Identification of the Major Facilitator Superfamily Efflux Pump KpsrMFS in Klebsiella pneumoniae That Is Down-Regulated in the Presence of Multi-Stress Factors
Abstract
:1. Introduction
2. Results
2.1. Identification of Genes That Had the Largest Fluctuations in Different Conditions
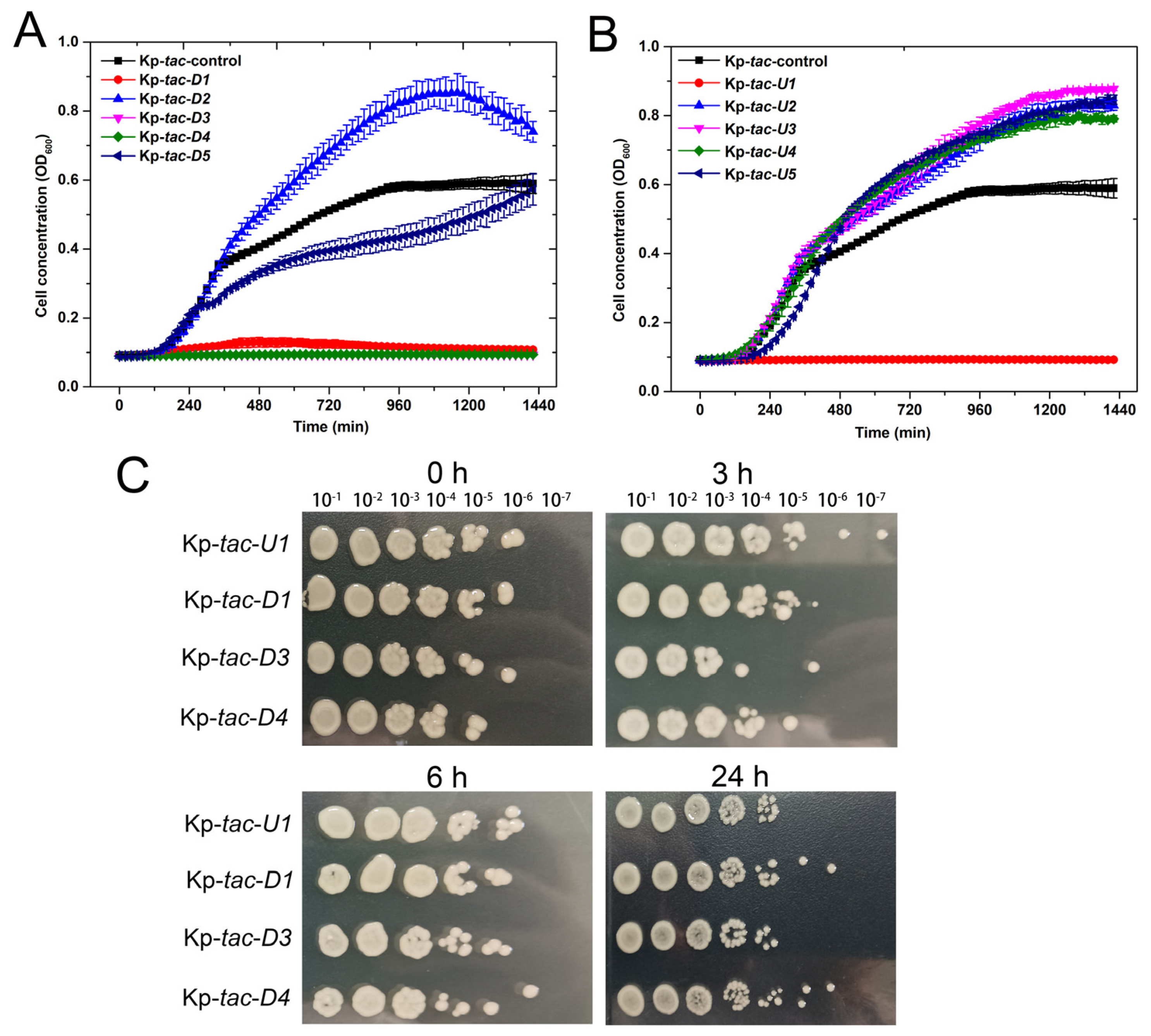
2.2. Overexpression of Kpsrmfs Resulted in Halted Cell Growth
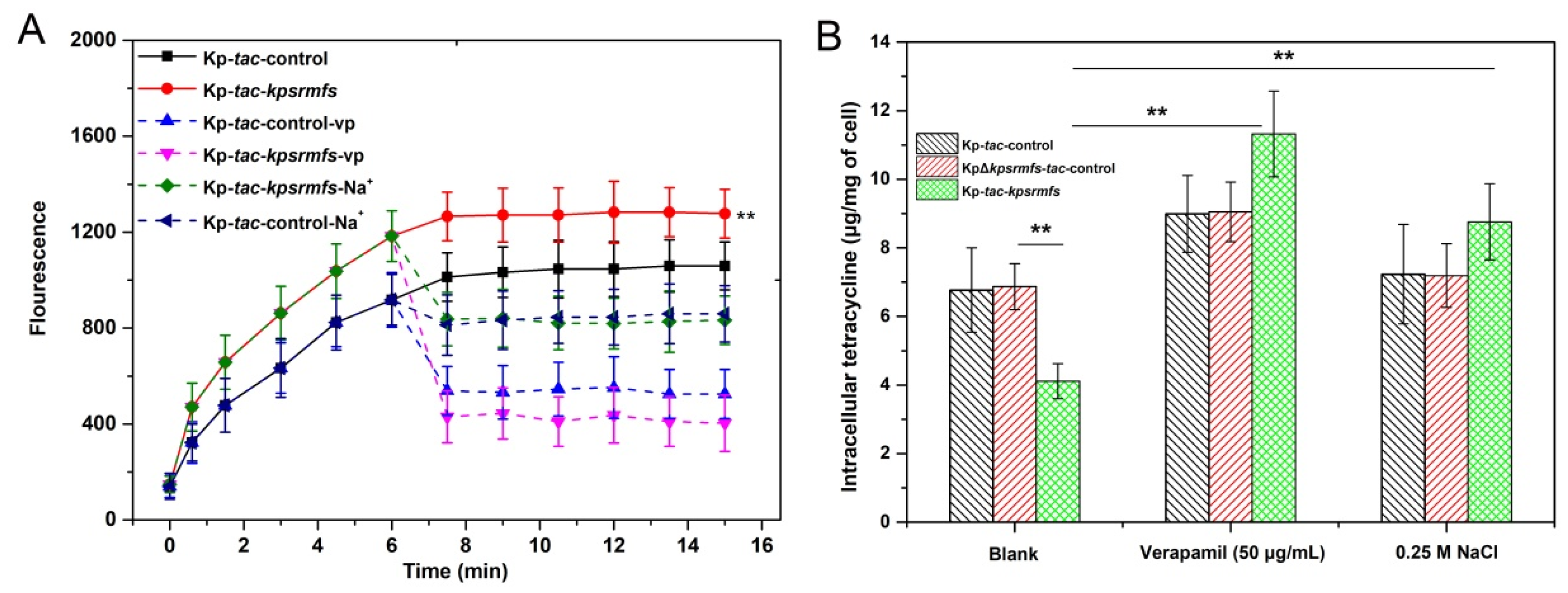
2.3. KpsrMFS Is an Active Efflux Pump for Tetracycline Export
2.4. Kpsrmfs Was Constitutively Expressed but Down-Regulated in the Presence of Multi-Stress Factors
2.5. Overexpression of Four Adjacent Genes of Kpsrmfs Resulted in Similar Phenotypes
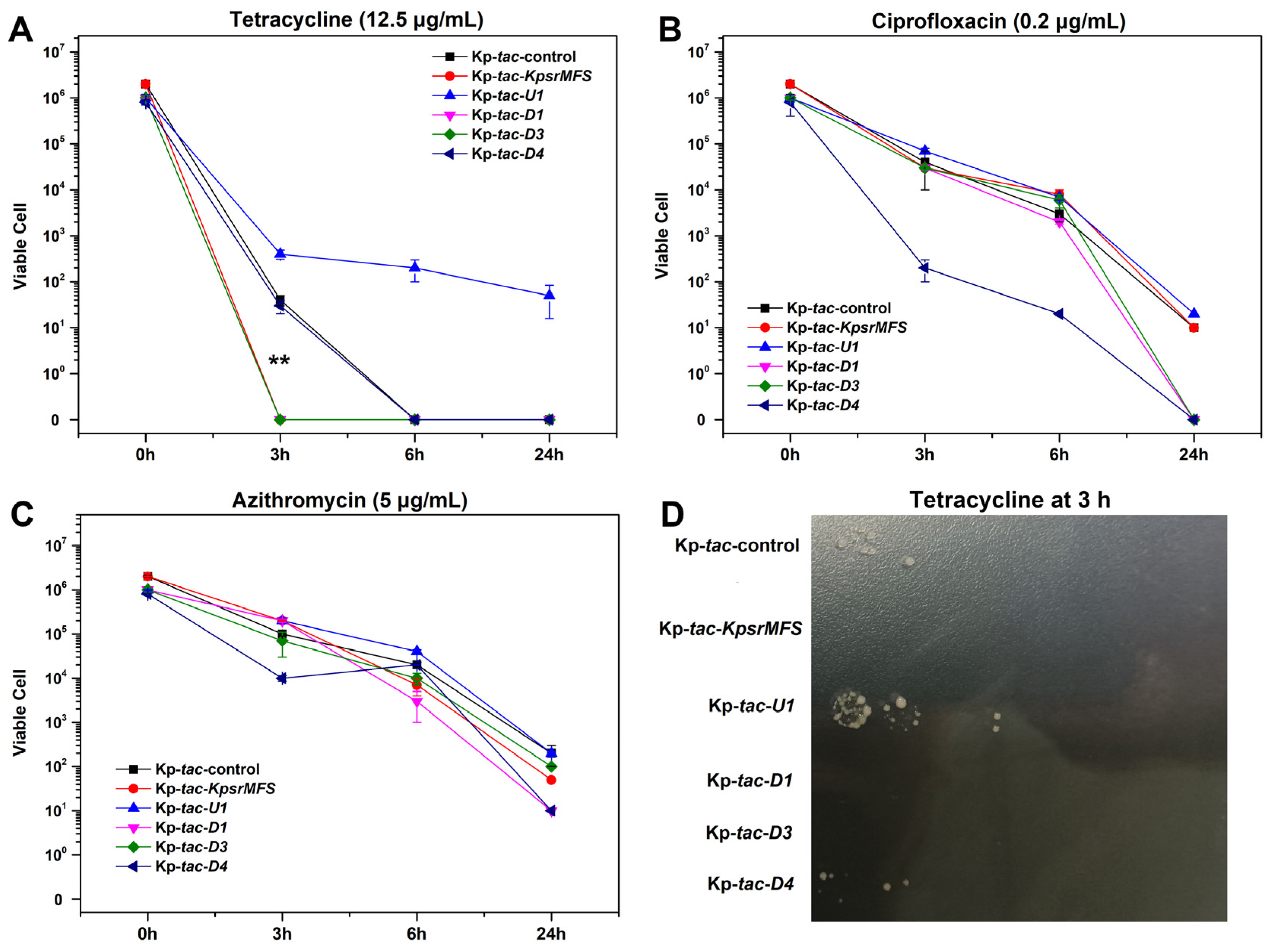
2.6. Overexpression of Kpsrmfs Conferred Slightly Reduced Tetracycline Tolerance in Killing Assay
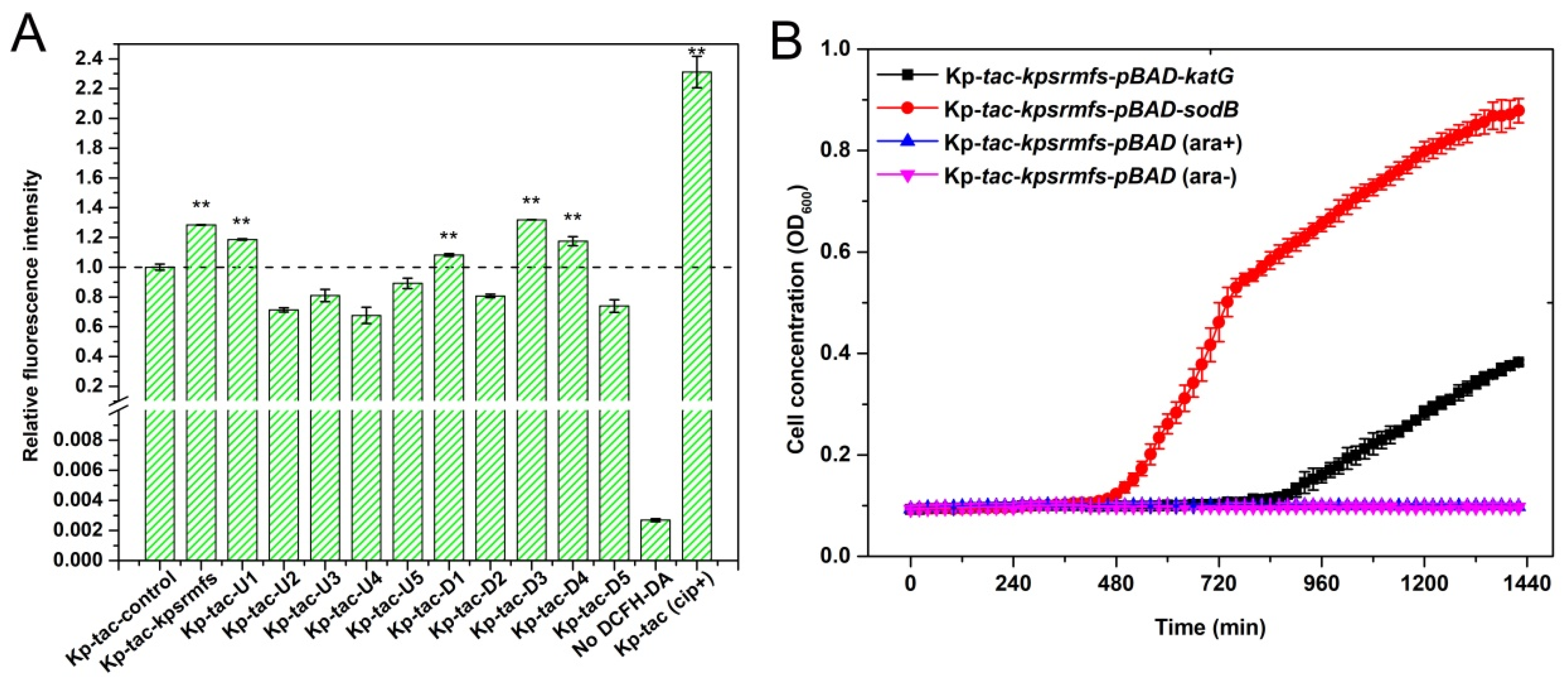
2.7. ROS Levels Were Significantly Elevated by Inducing Genes That Confer Halted Cell Growth
3. Discussion
4. Materials and Methods
4.1. Chemicals and Strains
4.2. Transcriptome Analysis
4.3. Next Generation Sequencing Data Analysis
4.4. Genetic Modifications
4.5. Determination of Minimum Inhibitory Concentration (MIC)
4.6. Growth Curve and Fluorescence Intensity Assay
4.7. Time-Dependent Killing Experiments
4.8. Gene Expression Analysis
4.9. Tetracycline Efflux and Accumulation Assay
4.10. Identification of Reactive Oxygen Species (ROS) Levels
4.11. Measurement of Catalase and Superoxide Dismutase Activities
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MFS | Major facilitator superfamily |
| ROS | Reactive oxygen species |
| ABC | ATP-binding cassette family |
| RND | Resistance/nodulation/cell division family |
| MATE | Multi-drug and toxic compound extrusion family |
| SMR | Small multi-drug resistance family |
| PACE | Proteobacterial antimicrobial compound efflux family |
| AbgT | p-Aminobenzoyl-glutamate transporter family |
| IPTG | Isopropyl β-D-1-thiogalactopyranoside |
| MHB | Mueller–Hinton Broth |
| LB | Luria–Bertani medium |
| MIC | Minimum inhibitory concentration |
| DCCM | Dynamical Cross-Correlation Matrix |
| CHL | Chloramphenicol |
References
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Choby, J.E.; Howard-Anderson, J.; Weiss, D.S. Hypervirulent Klebsiella pneumoniae—Clinical and Molecular Perspectives. J. Intern. Med. 2020, 287, 283–300. [Google Scholar] [CrossRef] [PubMed]
- Russo, T.A.; Marr, C.M. Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 2019, 32, e00001-19. [Google Scholar] [CrossRef] [PubMed]
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella pneumoniae: A Major Worldwide Source and Shuttle for Antibiotic Resistance. FEMS Microbiol. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef] [PubMed]
- Delmar, J.A.; Su, C.-C.; Yu, E.W. Bacterial Multidrug Efflux Transporters. Annu. Rev. Biophys. 2014, 43, 93–117. [Google Scholar] [CrossRef] [PubMed]
- Nolivos, S.; Cayron, J.; Dedieu, A.; Page, A.; Delolme, F.; Lesterlin, C. Role of AcrAB-TolC Multidrug Efflux Pump in Drug-Resistance Acquisition by Plasmid Transfer. Science 2019, 364, 778–782. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, H.; Ning, J.; Sajid, A.; Cheng, G.; Yuan, Z.; Hao, H. The Nature and Epidemiology of OqxAB, a Multidrug Efflux Pump. Antimicrob. Resist. Infect. Control 2019, 8, 44. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Nawaz, N.; Dermani, F.K.; Kohlan, A.K.; Saidijam, M.; Patching, S.G. Bacterial Multidrug Efflux Proteins: A Major Mechanism of Antimicrobial Resistance. Curr. Drug Targets 2018, 20, 16–28. [Google Scholar] [CrossRef]
- Teelucksingh, T.; Thompson, L.K.; Cox, G. The Evolutionary Conservation of Escherichia coli Drug Efflux Pumps Supports Physiological Functions. J. Bacteriol. 2020, 202, e00367-20. [Google Scholar] [CrossRef]
- Rizzi, A.; Roy, S.; Bellenger, J.-P.; Beauregard, P.B. Iron Homeostasis in Bacillus subtilis Requires Siderophore Production and Biofilm Formation. Appl. Environ. Microbiol. 2019, 85, e02439-18. [Google Scholar] [CrossRef]
- Alav, I.; Sutton, J.M.; Rahman, K.M. Role of Bacterial Efflux Pumps in Biofilm Formation. J. Antimicrob. Chemother. 2018, 73, 2003–2020. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Wu, C.; Gao, H.; Xu, C.; Dai, M.; Huang, L.; Hao, H.; Wang, X.; Cheng, G. Bacterial Multidrug Efflux Pumps at the Frontline of Antimicrobial Resistance: An Overview. Antibiotics 2022, 11, 520. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ge, X. Molecular Dynamics Investigation of MFS Efflux Pump MdfA Reveals an Intermediate State between Its Inward and Outward Conformations. Int. J. Mol. Sci. 2022, 24, 356. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Wan, M.; Wang, C.; Gao, X.; Yang, Q.; Partridge, S.R.; Wang, Y.; Zong, Z.; Doi, Y.; Shen, J.; et al. Emergence of a Plasmid-Encoded Resistance-Nodulation-Division Efflux Pump Conferring Resistance to Multiple Drugs, Including Tigecycline, in Klebsiella pneumoniae. mBio 2020, 11, e02930-19. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; van Veen, H.W.; Luisi, B.F. Assembly and Operation of Bacterial Tripartite Multidrug Efflux Pumps. Trends Microbiol. 2015, 23, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Alcalde-Rico, M.; Hernando-Amado, S.; Blanco, P.; Martínez, J.L. Multidrug Efflux Pumps at the Crossroad between Antibiotic Resistance and Bacterial Virulence. Front. Microbiol. 2016, 7, 1483. [Google Scholar] [CrossRef] [PubMed]
- Lamarche, M.G.; Déziel, E. MexEF-OprN Efflux Pump Exports the Pseudomonas quinolone Signal (PQS) Precursor HHQ (4-Hydroxy-2-Heptylquinoline). PLoS ONE 2011, 6, e24310. [Google Scholar] [CrossRef]
- Li, Y. A New Member of the Tripartite Multidrug Efflux Pumps, MexVW-OprM, in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2003, 52, 572–575. [Google Scholar] [CrossRef]
- Turner, W.J.; Dunlop, M.J. Trade-Offs in Improving Biofuel Tolerance Using Combinations of Efflux Pumps. ACS Synth. Biol. 2015, 4, 1056–1063. [Google Scholar] [CrossRef]
- Opperman, T.J.; Kwasny, S.M.; Kim, H.-S.; Nguyen, S.T.; Houseweart, C.; D’Souza, S.; Walker, G.C.; Peet, N.P.; Nikaido, H.; Bowlin, T.L. Characterization of a Novel Pyranopyridine Inhibitor of the AcrAB Efflux Pump of Escherichia coli. Antimicrob. Agents Chemother. 2014, 58, 722–733. [Google Scholar] [CrossRef]
- Srinivasan, V.B.; Rajamohan, G. KpnEF, a New Member of the Klebsiella pneumoniae Cell Envelope Stress Response Regulon, Is an SMR-Type Efflux Pump Involved in Broad-Spectrum Antimicrobial Resistance. Antimicrob. Agents Chemother. 2013, 57, 4449–4462. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Deng, Z.; Yan, A. Bacterial Multidrug Efflux Pumps: Mechanisms, Physiology and Pharmacological Exploitations. Biochem. Biophys. Res. Commun. 2014, 453, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Truong-Bolduc, Q.C.; Bolduc, G.R.; Okumura, R.; Celino, B.; Bevis, J.; Liao, C.-H.; Hooper, D.C. Implication of the NorB Efflux Pump in the Adaptation of Staphylococcus aureus to Growth at Acid pH and in Resistance to Moxifloxacin. Antimicrob. Agents Chemother. 2011, 55, 3214–3219. [Google Scholar] [CrossRef] [PubMed]
- Van Acker, H.; Coenye, T. The Role of Efflux and Physiological Adaptation in Biofilm Tolerance and Resistance. J. Biol. Chem. 2016, 291, 12565–12572. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cross, T.S.; Dörr, T. Analysis of AcrB in Klebsiella pneumoniae Reveals Natural Variants Promoting Enhanced Multidrug Resistance. Res. Microbiol. 2022, 173, 103901. [Google Scholar] [CrossRef] [PubMed]
- Cross, T.; Ransegnola, B.; Shin, J.-H.; Weaver, A.; Fauntleroy, K.; VanNieuwenhze, M.S.; Westblade, L.F.; Dörr, T. Spheroplast-Mediated Carbapenem Tolerance in Gram-negative Pathogens. Antimicrob. Agents Chemother. 2019, 63, e00756-19. [Google Scholar] [CrossRef] [PubMed]
- Truong-Bolduc, Q.C.; Wang, Y.; Hooper, D.C. Role of Staphylococcus aureus Tet38 in Transport of Tetracycline and Its Regulation in a Salt Stress Environment. J. Bacteriol. 2022, 204, e00142-22. [Google Scholar] [CrossRef]
- Skulj, M.; Okrslar, V.; Jalen, S.; Jevsevar, S.; Slanc, P.; Strukelj, B.; Menart, V. Improved Determination of Plasmid Copy Number Using Quantitative Real-Time PCR for Monitoring Fermentation Processes. Microb. Cell Factories 2008, 7, 6. [Google Scholar] [CrossRef]
- Li, Y.; Ge, X.-Z.; Tian, P.-F. Production of 1,3-Propanediol from Glycerol Using a New Isolate Klebsiella sp. AA405 Carrying Low Levels of Virulence Factors. Biotechnol. Biotechnol. Equip. 2017, 31, 725–732. [Google Scholar] [CrossRef]
- Wu, C.-J.; Chen, Y.; Li, L.-H.; Wu, C.-M.; Lin, Y.-T.; Ma, C.-H.; Yang, T.-C. Roles of SmeYZ, SbiAB, and SmeDEF Efflux Systems in Iron Homeostasis of Stenotrophomonas maltophilia. Microbiol. Spectr. 2022, 10, e02448-21. [Google Scholar] [CrossRef]
- Thomas, E.; Roman, E.; Claypool, S.; Manzoor, N.; Pla, J.; Panwar, S.L. Mitochondria Influence CDR1 Efflux Pump Activity, Hog1-Mediated Oxidative Stress Pathway, Iron Homeostasis, and Ergosterol Levels in Candida albicans. Antimicrob. Agents Chemother. 2013, 57, 5580–5599. [Google Scholar] [CrossRef] [PubMed]
- Vecchione, J.J.; Alexander, B.; Sello, J.K. Two Distinct Major Facilitator Superfamily Drug Efflux Pumps Mediate Chloramphenicol Resistance in Streptomyces coelicolor. Antimicrob. Agents Chemother. 2009, 53, 4673–4677. [Google Scholar] [CrossRef] [PubMed]
- Lamarche, M.G.; Wanner, B.L.; Crépin, S.; Harel, J. The Phosphate Regulon and Bacterial Virulence: A Regulatory Network Connecting Phosphate Homeostasis and Pathogenesis. FEMS Microbiol. Rev. 2008, 32, 461–473. [Google Scholar] [CrossRef] [PubMed]
- Wood, L.F.; Ohman, D.E. Identification of Genes in the σ 22 Regulon of Pseudomonas aeruginosa Required for Cell Envelope Homeostasis in Either the Planktonic or the Sessile Mode of Growth. mBio 2012, 3, e00094-12. [Google Scholar] [CrossRef] [PubMed]
- Skrt, M.; Jamnik, P.; Poklar Ulrih, N. Antioxidative Activity of Methanolic and Water Extracts from the Hyperthermophilic Archaeon Aeropyrum pernix K1. Acta Chim. Slov. 2018, 65, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Bonds, A.C.; Sampson, N.S. More than Cholesterol Catabolism: Regulatory Vulnerabilities in Mycobacterium tuberculosis. Curr. Opin. Chem. Biol. 2018, 44, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Aggett, R.; Mallette, E.; Gilbert, S.E.; Vachon, M.A.; Schroeter, K.L.; Kimber, M.S.; Seah, S.Y.K. The Steroid Side-Chain–Cleaving Aldolase Ltp2–ChsH2DUF35 Is a Thiolase Superfamily Member with a Radically Repurposed Active Site. J. Biol. Chem. 2019, 294, 11934–11943. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Choe, J. The X-Ray Crystal Structure of PA1374 from Pseudomonas aeruginosa, a Putative Oxidative-Stress Sensing Transcriptional Regulator. Biochem. Biophys. Res. Commun. 2013, 431, 376–381. [Google Scholar] [CrossRef]
- Newberry, K.J.; Fuangthong, M.; Panmanee, W.; Mongkolsuk, S.; Brennan, R.G. Structural Mechanism of Organic Hydroperoxide Induction of the Transcription Regulator OhrR. Mol. Cell 2007, 28, 652–664. [Google Scholar] [CrossRef]
- Li, Y.; He, R.; Ge, X. Glycerol Promotes Biomass Accumulation of Klebsiella pneumoniae by Activating dha Regulon. Process Biochem. 2022, 117, 68–75. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.-Y.; Dillies, M.-A. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ge, X. Discovering Interrelated Natural Mutations of Efflux Pump KmrA from Klebsiella pneumoniae That Confer Increased Multidrug Resistance. Protein Sci. 2022, 31, e4323. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, A.; Tauch, A.; Jäger, W.; Kalinowski, J.; Thierbach, G.; Pühler, A. Small Mobilizable Multi-Purpose Cloning Vectors Derived from the Escherichia coli Plasmids pK18 and pK19: Selection of Defined Deletions in the Chromosome of Corynebacterium Glutamicum. Gene 1994, 145, 69–73. [Google Scholar] [CrossRef]
- Lazarus, J.E.; Warr, A.R.; Kuehl, C.J.; Giorgio, R.T.; Davis, B.M.; Waldor, M.K. A New Suite of Allelic-Exchange Vectors for the Scarless Modification of Proteobacterial Genomes. Appl. Environ. Microbiol. 2019, 85, e00990-19. [Google Scholar] [CrossRef]
- Mortimer, P.G.S.; Piddock, L.J.V. A Comparison of Methods Used for Measuring the Accumulation of Quinolones by Enterobacteriaceae, Pseudomonas aeruginosa and Staphylococcus aureus. J. Antimicrob. Chemother. 1991, 28, 639–653. [Google Scholar] [CrossRef]
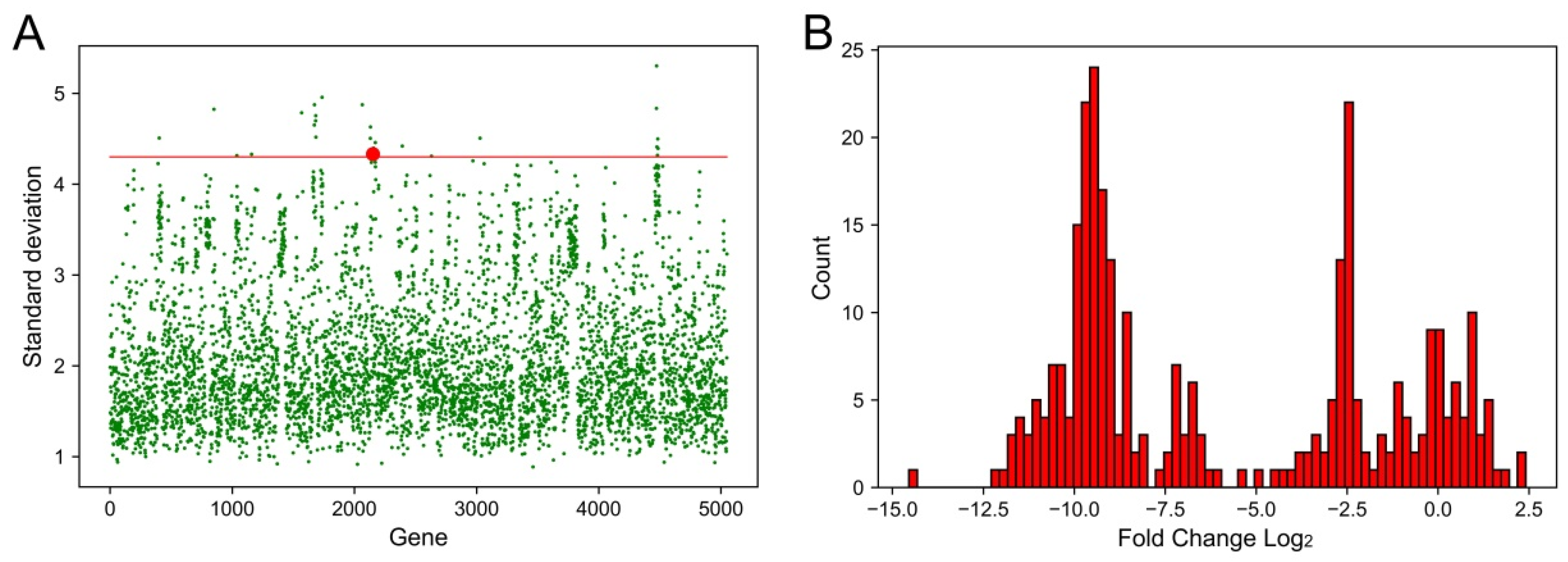



| Number | Gene ID | Annotation | Gene Name | Standard Deviation |
|---|---|---|---|---|
| 0 | I5G01_02020 | 50S ribosomal protein L22 | rplV | 4.51 |
| 1 | I5G01_04320 | Type 3 fimbria major subunit MrkA | mrkA | 4.83 |
| 2 | I5G01_05265 | S8 family peptidase | Non | 4.32 |
| 3 | I5G01_05895 | Helix-turn-helix domain-containing protein | Non | 4.33 |
| 4 | I5G01_08000 | Hypothetical protein | Non | 4.79 |
| 5 | I5G01_08515 | Mannose-1-phosphate guanylyltransferase/mannose-6-phosphate isomerase | Non | 4.65 |
| 6 | I5G01_08520 | O9 family phosphomannomutase RfbK1 | rfbK1 | 4.88 |
| 7 | I5G01_08570 | Glycosyltransferase | Non | 4.70 |
| 8 | I5G01_08575 | Hypothetical protein | Non | 4.76 |
| 9 | I5G01_08580 | Glycosyltransferase family 2 protein | Non | 4.52 |
| 10 | I5G01_08835 | Winged helix-turn-helix domain-containing protein | Non | 4.96 |
| 11 | I5G01_10545 | Major outer membrane lipoprotein | Non | 4.88 |
| 12 | I5G01_10880 | HAMP domain-containing histidine kinase CrrB | ccrB | 4.50 |
| 13 | I5G01_10885 | Response regulator | Non | 4.63 |
| 14 | I5G01_10985 | MFS transporter | KpsrMFS | 4.33 |
| 15 | I5G01_10995 | Tautomerase family protein | Non | 4.35 |
| 16 | I5G01_11005 | Chromate resistance protein | Non | 4.40 |
| 17 | I5G01_11075 | DeoR/GlpR family DNA-binding transcription regulator | Non | 4.46 |
| 18 | I5G01_12185 | LysE family translocator | Non | 4.42 |
| 19 | I5G01_15370 | EAL domain-containing protein | Non | 4.51 |
| 20 | I5G01_22760 | Polysaccharide pyruvyl transferase family protein | Non | 4.84 |
| 21 | I5G01_22765 | Glycosyltransferase | Non | 5.30 |
| 22 | I5G01_22775 | PAAR domain-containing protein | Non | 4.41 |
| 23 | I5G01_22800 | Hypothetical protein | Non | 4.32 |
| 24 | I5G01_22810 | AAA family ATPase | Non | 4.50 |
| 25 | I5G01_22825 | N-6 DNA methylase | Non | 4.39 |
| Strain | Kp-wt | KpΔkpsrmfs | Kp-pET-backbone | Kp-pET-Pstr-kpsrmfs |
|---|---|---|---|---|
| MIC (μg/mL) | ||||
| Meropenem | 0.0025 | 0.0025 | 0.00125 | 0.00125 |
| Chloramphenicol | 0.02 | 0.02 | 0.01 | 0.01 |
| Ciprofloxacin | 0.002 | <0.0004 | 0.0004 | 0.0004 |
| Tetracycline | 0.25 | 0.03125 | 0.0625 | 0.125 |
| Azithromycin | 0.1 | 0.025 | 0.05 | 0.1 |
| Rifampicin | 1 | 1 | 0.5 | 0.5 |
| Streptomycin | 0.125 | 0.125 | 0.0625 | 0.0625 |
| Hygromycin | 0.0625 | 0.0625 | 0.0625 | 0.0625 |
| Name | Annotation | Gene Name | Gene ID |
|---|---|---|---|
| U5 | 3-oxoacid CoA-transferase subunit B | Non | I5G01_10960 |
| U4 | CoA transferase subunit A | Non | I5G01_10965 |
| U3 | LysR family transcriptional regulator | Non | I5G01_10970 |
| U2 | Hypothetical protein | Non | I5G01_10975 |
| U1 | DUF535 family protein | Non | I5G01_10980 |
| KpsrMFS | MFS transporter | kpsrmfs | I5G01_10985 |
| D1 | Helix-turn-helix transcriptional regulator | Non | I5G01_10990 |
| D2 | Tautomerase family protein | Non | I5G01_10995 |
| D3 | MFS transporter | Non | I5G01_11000 |
| D4 | Chromate resistance protein | Non | I5G01_11005 |
| D5 | Chromate efflux transporter | chrA | I5G01_11010 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, W.; Jiang, M.; Li, Y.; Ge, X. Identification of the Major Facilitator Superfamily Efflux Pump KpsrMFS in Klebsiella pneumoniae That Is Down-Regulated in the Presence of Multi-Stress Factors. Int. J. Mol. Sci. 2024, 25, 1466. https://doi.org/10.3390/ijms25031466
He W, Jiang M, Li Y, Ge X. Identification of the Major Facilitator Superfamily Efflux Pump KpsrMFS in Klebsiella pneumoniae That Is Down-Regulated in the Presence of Multi-Stress Factors. International Journal of Molecular Sciences. 2024; 25(3):1466. https://doi.org/10.3390/ijms25031466
Chicago/Turabian StyleHe, Wei, Minzhi Jiang, Ying Li, and Xizhen Ge. 2024. "Identification of the Major Facilitator Superfamily Efflux Pump KpsrMFS in Klebsiella pneumoniae That Is Down-Regulated in the Presence of Multi-Stress Factors" International Journal of Molecular Sciences 25, no. 3: 1466. https://doi.org/10.3390/ijms25031466





