Mucosal Genes Encoding Clock, Inflammation and Their Mutual Regulators Are Disrupted in Pediatric Patients with Active Ulcerative Colitis
Abstract
1. Introduction
2. Results
2.1. Expression Level of Genes Encoding Clock, Inflammation and Their Mutual Regulators
2.2. Expression Pattern of Genes Encoding Clock, Inflammation and Their Mutual Regulators
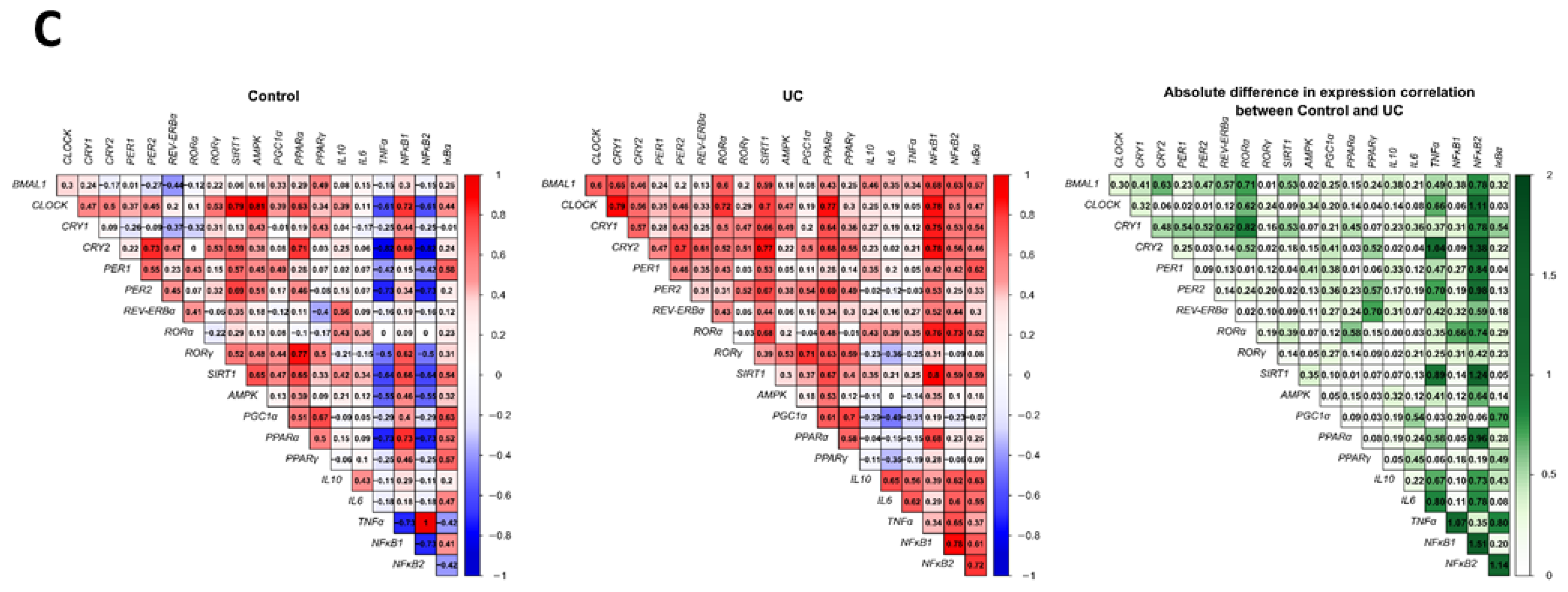
2.3. Correlation between Genes Encoding Clock, Inflammation and Their Mutual Regulators
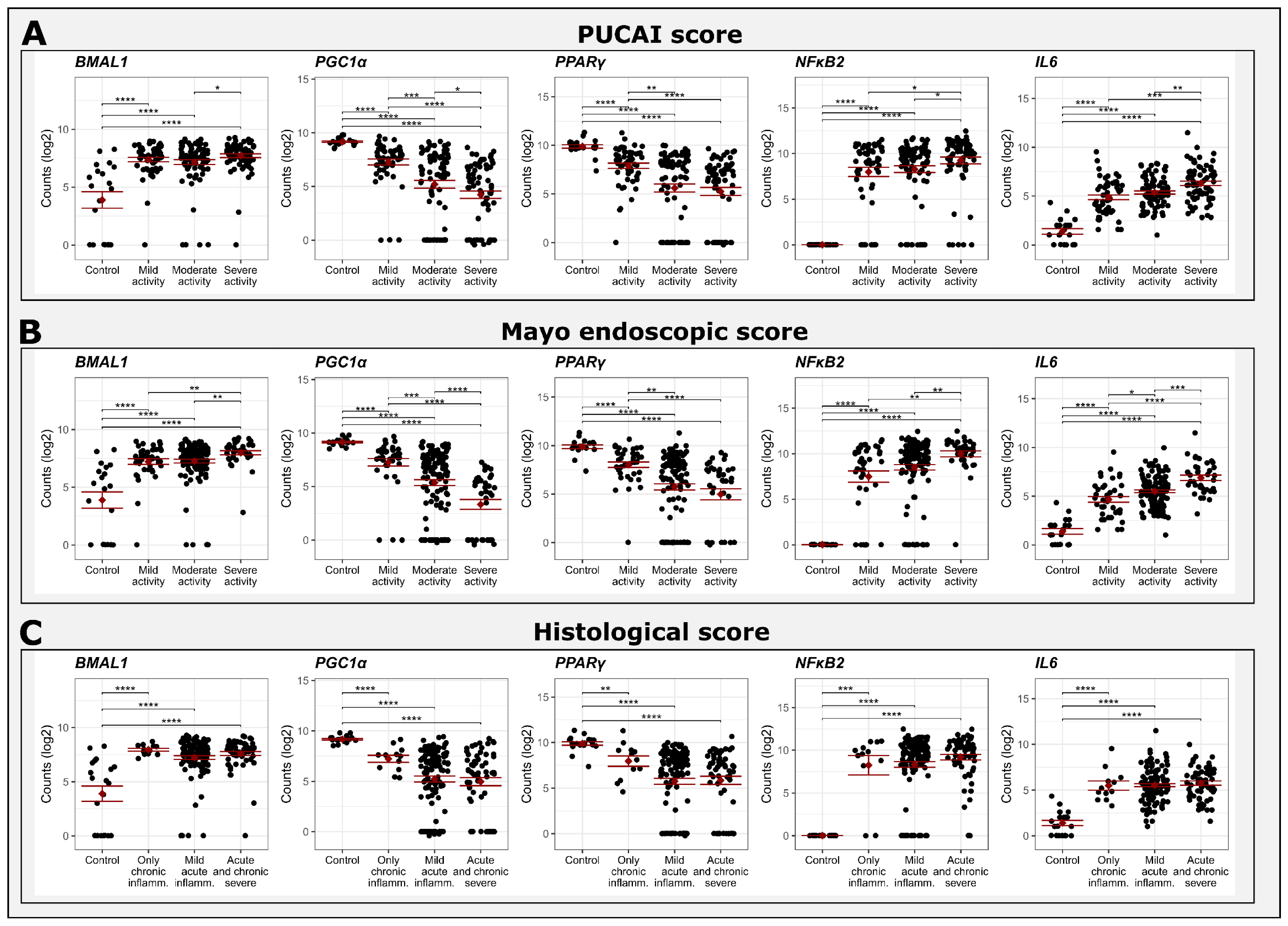
2.4. Comparison between Gene Expression and Clinical, Endoscopic and Histological Scores
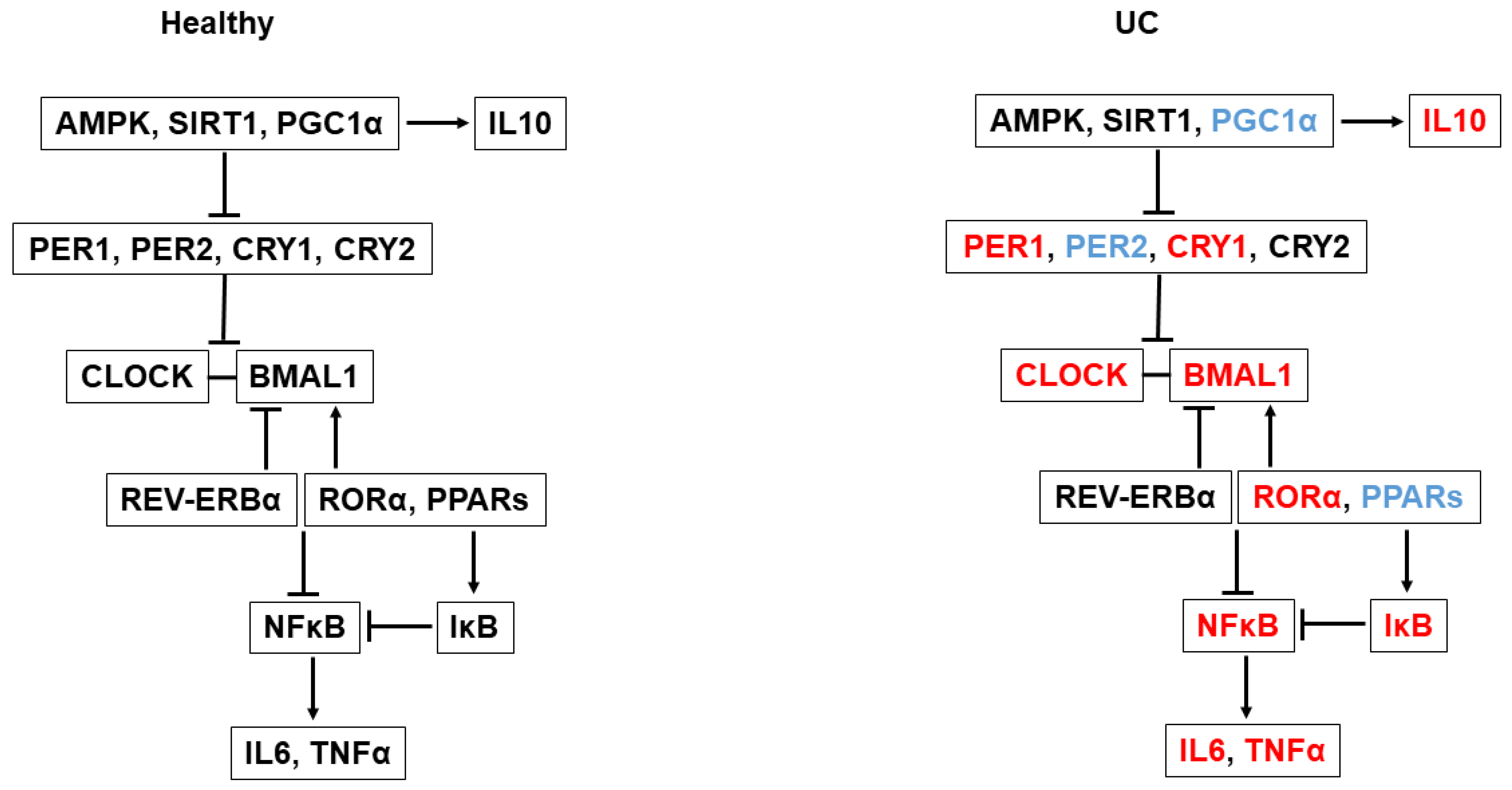
3. Discussion
4. Materials and Methods
4.1. Data Acquisition
4.2. Plotting and Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ungaro, R.; Mehandru, S.; Allen, P.B.; Peyrin-Biroulet, L.; Colombel, J.F. Ulcerative colitis. Lancet 2017, 389, 1756–1770. [Google Scholar] [CrossRef]
- Mosna, K.; Janega, P.; Sedlak, J.; Babal, P. Complex changes of circadian proteins expression in inflammatory bowel disease. Bratisl. Lek. Listy 2021, 122, 235–241. [Google Scholar] [CrossRef]
- Liu, X.; Yu, R.; Zhu, L.; Hou, X.; Zou, K. Bidirectional Regulation of Circadian Disturbance and Inflammation in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2017, 23, 1741–1751. [Google Scholar] [CrossRef]
- Palmieri, O.; Mazzoccoli, G.; Bossa, F.; Maglietta, R.; Palumbo, O.; Ancona, N.; Corritore, G.; Latiano, T.; Martino, G.; Rubino, R.; et al. Systematic analysis of circadian genes using genome-wide cDNA microarrays in the inflammatory bowel disease transcriptome. Chronobiol. Int. 2015, 32, 903–916. [Google Scholar] [CrossRef] [PubMed]
- Weintraub, Y.; Cohen, S.; Chapnik, N.; Ben-Tov, A.; Yerushalmy-Feler, A.; Dotan, I.; Tauman, R.; Froy, O. Clock Gene Disruption Is an Initial Manifestation of Inflammatory Bowel Diseases. Clin. Gastroenterol. Hepatol. 2020, 18, 115–122.e1. [Google Scholar] [CrossRef]
- Froy, O.; Garaulet, M. The Circadian Clock in White and Brown Adipose Tissue: Mechanistic, Endocrine, and Clinical Aspects. Endocr. Rev. 2018, 39, 261–273. [Google Scholar] [CrossRef]
- Allada, R.; Bass, J. Circadian Mechanisms in Medicine. N. Engl. J. Med. 2021, 384, 550–561. [Google Scholar] [CrossRef] [PubMed]
- Sobolewska-Wlodarczyk, A.; Wlodarczyk, M.; Szemraj, J.; Stec-Michalska, K.; Fichna, J.; Wisniewska-Jarosinska, M. Circadian rhythm abnormalities—Association with the course of inflammatory bowel disease. Pharmacol. Rep. 2016, 68, 847–851. [Google Scholar] [CrossRef]
- Hui, L.; Hua, F.; Diandong, H.; Hong, Y. Effects of sleep and sleep deprivation on immunoglobulins and complement in humans. Brain Behav. Immun. 2007, 21, 308–310. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, E.A.; Richardson, J.D.; Holman, G.D.; Tsintzas, K.; Thompson, D.; Betts, J.A. The causal role of breakfast in energy balance and health: A randomized controlled trial in obese adults. Am. J. Clin. Nutr. 2016, 103, 747–756. [Google Scholar] [CrossRef]
- Preitner, N.; Damiola, F.; Lopez-Molina, L.; Zakany, J.; Duboule, D.; Albrecht, U.; Schibler, U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 2002, 110, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.K.; Panda, S.; Miraglia, L.J.; Reyes, T.M.; Rudic, R.D.; McNamara, P.; Naik, K.A.; FitzGerald, G.A.; Kay, S.A.; Hogenesch, J.B. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 2004, 43, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, J.E.; Blaikley, J.; Beesley, S.; Matthews, L.; Simpson, K.D.; Boyce, S.H.; Farrow, S.N.; Else, K.J.; Singh, D.; Ray, D.W.; et al. The nuclear receptor REV-ERBalpha mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc. Natl. Acad. Sci. USA 2012, 109, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Pourcet, B.; Duez, H. Nuclear Receptors and Clock Components in Cardiovascular Diseases. Int. J. Mol. Sci. 2021, 22, 9721. [Google Scholar] [CrossRef] [PubMed]
- Delerive, P.; Monte, D.; Dubois, G.; Trottein, F.; Fruchart-Najib, J.; Mariani, J.; Fruchart, J.C.; Staels, B. The orphan nuclear receptor ROR alpha is a negative regulator of the inflammatory response. EMBO Rep. 2001, 2, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Raza, G.S.; Sodum, N.; Kaya, Y.; Herzig, K.H. Role of Circadian Transcription Factor Rev-Erb in Metabolism and Tissue Fibrosis. Int. J. Mol. Sci. 2022, 23, 12954. [Google Scholar] [CrossRef] [PubMed]
- Canaple, L.; Rambaud, J.; Dkhissi-Benyahya, O.; Rayet, B.; Tan, N.S.; Michalik, L.; Delaunay, F.; Wahli, W.; Laudet, V. Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor alpha defines a novel positive feedback loop in the rodent liver circadian clock. Mol. Endocrinol. 2006, 20, 1715–1727. [Google Scholar] [CrossRef]
- Liu, C.; Li, S.; Liu, T.; Borjigin, J.; Lin, J.D. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature 2007, 447, 477–481. [Google Scholar] [CrossRef]
- Wettstein, G.; Luccarini, J.M.; Poekes, L.; Faye, P.; Kupkowski, F.; Adarbes, V.; Defrene, E.; Estivalet, C.; Gawronski, X.; Jantzen, I.; et al. The new-generation pan-peroxisome proliferator-activated receptor agonist IVA337 protects the liver from metabolic disorders and fibrosis. Hepatol. Commun. 2017, 1, 524–537. [Google Scholar] [CrossRef]
- Puengel, T.; Liu, H.; Guillot, A.; Heymann, F.; Tacke, F.; Peiseler, M. Nuclear Receptors Linking Metabolism, Inflammation, and Fibrosis in Nonalcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2022, 23, 2668. [Google Scholar] [CrossRef]
- Croasdell, A.; Duffney, P.F.; Kim, N.; Lacy, S.H.; Sime, P.J.; Phipps, R.P. PPARgamma and the Innate Immune System Mediate the Resolution of Inflammation. PPAR Res. 2015, 2015, 549691. [Google Scholar] [CrossRef] [PubMed]
- Rius-Perez, S.; Torres-Cuevas, I.; Millan, I.; Ortega, A.L.; Perez, S. PGC-1alpha, Inflammation, and Oxidative Stress: An Integrative View in Metabolism. Oxid. Med. Cell Longev. 2020, 2020, 1452696. [Google Scholar] [CrossRef] [PubMed]
- Eide, E.J.; Woolf, M.F.; Kang, H.; Woolf, P.; Hurst, W.; Camacho, F.; Vielhaber, E.L.; Giovanni, A.; Virshup, D.M. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol. Cell. Biol. 2005, 25, 2795–2807. [Google Scholar] [CrossRef] [PubMed]
- Um, J.H.; Yang, S.; Yamazaki, S.; Kang, H.; Viollet, B.; Foretz, M.; Chung, J.H. Activation of 5′-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)-dependent degradation of clock protein mPER2. J. Biol. Chem. 2007, 282, 20794–20798. [Google Scholar] [CrossRef]
- Lamia, K.A.; Sachdeva, U.M.; DiTacchio, L.; Williams, E.C.; Alvarez, J.G.; Egan, D.F.; Vasquez, D.S.; Juguilon, H.; Panda, S.; Shaw, R.J.; et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 2009, 326, 437–440. [Google Scholar] [CrossRef] [PubMed]
- Asher, G.; Gatfield, D.; Stratmann, M.; Reinke, H.; Dibner, C.; Kreppel, F.; Mostoslavsky, R.; Alt, F.W.; Schibler, U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 2008, 134, 317–328. [Google Scholar] [CrossRef]
- Nakahata, Y.; Sahar, S.; Astarita, G.; Kaluzova, M.; Sassone-Corsi, P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 2009, 324, 654–657. [Google Scholar] [CrossRef]
- Nakahata, Y.; Kaluzova, M.; Grimaldi, B.; Sahar, S.; Hirayama, J.; Chen, D.; Guarente, L.P.; Sassone-Corsi, P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 2008, 134, 329–340. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Wu, Q. Sulforaphane protects intestinal epithelial cells against lipopolysaccharide-induced injury by activating the AMPK/SIRT1/PGC-1a pathway. Bioengineered 2021, 12, 4349–4360. [Google Scholar] [CrossRef]
- Parekh, P.J.; Oldfield Iv, E.C.; Challapallisri, V.; Ware, J.C.; Johnson, D.A. Sleep disorders and inflammatory disease activity: Chicken or the egg? Am. J. Gastroenterol. 2015, 110, 484–488. [Google Scholar] [CrossRef]
- Turner, D.; Ricciuto, A.; Lewis, A.; D’Amico, F.; Dhaliwal, J.; Griffiths, A.M.; Bettenworth, D.; Sandborn, W.J.; Sands, B.E.; Reinisch, W.; et al. STRIDE-II: An Update on the Selecting Therapeutic Targets in Inflammatory Bowel Disease (STRIDE) Initiative of the International Organization for the Study of IBD (IOIBD): Determining Therapeutic Goals for Treat-to-Target strategies in IBD. Gastroenterology 2021, 160, 1570–1583. [Google Scholar] [CrossRef] [PubMed]
- Haberman, Y.; Karns, R.; Dexheimer, P.J.; Schirmer, M.; Somekh, J.; Jurickova, I.; Braun, T.; Novak, E.; Bauman, L.; Collins, M.H.; et al. Ulcerative colitis mucosal transcriptomes reveal mitochondriopathy and personalized mechanisms underlying disease severity and treatment response. Nat. Commun. 2019, 10, 38. [Google Scholar] [CrossRef] [PubMed]
- Laurindo, L.F.; Santos, A.; Carvalho, A.C.A.; Bechara, M.D.; Guiguer, E.L.; Goulart, R.A.; Vargas Sinatora, R.; Araujo, A.C.; Barbalho, S.M. Phytochemicals and Regulation of NF-kB in Inflammatory Bowel Diseases: An Overview of In Vitro and In Vivo Effects. Metabolites 2023, 13, 96. [Google Scholar] [CrossRef]
- Tambuwala, M.M. Natural Nuclear Factor Kappa Beta Inhibitors: Safe Therapeutic Options for Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2016, 22, 719–723. [Google Scholar] [CrossRef] [PubMed]
- Oishi, K.; Shirai, H.; Ishida, N. CLOCK is involved in the circadian transactivation of peroxisome-proliferator-activated receptor alpha (PPARalpha) in mice. Biochem. J. 2005, 386, 575–581. [Google Scholar] [CrossRef]
- Inoue, I.; Shinoda, Y.; Ikeda, M.; Hayashi, K.; Kanazawa, K.; Nomura, M.; Matsunaga, T.; Xu, H.; Kawai, S.; Awata, T.; et al. CLOCK/BMAL1 is involved in lipid metabolism via transactivation of the peroxisome proliferator-activated receptor (PPAR) response element. J. Atheroscler. Thromb. 2005, 12, 169–174. [Google Scholar] [CrossRef]
- Cyr, P.; Bronner, S.M.; Crawford, J.J. Recent progress on nuclear receptor RORgamma modulators. Bioorg Med. Chem. Lett. 2016, 26, 4387–4393. [Google Scholar] [CrossRef]
- Wan, X.; Zhu, X.; Wang, H.; Feng, Y.; Zhou, W.; Liu, P.; Shen, W.; Zhang, L.; Liu, L.; Li, T.; et al. PGC1alpha protects against hepatic steatosis and insulin resistance via enhancing IL10-mediated anti-inflammatory response. FASEB J. 2020, 34, 10751–10761. [Google Scholar] [CrossRef]
- El-Ghannam, M.S.; Saad, M.A.; Nassar, N.N.; El-Yamany, M.F.; El-Bahy, A.A.Z. Linagliptin ameliorates acetic acid-induced colitis via modulating AMPK/SIRT1/PGC-1alpha and JAK2/STAT3 signaling pathway in rats. Toxicol. Appl. Pharmacol. 2022, 438, 115906. [Google Scholar] [CrossRef]
- Jiang, C.; Ting, A.T.; Seed, B. PPAR-gamma agonists inhibit production of monocyte inflammatory cytokines. Nature 1998, 391, 82–86. [Google Scholar] [CrossRef]
- Dubuquoy, L.; Jansson, E.A.; Deeb, S.; Rakotobe, S.; Karoui, M.; Colombel, J.F.; Auwerx, J.; Pettersson, S.; Desreumaux, P. Impaired expression of peroxisome proliferator-activated receptor gamma in ulcerative colitis. Gastroenterology 2003, 124, 1265–1276. [Google Scholar] [CrossRef]
- Curtis, A.M.; Bellet, M.M.; Sassone-Corsi, P.; O’Neill, L.A. Circadian clock proteins and immunity. Immunity 2014, 40, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Bromke, M.A.; Neubauer, K.; Kempinski, R.; Krzystek-Korpacka, M. Faecal Calprotectin in Assessment of Mucosal Healing in Adults with Inflammatory Bowel Disease: A Meta-Analysis. J. Clin. Med. 2021, 10, 2203. [Google Scholar] [CrossRef] [PubMed]
- Weintraub, Y.; Cohen, S.; Anafy, A.; Chapnik, N.; Tsameret, S.; Ben-Tov, A.; Yerushalmy-Feler, A.; Dotan, I.; Tauman, R.; Froy, O. Inverse Relationship Between Clock Gene Expression and Inflammatory Markers in Ulcerative Colitis Patients Undergoing Remission. Dig. Dis. Sci. 2023, 68, 2454–2462. [Google Scholar] [CrossRef] [PubMed]
- Pelia, R.; Venkateswaran, S.; Matthews, J.D.; Haberman, Y.; Cutler, D.J.; Hyams, J.S.; Denson, L.A.; Kugathasan, S. Profiling non-coding RNA levels with clinical classifiers in pediatric Crohn’s disease. BMC Med. Genomics 2021, 14, 194. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2—Elegant Graphics for Data Analysis, 2nd ed.; Springer: New York, NY, USA, 2016. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Labes, S.; Froy, O.; Tabach, Y.; Shamir, R.; Shouval, D.S.; Weintraub, Y. Mucosal Genes Encoding Clock, Inflammation and Their Mutual Regulators Are Disrupted in Pediatric Patients with Active Ulcerative Colitis. Int. J. Mol. Sci. 2024, 25, 1488. https://doi.org/10.3390/ijms25031488
Labes S, Froy O, Tabach Y, Shamir R, Shouval DS, Weintraub Y. Mucosal Genes Encoding Clock, Inflammation and Their Mutual Regulators Are Disrupted in Pediatric Patients with Active Ulcerative Colitis. International Journal of Molecular Sciences. 2024; 25(3):1488. https://doi.org/10.3390/ijms25031488
Chicago/Turabian StyleLabes, Sapir, Oren Froy, Yuval Tabach, Raanan Shamir, Dror S. Shouval, and Yael Weintraub. 2024. "Mucosal Genes Encoding Clock, Inflammation and Their Mutual Regulators Are Disrupted in Pediatric Patients with Active Ulcerative Colitis" International Journal of Molecular Sciences 25, no. 3: 1488. https://doi.org/10.3390/ijms25031488
APA StyleLabes, S., Froy, O., Tabach, Y., Shamir, R., Shouval, D. S., & Weintraub, Y. (2024). Mucosal Genes Encoding Clock, Inflammation and Their Mutual Regulators Are Disrupted in Pediatric Patients with Active Ulcerative Colitis. International Journal of Molecular Sciences, 25(3), 1488. https://doi.org/10.3390/ijms25031488






