High Frequencies of Genetic Variants in Patients with Atypical Femoral Fractures
Abstract
:1. Introduction
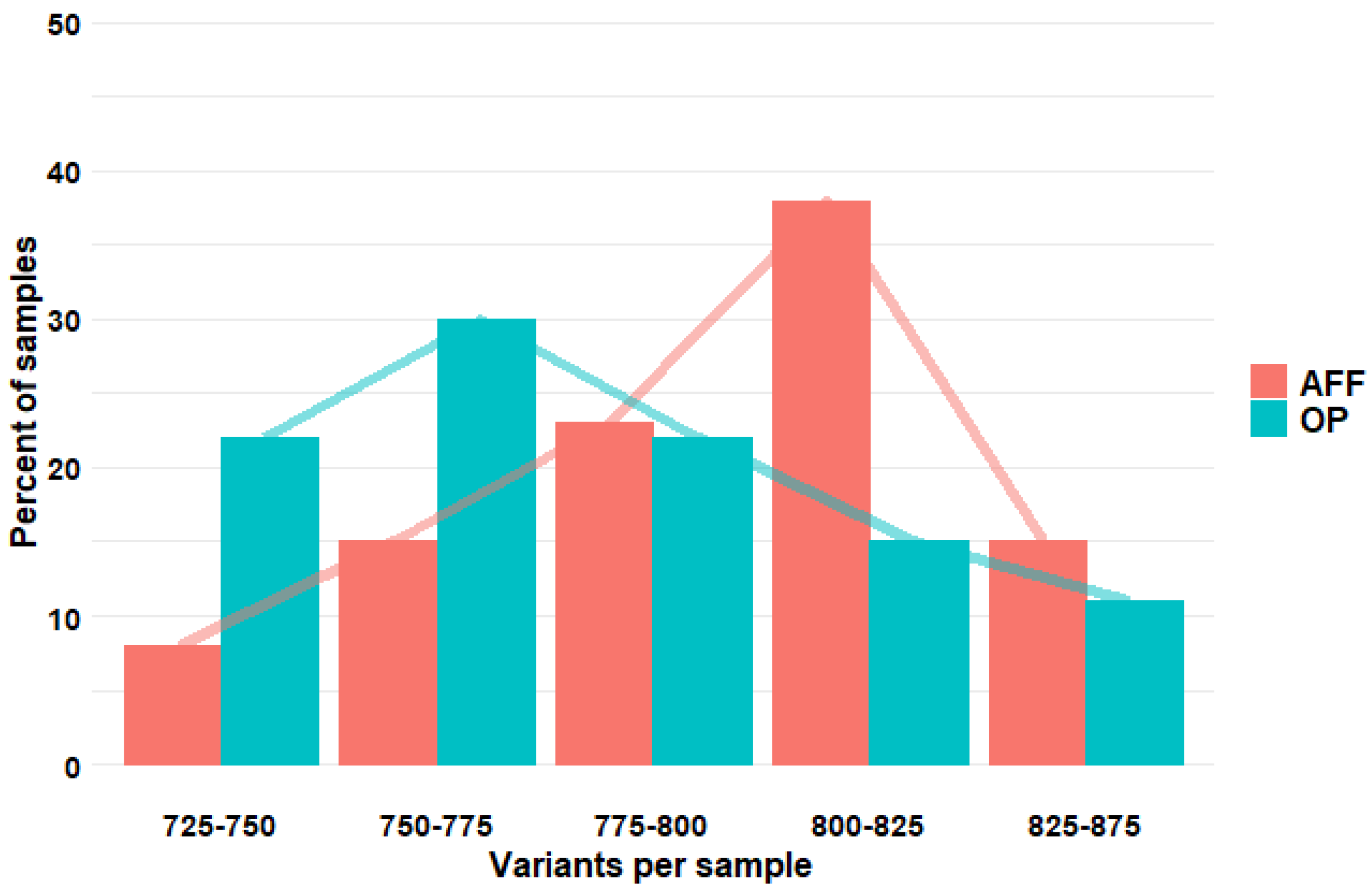
2. Results
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Whole Exome Sequencing
4.3. Gene Set Selection
4.4. Downloading of 1000G Data
4.5. Data Analysis and Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shane, E.; Burr, D.; Abrahamsen, B.; Adler, R.A.; Brown, T.D.; Cheung, A.M.; Cosman, F.; Curtis, J.R.; Dell, R.; Dempster, D.W.; et al. Atypical subtrochanteric and diaphyseal femoral fractures: Second report of a task force of the American Society for Bone and Mineral Research. J. Bone Miner. Res. 2014, 29, 1–23. [Google Scholar] [CrossRef]
- Zhou, W.; van Rooij, J.G.J.; Ebeling, P.R.; Verkerk, A.J.M.H.; Zillikens, M.C. The Genetics of Atypical Femur Fractures-a Systematic Review. Curr. Osteoporos. Rep. 2021, 19, 123–130. [Google Scholar] [CrossRef]
- Nguyen, H.H.; van de Laarschot, D.M.; Verkerk, A.J.M.H.; Milat, F.; Zillikens, M.C.; Ebeling, P.R. Genetic Risk Factors for Atypical Femoral Fractures (AFFs): A Systematic Review. JBMR Plus 2018, 2, 1–11. [Google Scholar] [CrossRef]
- Pérez-Núñez, I.; Pérez-Castrillón, J.L.; Zarrabeitia, M.T.; García-Ibarbia, C.; Martínez-Calvo, L.; Olmos, J.M.; Briongos, L.S.; Riancho, J.; Camarero, V.; Muñoz Vives, J.M.; et al. Exon array analysis reveals genetic heterogeneity in atypical femoral fractures. A pilot study. Mol. Cell. Biochem. 2015, 409, 45–50. [Google Scholar] [CrossRef]
- Roca-Ayats, N.; Balcells, S.; Garcia-Giralt, N.; Falcó-Mascaró, M.; Martínez-Gil, N.; Abril, J.F.; Urreizti, R.; Dopazo, J.; Quesada-Gómez, J.M.; Nogués, X.; et al. GGPS1 Mutation and Atypical Femoral Fractures with Bisphosphonates. N. Engl. J. Med. 2017, 376, 1794–1795. [Google Scholar] [CrossRef]
- Garcia-Giralt, N.; Roca-Ayats, N.; Abril, J.F.; Martinez-Gil, N.; Ovejero, D.; Castañeda, S.; Nogues, X.; Grinberg, D.; Balcells, S.; Rabionet, R. Gene Network of Susceptibility to Atypical Femoral Fractures Related to Bisphosphonate Treatment. Genes 2022, 13, 146. [Google Scholar] [CrossRef]
- Zhou, W.; Nguyen, H.H.; van de Laarschot, D.M.; Howe, T.S.; Koh, J.S.B.; Milat, F.; van Rooij, J.G.J.; Verlouw, J.A.M.; van der Eerden, B.C.J.; Stevenson, M.; et al. Whole Exome Sequencing in Two Southeast Asian Families with Atypical Femur Fractures. JBMR Plus 2022, 6, e10659. [Google Scholar] [CrossRef]
- Zhou, S.; Sosina, O.A.; Bovijn, J.; Laurent, L.; Sharma, V.; Akbari, P.; Forgetta, V.; Jiang, L.; Kosmicki, J.A.; Banerjee, N.; et al. Converging evidence from exome sequencing and common variants implicates target genes for osteoporosis. Nat. Genet. 2023, 55, 1277–1287. [Google Scholar] [CrossRef]
- Trajanoska, K.; Rivadeneira, F. The genetic architecture of osteoporosis and fracture risk. Bone 2019, 126, 2–10. [Google Scholar] [CrossRef]
- Maddirevula, S.; Alsahli, S.; Alhabeeb, L.; Patel, N.; Alzahrani, F.; Shamseldin, H.E.; Anazi, S.; Ewida, N.; Alsaif, H.S.; Mohamed, J.Y.; et al. Expanding the phenome and variome of skeletal dysplasia. Genet. Med. 2018, 20, 1609–1616. [Google Scholar] [CrossRef]
- Koide, H.; Holmbeck, K.; Lui, J.C.; Guo, X.C.; Driggers, P.; Chu, T.; Tatsuno, I.; Quaglieri, C.; Kino, T.; Baron, J.; et al. Mice Deficient in AKAP13 (BRX) Are Osteoporotic and Have Impaired Osteogenesis. J. Bone Miner. Res. 2015, 30, 1887–1895. [Google Scholar] [CrossRef]
- Li, L.; Zhao, D.; Zheng, W.; Wang, O.; Jiang, Y.; Xia, W.; Xing, X.; Li, M. A novel missense mutation in P4HB causes mild osteogenesis imperfecta. Biosci. Rep. 2019, 39, BSR20182118. [Google Scholar] [CrossRef]
- Rauch, F.; Fahiminiya, S.; Majewski, J.; Carrot-Zhang, J.; Boudko, S.; Glorieux, F.; Mort, J.S.; Bächinger, H.P.; Moffatt, P. Cole-Carpenter syndrome is caused by a heterozygous missense mutation in P4HB. Am. J. Hum. Genet. 2015, 96, 425–431. [Google Scholar] [CrossRef]
- Cotrina-Vinagre, F.J.; Rodríguez-García, M.E.; Martín-Hernández, E.; Durán-Aparicio, C.; Merino-López, A.; Medina-Benítez, E.; Martínez-Azorín, F. Characterization of a complex phenotype (fever-dependent recurrent acute liver failure and osteogenesis imperfecta) due to NBAS and P4HB variants. Mol. Genet. Metab. 2021, 133, 201–210. [Google Scholar] [CrossRef]
- Mullin, B.H.; Mamotte, C.; Prince, R.L.; Wilson, S.G. Influence of ARHGEF3 and RHOA knockdown on ACTA2 and other genes in osteoblasts and osteoclasts. PLoS ONE 2014, 9, e98116. [Google Scholar] [CrossRef]
- Arte, S.; Pöyhönen, M.; Myllymäki, E.; Ronkainen, E.; Rice, D.P.; Nieminen, P. Craniofacial and dental features of Axenfeld-Rieger syndrome patients with PITX2 mutations. Orthod. Craniofac. Res. 2023, 26, 320–330. [Google Scholar] [CrossRef]
- Ochoa, M.; Yang, A.; Kollias, C.; Bakir, C.; Carsen, S.; Lazier, J.; Innes, A.M.; Pagé, M.; Dawrant, J.; Robinson, M.E.; et al. From “ACAN” to “I CAN”: Restoring wellness in a boy with severe osteochondritis dissecans through diagnostic precision combined with optimal medical, surgical and rehabilitation management. Bone Rep. 2023, 18, 101663. [Google Scholar] [CrossRef]
- Ye, X.; Fang, D.; He, Y.; Yan, H.; Qiu, W.; Sun, Y. Dual diagnosis of osteogenesis imperfecta (OI) and short stature and advanced bone age with or without early-onset osteoarthritis and/or osteochondritis dissecans (SSOAOD) reveals a cumulative effect on stature caused by mutations in COL1A1 and ACAN genes. Eur. J. Med. Genet. 2020, 63, 104074. [Google Scholar] [CrossRef]
- Huang, H.; Jin, J.; Xiang, R.; Wang, X. Case report: A novel heterozygous frameshift mutation of ACAN in a Chinese family with short stature and advanced bone age. Front. Genet. 2023, 14, 1101695. [Google Scholar] [CrossRef]
- Martin, A.R.; Williams, E.; Foulger, R.E.; Leigh, S.; Daugherty, L.C.; Niblock, O.; Leong, I.U.S.; Smith, K.R.; Gerasimenko, O.; Haraldsdottir, E.; et al. PanelApp crowdsources expert knowledge to establish consensus diagnostic gene panels. Nat. Genet. 2019, 51, 1560–1565. [Google Scholar] [CrossRef]
- Del Real, Á.; Valero, C.; Olmos, J.M.; Hernández, J.L.; Riancho, J.A. Pharmacogenetics of Osteoporosis: A Pathway Analysis of the Genetic Influence on the Effects of Antiresorptive Drugs. Pharmaceutics 2022, 14, 776. [Google Scholar] [CrossRef] [PubMed]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Robinson, D.G.; Storey, J.D. The functional false discovery rate with applications to genomics. Biostatistics 2021, 22, 68. [Google Scholar] [CrossRef] [PubMed]
| GENE | Nº Variants | Alternative Allele Frequency (%) | FDR (p-Value) | ||
|---|---|---|---|---|---|
| Control OP | AFF | Allele | Genotype | ||
| ACAN | 16 | 47 | 59 | 9. × 10−3 | 2.85 × 10−2 |
| AKAP13 | 21 | 41 | 34 | 2.10 × 10−1 | 2.11 × 10−10 |
| APC | 7 | 61 | 68 | 8.88 × 10−1 | 3.48 × 10−2 |
| ARHGEF3 | 6 | 29 | 37 | 8.84 × 10−1 | 3.44 × 10−2 |
| CYP2D6 | 10 | 20 | 27 | 4.20 × 10−1 | 1.29 × 10−5 |
| NBN | 3 | 20 | 44 | 1.48 × 10−2 | 2.66 × 10−3 |
| NOTCH2 | 7 | 23 | 32 | 4.89 × 10−1 | 3.44 × 10−2 |
| P4HB | 3 | 19 | 26 | 1.00 | 2.91 × 10−2 |
| PITX2 | 2 | 8 | 35 | 9.82 × 10−3 | 2.85 × 10−2 |
| SPP1 | 3 | 37 | 44 | 1.00 | 2.85 × 10−2 |
| SUCO | 5 | 23 | 42 | 9.82 × 10−3 | 3.31 × 10−2 |
| UGT1A8 | 37 | 27 | 27 | 1.00 | 2.85 × 10−2 |
| GENE | Nº Variants | Alternative Allele Frequency (%) | FDR (p-Value) | ||
|---|---|---|---|---|---|
| Control IBS | AFF | Allele | Genotype | ||
| ACAN | 16 | 53 | 59 | 4.93 × 10−2 | 3.47 × 10−2 |
| AKAP13 | 21 | 47 | 34 | 6.08 × 10−8 | 5.62 × 10−23 |
| ARHGEF3 | 6 | 25 | 37 | 1.19 × 10−2 | 8.88 × 10−3 |
| P4HB | 3 | 14 | 26 | 2.21 × 10−2 | 8.38 × 10−3 |
| PITX2 | 2 | 18 | 35 | 2.21 × 10−2 | 3.47 × 10−2 |
| SUCO | 5 | 29 | 42 | 1.19 × 10−2 | 8.88 × 10−3 |
| UGT1A8 | 34 | 23 | 28 | 1.19 × 10−2 | 8.88 × 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
del Real, Á.; Cruz, R.; Sañudo, C.; Pérez-Castrillón, J.L.; Pérez-Núñez, M.I.; Olmos, J.M.; Hernández, J.L.; García-Ibarbia, C.; Valero, C.; Riancho, J.A. High Frequencies of Genetic Variants in Patients with Atypical Femoral Fractures. Int. J. Mol. Sci. 2024, 25, 2321. https://doi.org/10.3390/ijms25042321
del Real Á, Cruz R, Sañudo C, Pérez-Castrillón JL, Pérez-Núñez MI, Olmos JM, Hernández JL, García-Ibarbia C, Valero C, Riancho JA. High Frequencies of Genetic Variants in Patients with Atypical Femoral Fractures. International Journal of Molecular Sciences. 2024; 25(4):2321. https://doi.org/10.3390/ijms25042321
Chicago/Turabian Styledel Real, Álvaro, Raquel Cruz, Carolina Sañudo, José L. Pérez-Castrillón, María I. Pérez-Núñez, Jose M. Olmos, José L. Hernández, Carmen García-Ibarbia, Carmen Valero, and Jose A. Riancho. 2024. "High Frequencies of Genetic Variants in Patients with Atypical Femoral Fractures" International Journal of Molecular Sciences 25, no. 4: 2321. https://doi.org/10.3390/ijms25042321





