grain. 2017. Available online: http://www.aicpmip.res.in/Enhancing_Demand_of_Pearlmillet_as_Super_Grain.pdf (accessed on 10 February 2024).
- Chen, Z.; Chen, H.; Jiang, Y.; Wang, J.; Khan, A.; Li, P.; Cao, C. Metabolomic analysis reveals metabolites and pathways involved in grain quality traits of high-quality rice cultivars under a dry cultivation system. Food Chem. 2020, 326, 126845. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Wang, R.; Lu, W.; Zhu, J.; Zhang, H.; Xiong, Q.; Zhou, N. Metabolomics analysis of variation in grain quality of high-quality japonica rice. Agronomy 2024, 14, 430. [Google Scholar] [CrossRef]
- Shi, T.; Zhu, A.; Jia, J.; Hu, X.; Chen, J.; Liu, W.; Ren, X.; Sun, D.; Fernie, A.R.; Cui, F. Metabolomics analysis and metabolite-agronomic trait associations using kernels of wheat (Triticum aestivum) recombinant inbred lines. Plant J. 2020, 103, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Heiniö, R.; Lehtinen, P.; Oksman-Caldentey, K.; Poutanen, K. Differences between sensory profiles and development of rancidity during long-term storage of native and processed oat. Cereal Chem. 2002, 79, 367–375. [Google Scholar] [CrossRef]
- Franklin, L.M.; Mitchell, A.E. Review of the Sensory and Chemical Characteristics of Almond (Prunus dulcis) Flavor. J. Agric. Food Chem. 2019, 67, 2743–2753. [Google Scholar] [CrossRef]
- Moin, M.; Bommineni, P.R.; Tyagi, W. Exploration of the pearl millet phospholipase gene family to identify potential candidates for grain quality traits. BMC Genom. 2024, 25, 581. [Google Scholar] [CrossRef]
- Ghorbani Gorji, S.; Calingacion, M.; Smyth, H.E.; Fitzgerald, M. Effect of natural antioxidants on lipid oxidation in mayonnaise compared with BHA, the industry standard. Metabolomics 2019, 15, 106. [Google Scholar] [CrossRef]
- Taylor, J.R. Sorghum and Millets: Taxonomy, History, Distribution, and Production. In Sorghum and Millets; Elsevier: Amsterdam, The Netherlands, 2019; pp. 1–21. [Google Scholar]
- Sharma, M.; Yadav, D.N.; Singh, A.K.; Tomar, S.K. Rheological and functional properties of heat moisture treated pearl millet starch. J. Food Sci. Technol. 2015, 52, 6502–6510. [Google Scholar] [CrossRef]
- Nakayama, Y.; Saio, K.; Kito, M. Decomposition of phospholipids in soybean during storage. Cereal Chem. 1981, 58, 260–264. [Google Scholar]
- List, G.; Mounts, T.; Lanser, A. Factors promoting the formation of nonhydratable soybean phosphatides. J. Am. Oil Chem. 1992, 69, 443–446. [Google Scholar] [CrossRef]
- Kaur, A.; Neelam, K.; Kaur, K.; Kitazumi, A.; de Los Reyes, B.G.; Singh, K. Novel allelic variation in the phospholipase d alpha 1 gene (OsPLDα1) of wild oryza species implies to its low expression in rice bran. Sci. Rep. 2020, 10, 6571. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, W.; Martin, J.J.J.; Qadri, R.; Fu, X.; Feng, M.; Wei, L.; Zhang, A.; Yang, C.; Cao, H. Differential analysis of transcriptomic and metabolomic of free fatty acid rancidity process in oil palm (Elaeis guineensis) fruits of different husk types. Front. Plant Sci. 2023, 14, 1132024. [Google Scholar] [CrossRef] [PubMed]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An Overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef] [PubMed]
- Simunkova, M.; Barbierikova, Z.; Jomova, K.; Hudecova, L.; Lauro, P.; Alwasel, S.H.; Alhazza, I.; Rhodes, C.J.; Valko, M. Antioxidant vs. prooxidant properties of the flavonoid, kaempferol, in the Presence of Cu (II) Ions: A ROS-scavenging activity, Fenton reaction and DNA damage study. Int. J. Mol. Sci. 2021, 22, 1619. [Google Scholar] [CrossRef]
- Chaaban, H.; Ioannou, I.; Paris, C.; Charbonnel, C.; Ghoul, M. The photostability of flavanones, flavonols and flavones and evolution of their antioxidant activity. J. Photochem. Photobiol. A 2017, 336, 131–139. [Google Scholar] [CrossRef]

| Name | Molecular Formula | Exact Mass (Da) | Retention Time (min) | Observed Mass (Da) |
|---|---|---|---|---|
| Palmitic acid (PA, C16:0) | C16H32O2 | 256.24023 | 1.94 | 255.23 [M−H] |
| Stearic acid (SA, C18:0) | C18H36O2 | 284.27153 | 2.94 | 283.26 [M−H] |
| Linoleic acid (LIA, C18:2) | C18H32O2 | 280.24023 | 1.47 | 279.23 [M−H] |
| Linolenic acid (LNA, C18:3) | C18H30O2 | 278.22458 | 1.18 | 277.22 [M−H] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yogendra, K.; Sanivarapu, H.; Avuthu, T.; Gupta, S.K.; Durgalla, P.; Banerjee, R.; Raman, A.; Tyagi, W. Comparative Metabolomics to Unravel the Biochemical Mechanism Associated with Rancidity in Pearl Millet (Pennisetum glaucum L.). Int. J. Mol. Sci. 2024, 25, 11583. https://doi.org/10.3390/ijms252111583
Yogendra K, Sanivarapu H, Avuthu T, Gupta SK, Durgalla P, Banerjee R, Raman A, Tyagi W. Comparative Metabolomics to Unravel the Biochemical Mechanism Associated with Rancidity in Pearl Millet (Pennisetum glaucum L.). International Journal of Molecular Sciences. 2024; 25(21):11583. https://doi.org/10.3390/ijms252111583
Chicago/Turabian StyleYogendra, Kalenahalli, Hemalatha Sanivarapu, Tejaswi Avuthu, Shashi Kumar Gupta, Priyanka Durgalla, Roopa Banerjee, Anitha Raman, and Wricha Tyagi. 2024. "Comparative Metabolomics to Unravel the Biochemical Mechanism Associated with Rancidity in Pearl Millet (Pennisetum glaucum L.)" International Journal of Molecular Sciences 25, no. 21: 11583. https://doi.org/10.3390/ijms252111583
APA StyleYogendra, K., Sanivarapu, H., Avuthu, T., Gupta, S. K., Durgalla, P., Banerjee, R., Raman, A., & Tyagi, W. (2024). Comparative Metabolomics to Unravel the Biochemical Mechanism Associated with Rancidity in Pearl Millet (Pennisetum glaucum L.). International Journal of Molecular Sciences, 25(21), 11583. https://doi.org/10.3390/ijms252111583







