Abstract
Metabolic bone diseases cover a broad spectrum of disorders that share alterations in bone metabolism that lead to a defective skeleton, which is associated with increasing morbidity, disability, and mortality. There is a close connection between the etiology of metabolic bone diseases and genetic factors, with TP53 being one of the genes associated therewith. The single nucleotide polymorphism (SNP) Arg72Pro of TP53 is a genetic factor associated with several pathologies, including cancer, stroke, and osteoporosis. Here, we aim to analyze the influence of the TP53 Arg72Pro SNP on bone mass in humanized Tp53 Arg72Pro knock-in mice. This work reports on the influence of the TP53 Arg72Pro polymorphism in bone microarchitecture, OPG expression, and apoptosis bone status. The results show that the proline variant of the TP53 Arg72Pro polymorphism (Pro72-p53) is associated with deteriorated bone tissue, lower OPG/RANK ratio, and lower apoptosis in bone tissue. In conclusion, the TP53 Arg72Pro polymorphism modulates bone microarchitecture and may be a genetic biomarker that can be used to identify individuals with an increased risk of suffering metabolic bone alterations.
1. Introduction
Metabolic bone diseases cover a broad spectrum of disorders that share alterations in bone metabolism that lead to a defective skeleton [1]. The most prevalent metabolic bone disease worldwide is osteoporosis (OMIM: 166710), which is a chronic, progressive, systemic disease associated with reduced bone mineral density (BMD) and alterations in the microarchitecture of bone tissue [2,3]. The underlying mechanism associated with osteoporosis is an imbalance in bone formation and resorption which leads to a reduction in BMD and increased bone fragility [1,4]. Bone fragility fractures are the most relevant clinical complication of osteoporosis [5,6] and have become a public health issue that increases morbidity, disability, and mortality [7]. The identification of risk biomarkers for osteoporotic fracture is key to the establishment of early treatment and to avoid the appearance of the first fracture or re-fractures. Avoiding the first fracture is important to stop the cascade of subsequent fractures. In this sense, the use of genetic biomarkers could be a useful tool to identify individuals at higher risk.
The most accepted hypothesis of the etiopathogenic of osteoporosis and bone fragility fracture is the combined action of environmental and genetic factors. Many risk factors are involved, including age, physical activity, medication use, and coexisting diseases. But one of the most important is a positive family history, as it emphasizes the relevance of genetic predisposition in the pathogenesis of osteoporosis and bone fracture [8,9,10,11]. Several hundreds of genetic loci have been associated with osteoporosis, low BMD, and fragility fractures [12,13]. One of the genes so far associated therewith is TP53 [14], the Arg72Pro missense variant which has been linked to an increased risk of osteoporosis. The proline allele has been associated more frequently with patients with osteoporosis, suggesting that the TP53 p.Arg72Pro variant may be a risk factor for osteoporosis [15]. The TP53 gene encodes protein p53 with 393 amino acids. The principal role of p53 is to promote cell cycle arrest, apoptosis, and cell senescence [16]. The p53 protein is also involved in bone metabolism; it regulates osteoblast differentiation, bone formation, osteoblast-dependent osteoclast differentiation, and bone remodeling [17,18]. The Arg72Pro polymorphism results in a change in the structure of the p53 protein in a proline-rich domain. This genetic variant is located in a domain involved in the apoptotic role of p53 [19,20,21]. In this sense, it has been reported that the arginine allele of the Arg72Pro polymorphism is associated with more apoptosis induction than the proline variant [19,20,21]. The p. Arg72pro polymorphism in the p53 protein has been frequently associated with the pathophysiology of a variety of diseases including bone metabolism alterations. According to the results from Jia et al. [15], the TP53 p. Arg72Pro missense variant could be a genetic biomarker of osteoporosis risk.
In such a scenario, and with an existing need to better understand the genetic factors determining osteoporosis and fragility fractures, this study aimed to obtain a deeper understanding of the pathophysiological bone consequences of the TP53 Arg72Pro missense genetic variant using a humanized mouse model. The putative rol of the TP53 p.Arg72Pro polymorphism in the susceptibility of suffering bone fragility or osteoporosis will be evaluated.
2. Results
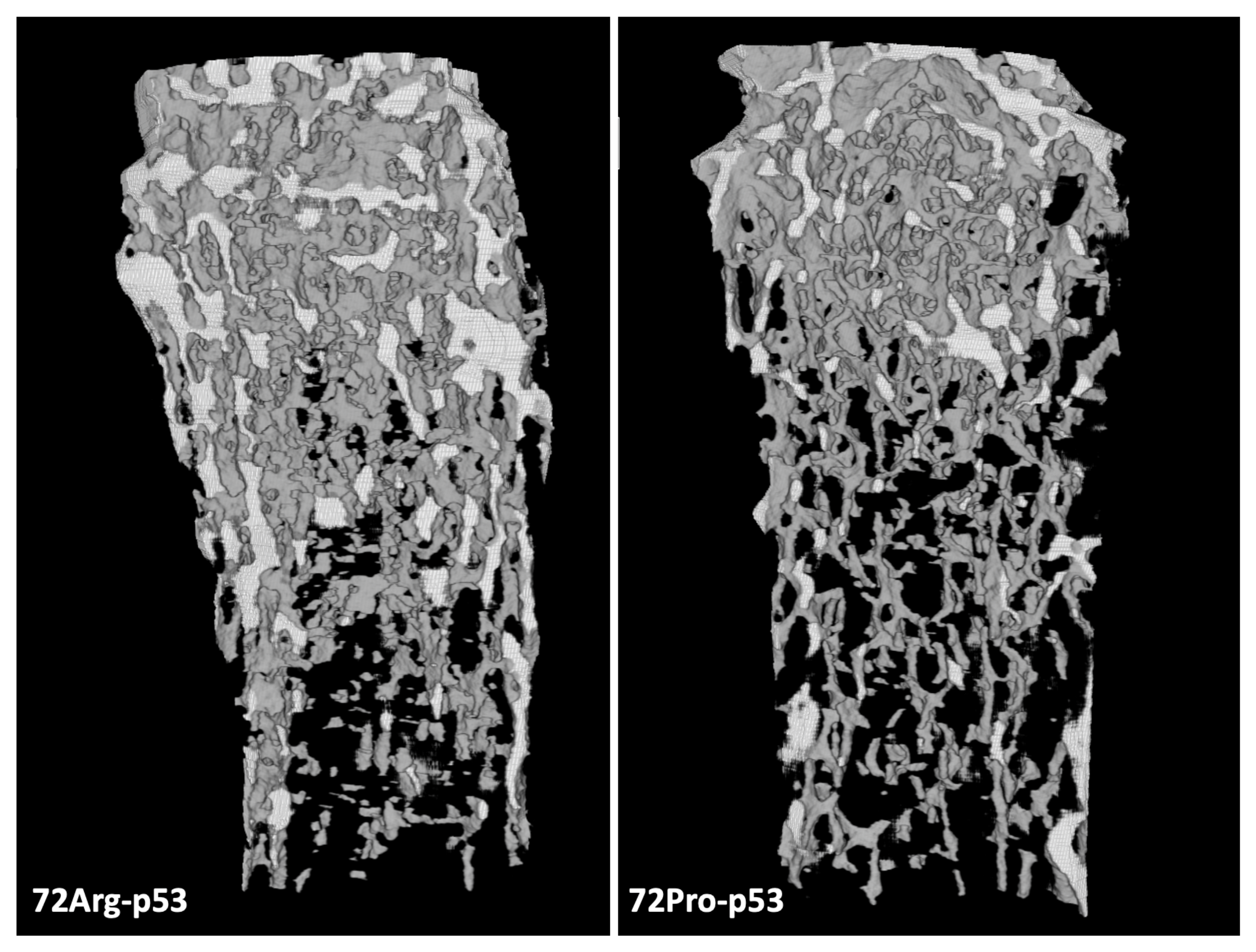
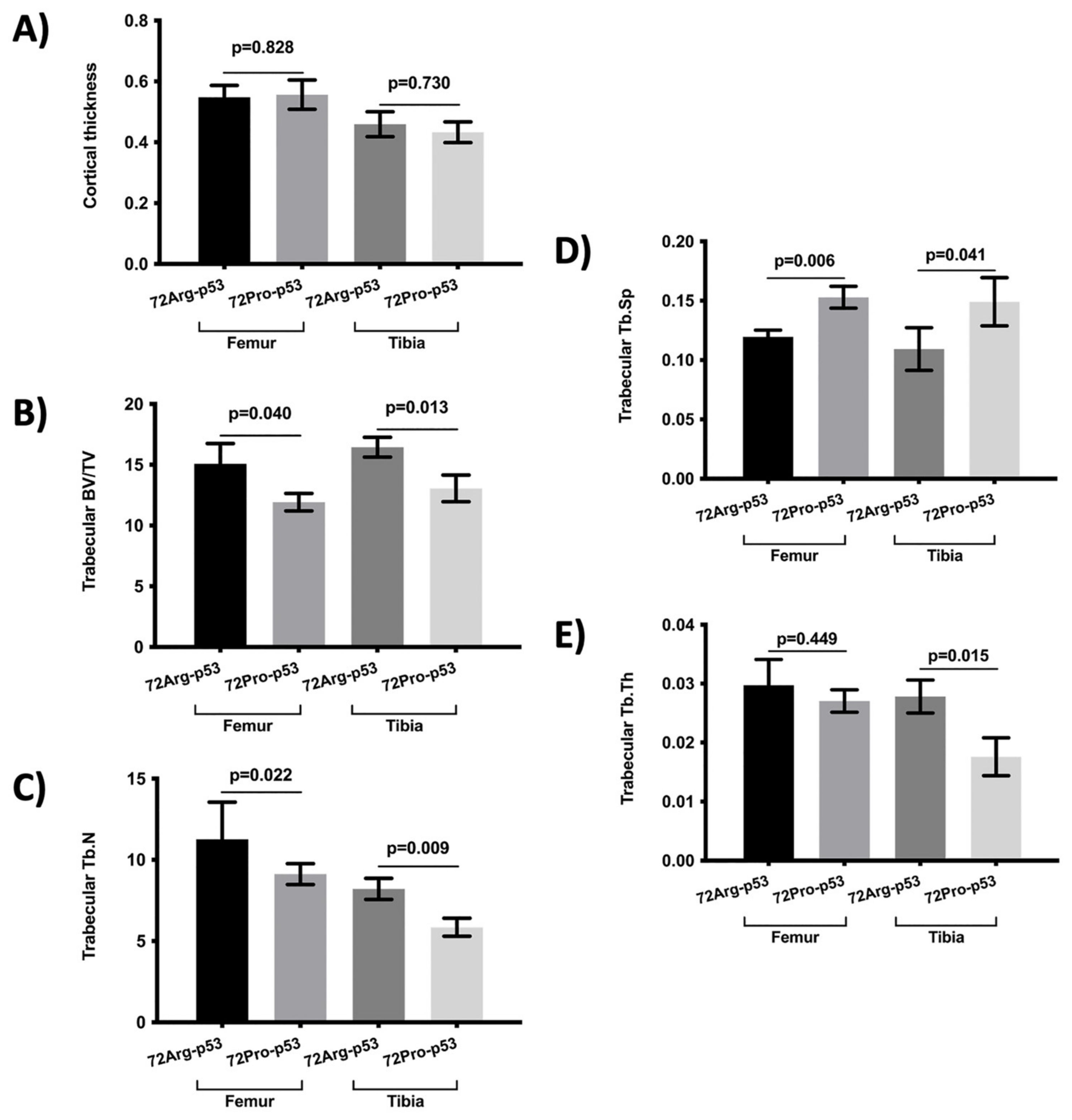
The results from the micro-computed tomography (μCT) showed that the bone from 72Pro-p53 mice was more deteriorated than that of 72Arg-p53 mice. A representative comparison of femur trabecular areas between 72Arg- and 72Pro-p53 mice is shown in Figure 1, with deterioration shown in the trabecular area in the 72Pro-p53 mice. The bone histomorphometry parameters were evaluated in humanized 72Arg-p53 and 72Pro-p53 mice (Figure 2). The results showed that the 72Pro-p53 mice had a lower trabecular bone mass, both at the femur and the tibia, with the lower femur and tibia bone volume over total volume (BV/TV), femur and tibia trabecular separation (Tb.Sp), and tibia trabecular thickness (Tb.Th) included. In addition, the 72Pro-p53 mice had lower femur and tibia trabecular number (Tb.N) than the 72Arg-p53 mice (Figure 2). The analysis of the cortical thickness did not show statistical differences between both genotypes. These results are also shown in Supplementary Table S1.

Figure 1.
Micro-computed tomography (μCT) results for the femur trabecular area of 72Arg-p53 and 72Pro-p53 mice.
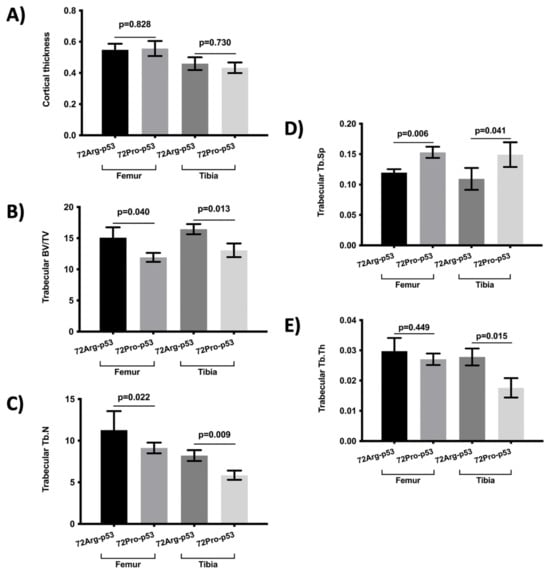
Figure 2.
Comparison of bone morphometry parameters between 72Arg-p53 and 72Pro-p53 mice in the femur and tibia: (A) cortical thickness, (B) trabecular BV/TV, (C) Tb.N, (D) Tb.Sp, and (E) Tb.Th. Variables are presented as the mean (standard deviation). Bars represent the mean values and their respective standard deviation. BV/TV: percent bone volume; Tb.Th: trabecular thickness; Tb.Sp: trabecular separation and Tb.N: trabecular number.
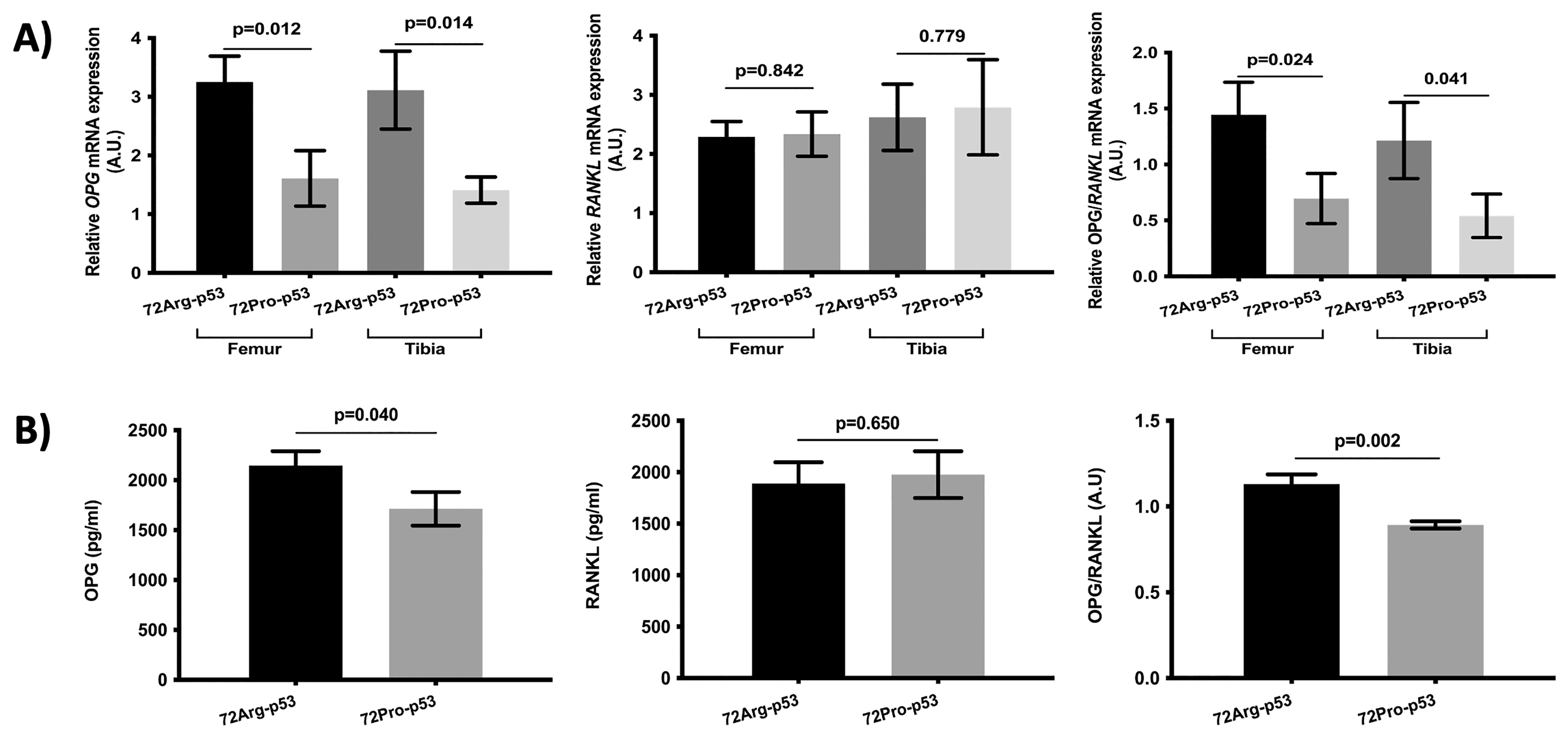
The results of the analysis of osteoclast maturation- and activation-related genes showed lower relative expression of osteoprotegerin (OPG) in the femur and tibia bone tissue in the 72Pro-p53 mice. Also, the OPG/RANKL ratio was lower in the femur and tibia of the 72Pro-p53 mice (Figure 3A). In the case of plasma, the OPG protein levels and OPG/RANKL plasma protein ratio were lower in the 72Pro-p53 mice than in the 72Arg-p53 mice (Figure 3B).

Figure 3.
Comparison of the OPG and RANKL analyses between the 72Arg-p53 and 72Pro-p53 mice. Relative mRNA expression of the OPG gene and the RANKL gene and the OPG/RANKL ratio in the femur and tibia bone tissue (A). OPG and RANKL plasma protein levels and OPG/RANKL plasma ratio (B). Bars represent mean values and their respective standard deviation. A.U.: arbitrary units.
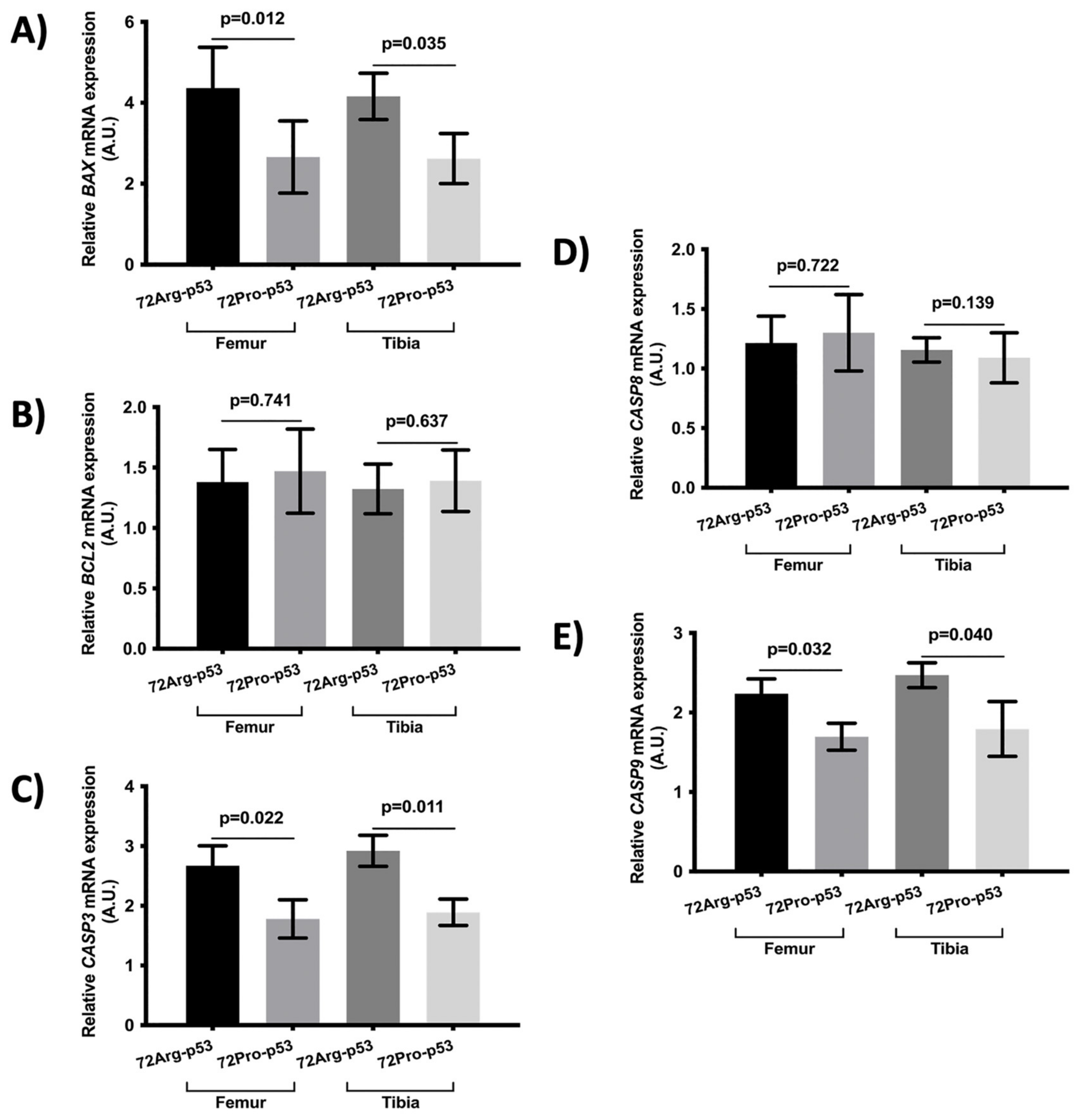
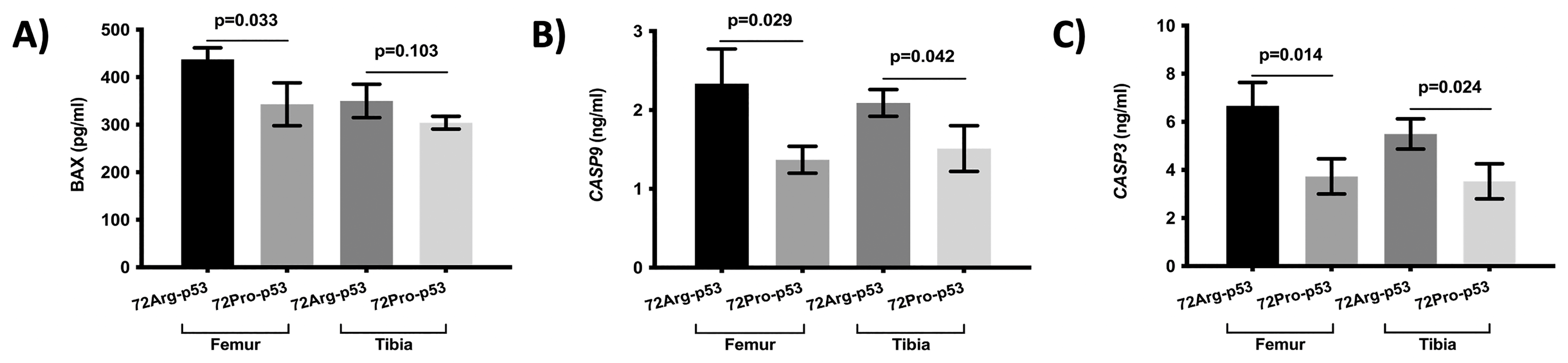
The results also showed statistical differences in the expression of genes involved in apoptosis between 72Arg-p53 and 72Pro-p53 mice. Relative mRNA quantification of gene expression involved in apoptosis showed lower relative expression of BAX, CASP3, and CASP9 in the femur and tibia of the 72Pro-p53 mice. The expression of these genes was higher in the 72Arg-p53 mice (Figure 4). The relative expression of BCL2 and CASP8 did not show statistical differences between the 72Arg-p53 and 72Pro-p53 mice (Figure 4). In addition, the expression of apoptosis-related proteins in the femur and tibia bone tissue was also analyzed. The results showed that the protein expression of BAX, CASP3, and CASP9 was lower in the femur bone tissue of the 72Pro-p53 mice than that of the 72Arg-p53 mice. The expression of CASP3 and CASP9 proteins was also lower in the tibia bone tissue of the 72Pro-p53 mice (Figure 5). The relative expression of genes involved in inflammation and oxidative stress did not yield statistical differences (Supplementary Figures S1 and S2).
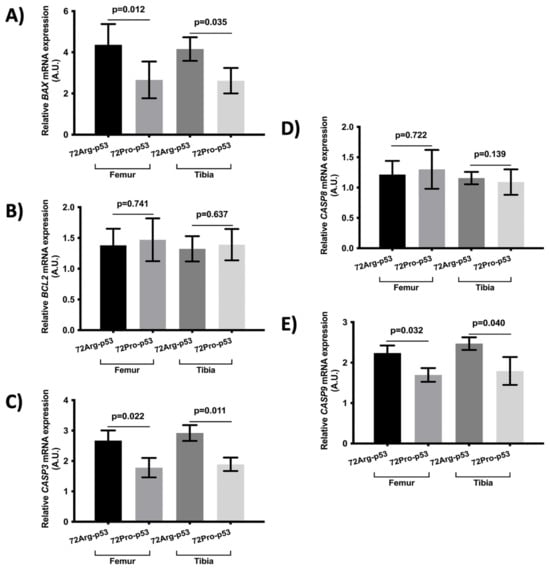
Figure 4.
Relative mRNA expression of apoptosis-related genes in the femur and tibia bone tissue. Relative mRNA expression of the BAX gene (A), BCL2 gene (B), CASP3 gene (C), CASP8 gene (D), and CASP9 gene (E). Bars represent mean values and their respective standard deviation. A.U.: arbitrary units.
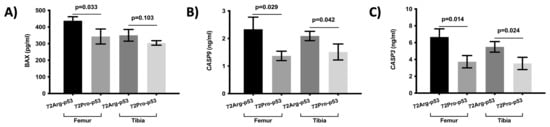
Figure 5.
Expression of apoptosis-related proteins in the femur and tibia bone tissue. Protein expression of BAX (A), CASP9 (B), and CASP3 (C). Bars represent mean values and their respective standard deviation.
3. Discussion
Osteoporosis, characterized by low BMD and alteration in bone microarchitecture, is the most common metabolic bone disease [2,3]. Our results are in line with the concept that genetic predisposition could be crucial in its etiology. The results show that the proline variant of the TP53 Arg72Pro polymorphism is associated with deteriorated bone tissue, reinforcing the hypothesis that the TP53 gene could be involved in determining individual osteoporosis susceptibility [14,15]. These results indicate that this genetic variant could be a good indicator of osteoporosis risk. Morphologically, it has been associated with a deterioration in bone microarchitecture and there is a biological explanation for this: lower apoptosis and decreased osteoprotegerin activity. These data could imply that this higher-risk subgroup could benefit from treatment with bisphosphonates or denosumab, drugs that inhibit proliferation and increase osteoclast apoptosis.
Protein p53 is a tumor suppressor, that is, a stress sensor that induces apoptosis, cell cycle arrest, or senescence [16]. It has also been published that p53 plays a key role in bone metabolism [17,18]. Protein p53 could be involved in osteogenesis [22]; it modulates osteoblastic and osteoclastic differentiation, increasing the Notch signaling pathway [17,23]. P53 has also been associated with the regulation of OPG synthesis; it has been reported that there is a negative correlation between TP53 gene modifications and OPG regulation [24]. OPG is a soluble member of the tumor necrosis factor receptor superfamily. It is a decoy receptor for RANKL because OPG inhibits osteoclastic bone resorption by interfering with the binding of RANKL to RANK [25,26]. Multiple single nucleotide polymorphisms (SNPs) have been identified in TP53 [27,28]; one of the most studied is the Arg72Pro variant. This genetic polymorphism has been associated with various cancers, inflammatory diseases, and stroke [29,30,31,32]. Also, the TP53 Arg72Pro genetic variant has been found to be involved in osteoporosis; the proline variant has been associated with an increased risk of suffering osteoporosis [15]. In this sense, our results showed that the proline allele of the TP53 Arg72Pro polymorphism is associated with a higher level of deterioration of bone tissue in the trabecular femur and tibia. In addition, we report that the proline allele has been found to be associated with lower OPG gene expression and a lower s/RANKL ratio in the femur and tibia, lower OPG protein plasma levels, and a lower OPG/RANKL plasma protein ratio. The crucial role of protein p53 in the regulation of OPG status [24] and therefore bone metabolism has been reported [18]. Our results support the hypothesis that the TP53 Arg72Pro variant could be associated with negative regulation of OPG and, therefore, increased osteoclastogenesis and, consequently, a more impaired bone microarchitecture.
Also, P53 has a crucial role in apoptosis induction. Arg72Pro SNP is in a proline-rich domain involved in the apoptotic role of p53 [19,20,21]. In this sense, it has been reported that the arginine allele of the Arg72Pro polymorphism is associated with more apoptosis induction than the proline variant [19,20,21]. Our report showed that the arginine variant could be associated with more apoptosis induction in bone tissue, specifically with the intrinsic apoptotic pathway. The results associated the arginine variant of the Arg72Pro TP53 polymorphism with altered expression of the BAX, CASP3, and CASP9 genes and proteins in the bone tissue. Although genetic factors associated with inflammation and oxidative stress have been associated with bone osteoporotic fracture [33,34], our results did not show modifications in inflammation and oxidative stress gene expression in bone tissue with regard to the TP53 Arg72Pro polymorphism. Many risk factors have been associated with alterations in bone metabolism and the risk of suffering bone metabolism diseases [6]. Our hypothesis is that the metabolic bone response to these risk factors could be different depending on the TP53 Arg72Pro genetic variant. The results of this work indicate that the proline allele of the TP53 Arg72Pro polymorphism is associated with decreased apoptotic function, lower OPG/RANKL ratio, and worsened bone microarchitecture. It could be speculated that reduced levels of apoptosis may be followed by a more aggressive cellular response and thus more bone alterations. On the other hand, and in addition, the proline allele of TP53 Arg72Pro SNP has been associated with increased activation of the NF-kB pathway [35], which is crucial in osteoclastogenesis [36,37]. Also, p53 is involved in the regulation of OPG [24] which has a crucial role in osteoclastic bone resorption [25,26]. In this sense, we report an association between the proline allele of the TP53 Arg72Pro variant and OPG expression; the proline variant was associated with lower gene expression and lower OPG/RANKL ratio. Hence, the proline variant of the TP53 Arg72Pro polymorphism could be associated with more osteoclast maturation and activation and therefore more bone resorption. This mutation may have special significance in the elderly population. The decrease in apoptosis increases the percentage of senescent cells in bone tissue, which are viable cells but with an irreversible arrest of the cell cycle. The p53/21 metabolic pathway plays a role in this arrest. Clinical–functional repercussions increase morbidity and mortality among this population [38].
The main limitation of the study is that we did not evaluate the influence of the TP53 Arg72Pro genetic variant in bone metabolism under conditions of bone injury and that we only analyzed bone quality using μCT. Another limitation could be the lack of in vitro studies to analyze the consequence of the mutation, but we consider that this is not necessary since we observed the effect of the mutation in an in vivo model. On the other hand, and as a main strength, this work summarizes, for the first time, the influence of the TP53 Arg72Pro polymorphism in OPG gene expression, OPG/RANKL ratio, bone microarchitecture, and apoptosis bone status.
In conclusion, we described the influence of the TP53 Arg72Pro polymorphism in bone microarchitecture, reinforcing the hypothesis that the TP53 Arg72Pro genetic variant could be crucial in osteoporosis risk. The TP53 Arg72Pro missense variant could be a genetic biomarker to identify individuals with an increased risk of suffering osteoporosis.
4. Materials and Methods
4.1. Animals
Humanized TP53 Arg72Pro knock-in (KI) mice were used. No differences in whole skeleton, body weight, and survival between the two genotypes were described. The animals were bred at the Animal Welfare and Research Service of the University of Valladolid in accordance with Spanish law (RD 53/2013). All experiments were carried out in compliance with the applicable international rules and policies—European Union Directive for Protection of Vertebrates Used for Experimental and Other Scientific Ends (2010/63/EU)—and were reviewed and approved by the University of Valladolid Institutional Committee for Animal Care and Use.
4.2. Micro-Computed Tomography (μCT)
The bone histomorphometry was analyzed using μCT. The femur and tibia specimens were scanned using a high-energy micro-computed tomography system 527 (SkyScan 11732, Bruker Micro-CT, Kontich, Belgium) and Sky-scan 1172 µCT data acquisition software. Since the aim was to maximize the resolution of the samples, the pixel size was reduced to the minimum, with it reaching a pixel size of 6.7 µm and a voxel size of 300.76 µm3. Scanning was performed at 50 kV and an Al 0.5 mm filter was used to reduce noise during scanning. During the reconstruction, parameters were used to correct possible beam hardening, ring artifacts, and misalignment problems. Maximum and minimum values for the attenuation coefficient were established. The minimum value was set at 0. For the maximum value, the critical section of all scans, the one with the maximum attenuation coefficient value, was selected at the operator’s discretion. Once this section was defined, the maximum value within the histogram was determined and a margin of error of 10% was applied. Finally, the cortical and trabecular areas of the tibia and femur were analyzed. The trabecular bone analysis was performed on the distal femur and proximal tibia areas. The regions of interest covered a total of 3 mm, specifically 2 mm below the growth plate. For the analysis of the cortical area, 3 mm of the central regions of the femur and tibia were selected at a distance of 15 mm from the growth plate of the tibia and femur. The regions of interest for delineation in each image were fully automated and assessed, as described by Bruker’s instructions [39,40]. The scan parameters have been included in Supplementary Table S2.
The structural parameter of the cortical bone analyzed was cortical thickness. In the trabecular bone, bone volume over total volume (BV/TV), trabecular number (Tb.N), trabecular thickness (Tb.Th), and trabecular separation (Tb.Sp) were analyzed. The BV/TV parameter indicates the ratio of bone tissue within the whole sample, and Tb.N, Tb.Th, and Tb.Sp determine the quality of the trabecular bone.
4.3. Sample Processing and RNA Extraction
Total RNA was extracted from bone tissue using the RNA easy Mini Kit (QIAGEN, Barcelona, Spain), following the manufacturer’s instructions. Bone tissue was submerged in RNA-stabilizing solution (RNAlater, Invitrogen, Waltham, MA, USA) and stored at −80 °C until RNA extraction. Bone tissue was homogenized with polytron tissue homogenizer using Trizol reagent (Invitrogen). Finally, RNA was extracted using a phenol–chloroform mixture, precipitated in ethanol, and purified using RNase-free columns. RNA quantity and purity were determined according to absorbance in a spectrophotometer (NanoDrop 2000, Thermo, Waltham, MA, USA).
4.4. Reverse Transcription and Real-Time Quantitative PCR
Complementary DNA (cDNA) was synthesized by means of reverse transcription using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). The relative quantitative real-time polymerase chain reaction (qPCR) was performed using SYBR Green PCR master mix (Applied Biosystems) and mice-specific primer sets (Supplementary Table S3). Relative mRNA expression was analyzed for apoptosis, inflammation, and oxidative stress-related genes. The qPCR experiments were conducted using the Applied Biosystems 7500 Real-Time PCR System under the following conditions: 95 °C, 10 min; 40 cycles of 95 °C, 15 s; 60 °C, 1 min; and a final melting curve step. The GAPDH gene was used as a housekeeping gene for the normalization of the expression level of mRNA. The threshold cycle was determined for each reaction, and gene expression was quantified using the 2−ΔΔCt method [41]. All qPCR reactions were performed in triplicate.
4.5. Enzyme-Linked Immunosorbent Assay
The plasma levels of OGP and RANKL proteins (involved in bone metabolism) were measured using the enzyme-linked immunosorbent assay (ELISA) method, following the instructions of the manufacturer. Bone tissue levels of BAX, CASP3, and CASP9 proteins (involved in apoptosis) were also measured. The samples were measured in triplicate. Absorbance was determined using a spectrophotometer ELx800 Universal Microplate Reader (Bio-Tek Instruments Inc., Winooski, VT, USA) at 450 nm with a wavelength correction of 620 nm.
4.6. Statistical Analysis
Continuous variables were expressed as the mean (standard deviation). The Kolmogorov–Smirnov test was used to analyze the distribution of continuous variables. As for normally distributed variables, the analysis of variance t-test was applied. In the case of non-normally distributed variables, the groups were compared using the Mann–Whitney U-test (two groups) or the Kruskal–Wallis test (more than two groups). A p-value < 0.05 was considered significant. All analyses were performed using the SPSS version 22.0 statistical package (SPSS, Chicago, IL, USA).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25031395/s1.
Author Contributions
Conceptualization: R.U.-M. and J.L.P.-C.; methodology: R.U.-M., N.G.-C., S.P.-I., J.M.F.-G., Á.d.R., D.F., R.L., F.M.-R., J.A.R., Á.A. and J.L.P.-C.; formal analysis: R.U.-M., N.G.-C. and J.L.P.-C.; investigation: R.U.-M., N.G.-C., S.P.-I., J.M.F.-G., Á.d.R., D.F., R.L., F.M.-R., J.A.R., Á.A. and J.L.P.-C.; resources: R.U.-M.; writing—original draft preparation: R.U.-M.; writing—review and editing: R.U.-M., N.G.-C., S.P.-I., J.M.F.-G., Á.d.R., D.F., R.L., F.M.-R., J.A.R., Á.A. and J.L.P.-C.; supervision: R.U.-M. and J.L.P.-C.; project administration: R.U.-M. and J.L.P.-C.; funding acquisition, R.U.-M., S.P.-I., Á.A. and J.L.P.-C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Ministerio de Ciencia e Innovación (Spain Government; PID2020-114585RA-I00 to RUM and SPI) and Instituto de Salud Carlos III (Spain; PI21/00727 to AA).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
The animals were bred at the Animal Welfare and Research Service of the University of Valladolid in accordance with Spanish law (RD 53/2013) (protocol code: 9809595, date of approval: July 2021). All experiments were carried out in compliance with the applicable international rules and policies—European Union Directive for Protection of Vertebrates Used for Experimental and Other Scientific Ends (2010/63/EU)—and were reviewed and approved by the University of Valladolid Institutional Committee for Animal Care and Use.
Data Availability Statement
All of the results are in the main manuscript.
Conflicts of Interest
The authors declare no conflicts of interests. The funders had no role in the design of the study; in the collection, analysis, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
References
- Weitzmann, M.N.; Ofotokun, I. Physiological and pathophysiological bone turnover—Role of the immune system. Nat. Rev. Endocrinol. 2016, 12, 518–532. [Google Scholar] [CrossRef] [PubMed]
- Kanis, J.A. Diagnosis of osteoporosis. Osteoporos. Int. 1997, 7 (Suppl. 3), S108–S116. [Google Scholar] [CrossRef]
- Yang, T.L.; Shen, H.; Liu, A.; Dong, S.S.; Zhang, L.; Deng, F.Y.; Zhao, Q.; Deng, H.W. A road map for understanding molecular and genetic determinants of osteoporosis. Nat. Rev. Endocrinol. 2020, 16, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, M. Skeletal remodeling in health and disease. Nat. Med. 2007, 13, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Siris, E.S.; Miller, P.D.; Barrett-Connor, E.; Faulkner, K.G.; Wehren, L.E.; Abbott, T.A.; Berger, M.L.; Santora, A.C.; Sherwood, L.M. Identification and fracture outcomes of undiagnosed low bone mineral density in postmenopausal women: Results from the National Osteoporosis Risk Assessment. JAMA 2001, 286, 2815–2822. [Google Scholar] [CrossRef]
- Kanis, J.A. Diagnosis of osteoporosis and assessment of fracture risk. Lancet 2002, 359, 1929–1936. Available online: https://pubmed.ncbi.nlm.nih.gov/12057569/ (accessed on 26 May 2023). [CrossRef] [PubMed]
- Adachi, J.D.; Adami, S.; Gehlbach, S.; Anderson, F.A.; Boonen, S.; Chapurlat, R.D.; Compston, J.E.; Cooper, C.; Delmas, P.; Diez-Perez, A.; et al. Impact of prevalent fractures on quality of life: Baseline results from the global longitudinal study of osteoporosis in women. Mayo Clin. Proc. 2010, 85, 806–813. [Google Scholar] [CrossRef]
- Kanis, J.A.; Johansson, H.; Odén, A.; Johnell, O.; De Laet, C.E.; Eisman, J.A.; McCloskey, E.V.; Mellstrom, D.; Melton, L.J., III; Pols, H.A.; et al. A family history of fracture and fracture risk: A meta-analysis. Bone 2004, 35, 1029–1037. [Google Scholar] [CrossRef]
- Ralston, S.H.; Uitterlinden, A.G. Genetics of Osteoporosis. Endocr. Rev. 2010, 31, 629–662. [Google Scholar] [CrossRef]
- Trajanoska, K.; Rivadeneira, F. The genetic architecture of osteoporosis and fracture risk. Bone 2019, 126, 2–10. [Google Scholar] [CrossRef]
- Mitek, T.; Nagraba, Ł.; Deszczyński, J.; Stolarczyk, M.; Kuchar, E.; Stolarczyk, A. Genetic Predisposition for Osteoporosis and Fractures in Postmenopausal Women. Adv. Exp. Med. Biol. 2019, 1211, 17–24. [Google Scholar] [PubMed]
- Koromani, F.; Trajanoska, K.; Rivadeneira, F.; Oei, L. Recent Advances in the Genetics of Fractures in Osteoporosis. Front. Endocrinol. 2019, 10, 337. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Bai, W.; Zheng, H. Twelve years of GWAS discoveries for osteoporosis and related traits: Advances, challenges and applications. Bone Res. 2021, 9, 23. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; You, X.; Zhou, H.; Kang, A.; He, W.; Li, Z.; Li, B.; Xia, J.; Zhu, H.; Zhao, Y.; et al. p53 plays a central role in the development of osteoporosis. Aging 2020, 12, 10473–10487. [Google Scholar] [CrossRef] [PubMed]
- Jia, F.; Sun, R.; Li, J.; Li, Q.; Chen, G.; Fu, W. Interactions of Pri-miRNA-34b/c and TP53 Polymorphisms on the Risk of Osteoporosis. Genet. Test. Mol. Biomark. 2016, 20, 398–401. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Lane, D.; Levine, A.J. Surfing the p53 network. Nature 2000, 408, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Kua, H.Y.; Hu, Y.; Guo, K.; Zeng, Q.; Wu, Q.; Ng, H.H.; Karsenty, G.; de Crombrugghe, B.; Yeh, J.; et al. p53 functions as a negative regulator of osteoblastogenesis, osteoblast-dependent osteoclastogenesis, and bone remodeling. J. Cell Biol. 2006, 172, 115–125. [Google Scholar] [CrossRef]
- Liu, H.; Li, B. p53 control of bone remodeling. J. Cell. Biochem. 2010, 111, 529–534. [Google Scholar] [CrossRef]
- Sakamuro, D.; Sabbatini, P.; White, E.; Prendergast, G.C. The polyproline region of p53 is required to activate apoptosis but not growth arrest. Oncogene 1997, 15, 887–898. [Google Scholar] [CrossRef]
- Bonafé, M.; Salvioli, S.; Barbi, C.; Trapassi, C.; Tocco, F.; Storci, G.; Invidia, L.; Vannini, I.; Rossi, M.; Marzi, E.; et al. The different apoptotic potential of the p53 codon 72 alleles increases with age and modulates in vivo ischaemia-induced cell death. Cell Death Differ. 2004, 11, 962–973. [Google Scholar] [CrossRef][Green Version]
- Dumont, P.; Leu, J.I.J.; Della Pietra, A.C.; George, D.L.; Murphy, M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat. Genet. 2003, 33, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Qi, M.; Konermann, A.; Zhang, L.; Jin, F.; Jin, Y. The p53/miR-17/Smurf1 pathway mediates skeletal deformities in an age-related model via inhibiting the function of mesenchymal stem cells. Aging 2015, 7, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.; Espinoza, I.; Pannuti, A.; Romero, D.; Martinez, L.; Caskey, M.; Stanculescu, A.; Bocchetta, M.; Rizzo, P.; Band, V.; et al. p53 Modulates Notch Signaling in MCF-7 Breast Cancer Cells by Associating with the Notch Transcriptional Complex via MAML1. J. Cell. Physiol. 2015, 230, 3115–3127. [Google Scholar] [CrossRef] [PubMed]
- Velletri, T.; Huang, Y.; Wang, Y.; Li, Q.; Hu, M.; Xie, N.; Yang, Q.; Chen, X.; Chen, Q.; Shou, P.; et al. Loss of p53 in mesenchymal stem cells promotes alteration of bone remodeling through negative regulation of osteoprotegerin. Cell Death Differ. 2020, 28, 156–169. [Google Scholar] [CrossRef]
- Simonet, W.S.; Lacey, D.L.; Dunstan, C.R.; Kelley, M.C.; Chang, M.S.; Lüthy, R.; Nguyen, H.Q.; Wooden, S.; Bennett, L.; Boone, T.; et al. Osteoprotegerin: A novel secreted protein involved in the regulation of bone density. Cell 1997, 89, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Boyle, W.J.; Simonet, W.S.; Lacey, D.L. Osteoclast differentiation and activation. Nature 2003, 423, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Olivier, M.; Eeles, R.; Hollstein, M.; Khan, M.A.; Harris, C.C.; Hainaut, P. The IARC TP53 database: New online mutation analysis and recommendations to users. Hum. Mutat. 2002, 19, 607–614. [Google Scholar] [CrossRef]
- Pietsch, E.C.; Humbey, O.; Murphy, M.E. Polymorphisms in the p53 pathway. Oncogene 2006, 25, 1602–1611. [Google Scholar] [CrossRef]
- Tian, X.; Dai, S.; Sun, J.; Jiang, S.; Jiang, Y. The association between the TP53 Arg72Pro polymorphism and colorectal cancer: An updated meta-analysis based on 32 studies. Oncotarget 2016, 8, 1156–1165. [Google Scholar] [CrossRef]
- Gomez-Sanchez, J.C.; Delgado-Esteban, M.; Rodriguez-Hernandez, I.; Sobrino, T.; de la Ossa, N.P.; Reverte, S.; Bolaños, J.P.; Gonzalez-Sarmiento, R.; Castillo, J.; Almeida, A. The human Tp53 Arg72Pro polymorphism explains different functional prognosis in stroke. J. Exp. Med. 2011, 208, 429–437. [Google Scholar] [CrossRef]
- Leu, J.I.J.; Murphy, M.E.; George, D.L. The p53 Codon 72 Polymorphism Modifies the Cellular Response to Inflammatory Challenge in the Liver. J. Liver 2013, 2, 117. [Google Scholar] [PubMed]
- Diakite, B.; Kassogue, Y.; Dolo, G.; Wang, J.; Neuschler, E.; Kassogue, O.; Keita, M.L.; Traore, C.B.; Kamate, B.; Dembele, E.; et al. p.Arg72Pro polymorphism of P53 and breast cancer risk: A meta-analysis of case-control studies. BMC Med. Genet. 2020, 21, 206. [Google Scholar] [CrossRef] [PubMed]
- Usategui-Martín, R.; Lendinez-Tortajada, V.; Pérez-Castrillón, J.L.; Briongos-Figuero, L.; Abadía-Otero, J.; Martín-Vallejo, J.; Lara-Hernandez, F.; Chaves, F.J.; García-Garcia, A.B.; Martín-Escudero, J.C. Polymorphisms in genes involved in inflammation, the NF-kB pathway and the renin-angiotensin-aldosterone system are associated with the risk of osteoporotic fracture. The Hortega Follow-up Study. Bone 2020, 138, 115477. [Google Scholar] [CrossRef] [PubMed]
- Usategui-Martín, R.; Pérez-Castrillón, J.L.; Mansego, M.L.; Lara-Hernández, F.; Manzano, I.; Briongos, L.; Abadía-Otero, J.; Martín-Vallejo, J.; García-García, A.B.; Martín-Escudero, J.C.; et al. Association between genetic variants in oxidative stress-related genes and osteoporotic bone fracture. The Hortega follow-up study. Gene 2022, 809, 146036. [Google Scholar] [CrossRef]
- Frank, A.K.; Julia, I.; Leu, J.; Zhou, Y.; Devarajan, K.; Nedelko, T.; Klein-Szanto, A.; Hollstein, M.; Murphy, M.E. The Codon 72 Polymorphism of p53 Regulates Interaction with NF-κB and Transactivation of Genes Involved in Immunity and Inflammation. Mol. Cell. Biol. 2011, 31, 1201–1213. [Google Scholar] [CrossRef] [PubMed]
- Novack, D.V. Role of NF-κB in the skeleton. Cell Res. 2011, 21, 169–182. [Google Scholar] [CrossRef]
- Boyce, B.F.; Xiu, Y.; Li, J.; Xing, L.; Yao, Z. NF-κB-Mediated Regulation of Osteoclastogenesis. Endocrinol. Metab. 2015, 30, 35–44. [Google Scholar] [CrossRef]
- Farr, J.N.; Khosla, S. Cellular senescence in bone. Bone 2019, 121, 121–133. [Google Scholar] [CrossRef]
- Campbell, G.M.; Sophocleous, A. Quantitative analysis of bone and soft tissue by micro-computed tomography: Applications to ex vivo and in vivo studies. BoneKEy Rep. 2014, 3, 564. [Google Scholar] [CrossRef]
- Van’t Hof, R.J. Analysis of bone architecture in rodents using microcomputed tomography. Methods Mol. Biol. 2012, 816, 461–476. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).