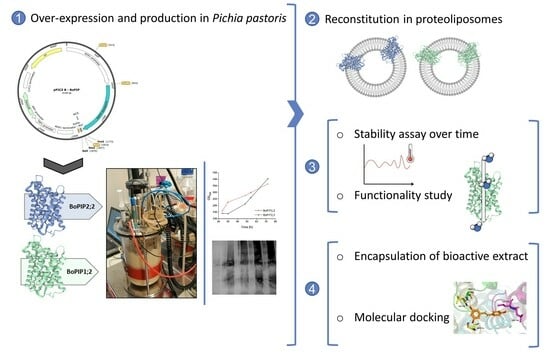
Brassica oleracea L. var. italica Aquaporin Reconstituted Proteoliposomes as Nanosystems for Resveratrol Encapsulation
Abstract
:1. Introduction
2. Results
2.1. BoPIP1;2 and BoPIP2;2 Production in P. pastoris: Cell Yield and Membrane Recovery
2.2. Membrane Protein Solubilization and AQP Purification
2.3. Reconstitution of BoPIP1;2 and BoPIP2;2 in Liposomes
2.4. Encapsulation of Resveratrol Extract in BoPIP2;2 Proteoliposomes
2.5. Molecular Docking of Resveratrol and Integrin with PIP2 Aquaporin
3. Discussion
4. Materials and Methods
4.1. Recombinant Protein Overproduction
4.1.1. Plasmid Construction and Cloning
4.1.2. Small-and Large-Scale Expression
4.2. AQPs Purification from Pichia pastoris
4.2.1. Membrane Pichia pastoris Preparation
4.2.2. Detergent Screening
4.2.3. Protein Solubilization and Ni-NTA Affinity Chromatography
4.3. AQPs Reconstitution into Proteoliposomes
4.4. Resveratrol Extract Encapsulation in Liposomes and BoPIP2;2 Proteoliposomes
4.5. Molecular Docking of Resveratrol and Integrin with AQP
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Finn, R.N.; Cerdá, J. Evolution and Functional Diversity of Aquaporins. Biol. Bull. 2015, 229, 6–23. [Google Scholar] [CrossRef] [PubMed]
- Martínez Ballesta, M.C.; García-Gomez, P.; Yepes-Molina, L.; Guarnizo, A.L.; Teruel, J.A.; Carvajal, M. Plasma Membrane Aquaporins Mediates Vesicle Stability in Broccoli. PLoS ONE 2018, 13, e0192422. [Google Scholar] [CrossRef] [PubMed]
- Martínez Ballesta, M.C.; Pérez-Sánchez, H.; Moreno, D.A.; Carvajal, M. Plant Plasma Membrane Aquaporins in Natural Vesicles as Potential Stabilizers and Carriers of Glucosinolates. Colloids Surf. B Biointerfaces 2016, 143, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Yepes-Molina, L.; Martínez-Ballesta, M.C.; Carvajal, M. Plant Plasma Membrane Vesicles Interaction with Keratinocytes Reveals Their Potential as Carriers. J. Adv. Res. 2020, 23, 101–111. [Google Scholar] [CrossRef] [PubMed]
- van Rooijen, N.; van Nieuwmegen, R. Liposomes in Immunology: Multilamellar Phosphatidylcholine Liposomes as a Simple, Biodegradable and Harmless Adjuvant without Any Immunogenic Activity of Its Own. Immunol. Commun. 1980, 9, 243–256. [Google Scholar] [CrossRef]
- Schwendener, R.A. Liposomes as Vaccine Delivery Systems: A Review of the Recent Advances. Ther. Adv. Vaccines 2014, 2, 159–182. [Google Scholar] [CrossRef]
- Karbalaei, M.; Rezaee, S.A.; Farsiani, H. Pichia Pastoris: A Highly Successful Expression System for Optimal Synthesis of Heterologous Proteins. J. Cell Physiol. 2020, 235, 5867–5881. [Google Scholar] [CrossRef]
- Kurtzman, C.P. Biotechnological Strains of Komagataella (Pichia) Pastoris Are Komagataella phaffii as Determined from Multigene Sequence Analysis. J. Ind. Microbiol. Biotechnol. 2009, 36, 1435–1438. [Google Scholar] [CrossRef]
- Romanos, M.; Scorer, C.; Sreekrishna, K.; Clare, J. The Generation of Multicopy Recombinant Strains. Methods Mol. Biol. 1998, 103, 55–72. [Google Scholar] [CrossRef]
- Nordén, K.; Agemark, M.; Danielson, J.J.T.; Alexandersson, E.; Kjellbom, P.; Johanson, U. Increasing Gene Dosage Greatly Enhances Recombinant Expression of Aquaporins in Pichia Pastoris. BMC Biotechnol. 2011, 11, 47. [Google Scholar] [CrossRef]
- Kirscht, A.; Survery, S.; Kjellbom, P.; Johanson, U. Increased Permeability of the Aquaporin SoPIP2;1 by Mercury and Mutations in Loop a. Front. Plant Sci. 2016, 7, 1249. [Google Scholar] [CrossRef]
- Wang, H.; Schoebel, S.; Schmitz, F.; Dong, H.; Hedfalk, K. Characterization of Aquaporin-Driven Hydrogen Peroxide Transport. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183065. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Chung, T.S.; Jeyaseelan, K.; Armugam, A. Stabilization and Immobilization of Aquaporin Reconstituted Lipid Vesicles for Water Purification. Colloids Surf. B Biointerfaces 2013, 102, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Gambini, J.; Inglés, M.; Olaso, G.; Lopez-Grueso, R.; Bonet-Costa, V.; Gimeno-Mallench, L.; Mas-Bargues, C.; Abdelaziz, K.M.; Gomez-Cabrera, M.C.; Vina, J.; et al. Properties of Resveratrol: In Vitro and In Vivo Studies about Metabolism, Bioavailability, and Biological Effects in Animal Models and Humans. Oxid. Med. Cell Longev. 2015, 2015, 837042. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Robles, A.; Antunes-Ricardo, M.; Guajardo-Flores, D. Encapsulation of Phenolic Compounds with Liposomal Improvement in the Cosmetic Industry. Int. J. Pharm. 2021, 593, 120125. [Google Scholar] [CrossRef]
- Gonda, A.; Kabagwira, J.; Senthil, G.N.; Wall, N.R. Internalization of Exosomes through Receptor-Mediated Endocytosis. Mol. Cancer Res. 2019, 17, 337–347. [Google Scholar] [CrossRef]
- Chen, Y.; Rice, W.; Gu, Z.; Li, J.; Huang, J.; Brenner, M.B.; Van Hoek, A.; Xiong, J.; Gundersen, G.G.; Norman, J.C.; et al. Aquaporin 2 Promotes Cell Migration and Epithelial Morphogenesis. J. Am. Soc. Nephrol. 2012, 23, 1506–1517. [Google Scholar] [CrossRef]
- Nicolas-Espinosa, J.; Carvajal, M. Genome-wide Identification and Biological Relevance of Broccoli Aquaporins. Plant Genome 2022, 15, e20262. [Google Scholar] [CrossRef]
- Kozak, M. Point Mutations Close to the AUG Initiator Codon Affect the Efficiency of Translation of Rat Preproinsulin in Vivo. Nature 1984, 308, 241–246. [Google Scholar] [CrossRef]
- Oberg, F.; Hedfalk, K. Recombinant Production of the Human Aquaporins in the Yeast Pichia Pastoris (Invited Review). Mol. Membr. Biol. 2013, 30, 15–31. [Google Scholar] [CrossRef]
- Al-Jubair, T.; Steffen, J.H.; Missel, J.W.; Kitchen, P.; Salman, M.M.; Bill, R.M.; Gourdon, P.; Törnroth-Horsefield, S. High-Yield Overproduction and Purification of Human Aquaporins from Pichia Pastoris. STAR Protoc. 2022, 3, 101298. [Google Scholar] [CrossRef]
- Sabir, F.; Leandro, M.J.; Martins, A.P.; Loureiro-Dias, M.C.; Moura, T.F.; Soveral, G.; Prista, C. Exploring Three PIPs and Three TIPs of Grapevine for Transport of Water and Atypical Substrates through Heterologous Expression in Aqy-Null Yeast. PLoS ONE 2014, 9, e102087. [Google Scholar] [CrossRef] [PubMed]
- Bienert, M.D.; Diehn, T.A.; Richet, N.; Chaumont, F.; Bienert, G.P. Heterotetramerization of plant PIP1 and PIP2 aquaporins is an evolutionary ancient feature to guide PIP1 plasma membrane localization and function. Front. Plant Sci. 2018, 9, 382. [Google Scholar] [CrossRef] [PubMed]
- Yaneff, A.; Vitali, V.; Amodeo, G. PIP1 Aquaporins: Intrinsic Water Channels or PIP2 Aquaporin Modulators? FEBS Lett. 2015, 589, 3508–3515. [Google Scholar] [CrossRef]
- Qi, S.; Wang, R.; Chaitra, G.K.M.; Torres, J.; Hu, X.; Fane, A.G. Aquaporin-Based Biomimetic Reverse Osmosis Membranes: Stability and Long Term Performance. J. Memb. Sci. 2016, 508, 94–103. [Google Scholar] [CrossRef]
- Rosa, M.; Roberts, C.J.; Rodrigues, M.A. Connecting High-Temperature and Lowtemperature Protein Stability and Aggregation. PLoS ONE 2017, 12, e0176748. [Google Scholar] [CrossRef]
- Ratz-Łyko, A.; Arct, J. Resveratrol as an Active Ingredient for Cosmetic and Dermatological Applications: A Review. J. Cosmet. Laser Ther. 2019, 21, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.G.M.; Ming, L.C.; Lee, K.S.; Yuen, K.H. Influence of the Encapsulation Efficiency and Size of Liposome on the Oral Bioavailability of Griseofulvin-Loaded Liposomes. Pharmaceutics 2016, 8, 25. [Google Scholar] [CrossRef]
- Ghorbani Gorji, E.; Rocchi, E.; Schleining, G.; Bender-Bojalil, D.; Furtmüller, P.G.; Piazza, L.; Iturri, J.J.; Toca-Herrera, J.L. Characterization of Resveratrol-Milk Protein Interaction. J. Food Eng. 2015, 167, 217–225. [Google Scholar] [CrossRef]
- French, K.C.; Antonyak, M.A.; Cerione, R.A. Extracellular Vesicle Docking at the Cellular Port: Extracellular Vesicle Binding and Uptake. Semin. Cell Dev. Biol. 2017, 67, 48–55. [Google Scholar] [CrossRef]
- Karlsson, M.; Fotiadis, D.; Sjövall, S.; Johansson, I.; Hedfalk, K.; Engel, A.; Kjellbom, P. Reconstitution of Water Channel Function of an Aquaporin Overexpressed and Purified from Pichia Pastoris. FEBS Lett. 2003, 537, 68–72. [Google Scholar] [CrossRef]
- Stratton, J.; Chiruvolu, V.; Meagher, M. High Cell-Density Fermentation. In Pichia Protocols; Springer: Berlin/Heidelberg, Germany, 1998; pp. 107–120. [Google Scholar]
- Bushell, M.E.; Rowe, M.; Avignone-Rossa, C.A.; Wardell, J.N. Cyclic Fed-Batch Culture for Production of Human Serum Albumin in Pichia Pastoris. Biotechnol. Bioeng. 2003, 82, 678–683. [Google Scholar] [CrossRef]
- Fotiadis, D.; Jenö, P.; Mini, T.; Wirtz, S.; Müller, S.A.; Fraysse, L.; Kjellbom, P.; Engel, A. Structural Characterization of Two Aquaporins Isolated from Native Spinach Leaf Plasma Membranes. J. Biol. Chem. 2001, 276, 1707–1714. [Google Scholar] [CrossRef]
- Bearden, J.C. Quantitation of Submicrogram Quantities of Protein by an Improved Protein-Dye Binding Assay. BBA-Protein Struct. 1978, 533, 525–529. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Springer: Berlin/Heidelberg, Germany, 2005; pp. 571–607. [Google Scholar]
- Maurel, C. Aquaporins and Water Permeability of Plant Membranes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 399–429. [Google Scholar] [CrossRef] [PubMed]
- Blois, M.S. Antioxidant Determinations by the Use of a Stable Free Radical. Nature 1958, 181, 1199–1200. [Google Scholar] [CrossRef]
- Frick, A.; Järvå, M.; Ekvall, M.; Uzdavinys, P.; Nyblom, M.; Törnroth-Horsefield, S. Mercury Increases Water Permeability of a Plant Aquaporin through a Non-Cysteine-Related Mechanism. Biochem. J. 2013, 454, 491–499. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Huey, R.; Morris, G.M.; Olson, A.J.; Goodsell, D.S. A Semiempirical Free Energy Force Field with Charge-Based Desolvation. J. Comput. Chem. 2007, 28, 1145–1152. [Google Scholar] [CrossRef]
- Schrödinger, LLC. The PyMOL Molecular Graphics System, Version 2.3; Schrödinger, LLC.: New York, NY, USA, 2015. [Google Scholar]
- Bálint, M.; Jeszenői, N.; Horváth, I.; van der Spoel, D.; Hetényi, C. Systematic Exploration of Multiple Drug Binding Sites. J. Cheminform 2017, 9, 65. [Google Scholar] [CrossRef]
- van Zundert, G.C.P.; Rodrigues, J.P.G.L.M.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A.M.J.J. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.C.; Rodrigues, J.P.; Kastritis, P.L.; Bonvin, A.M.; Vangone, A. PRODIGY: A Web Server for Predicting the Binding Affinity of Protein–Protein Complexes. Bioinformatics 2016, 32, 3676–3678. [Google Scholar] [CrossRef] [PubMed]




| Size (nm) | PDI (0–1) | Rate Constant (s−1) | Pf (µm s−1) | |
|---|---|---|---|---|
| Liposomes | 296.95 ± 36.20 a | 0.34 ± 0.04 a | 4.21 ± 0.62 a | 115.75 ± 17.10 a |
| BoPIP1;2 proteoliposomes | 255.63 ± 20.62 a | 0.32 ± 0.01 a | 3.76 ± 0.25 a | 89.11 ± 5.93 a |
| BoPIP2;2 proteoliposomes | 278.80 ± 37.50 a | 0.33 ± 0.02 a | 9.66 ± 0.79 b | 249.38 ± 20.46 b |
| EE (%) | Size (nm) | PDI | DPPH (µM TE g−1) | |
|---|---|---|---|---|
| Free resveratrol extract | / | / | / | 1578.34 ± 167.27 a |
| Liposomes | / | 218.93 ± 7.99 b | 0.46 ± 0.02 ab | / |
| BoPIP2;2 proteoliposomes | / | 264.83 ± 8.05 ab | 0.53 ± 0.03 a | / |
| Liposomes with resveratrol extract | 23.17 ± 3.51 b | 223.10 ± 7.56 b | 0.22 ± 0.05 c | 1624.84 ± 121.88 a |
| BoPIP2;2 proteoliposomes with resveratrol extract | 52.31 ± 3.35 a | 358.33 ± 48.4 a | 0.36 ± 0.01 b | 1426.92 ± 118.92 a |
| Pose # | ΔG (kcal/mol) | Kd (nM) | Amino Acid Residues within 2.5 Å of the Ligand | ||||
|---|---|---|---|---|---|---|---|
| 1 | −5.58 | 80 | GLU65A | CYS69A | SER71A | SER71C | |
| 2 | −5.34 | 120 | LYS64A | LYS138A | ALA139A | LYS142A | ASN160D |
| 3 | −5.19 | 160 | LYS64B | LYS142B | ASN160C | THR163C | |
| 4 | −4.97 | 230 | SER154B | LYS64D | GLY70D | ||
| 5 | −4.97 | 230 | GLY61A | LYS64A | THR66A | SER154D | |
| 6 | −4.94 | 240 | ASN160A | THR163A | ALA139C | LYS142C | |
| 7 | −4.91 | 250 | LYS64B | GLU65B | ALA152C | SER154C | |
| 8 | −4.89 | 260 | ASN160B | THR163B | LYS64D | ALA139D | |
| 9 | −4.87 | 270 | VAL68A | VAL67D | CYS69D | GLY70D | |
| 10 | −4.81 | 300 | VAL67B | CYS69B | SER71B | GLU65D | |
| 11 | −4.80 | 300 | ALA152A | GLY218A | ARG225A | GLU65C | |
| 12 | −4.75 | 330 | GLU65B | GLU65C | VAL67C | GLY70C | |
| 13 | −4.49 | 510 | HIS62B | SER63B | PHE148B | GLY218B | ARG225B |
| Integrin | Aquaporin | Distance (Å) |
|---|---|---|
| ARG220A | VAL155A | 3.37 |
| ARG220A | LYS237A | 1.71 |
| SER224A | GLN147A | 2.18 |
| TYR226A | VAL67C | 3.42 |
| ASN256A | VAL68C | 3.29 |
| ARG271A | VAL155A | 3.37 |
| ARG271A | GLY158A | 3.21 |
| ARG271A | TYR159A | 2.71 |
| ARG271A | LYS237A | 3.00 |
| SER272A | GLY158A | 2.49 |
| TYR274A | GLY143C | 3.46 |
| TYR274A | GLN147C | 2.86 |
| ASN275A | THR66C | 2.43 |
| ASN275A | GLN147C | 3.24 |
| ALA332A | ASN146C | 2.35 |
| ILE334A | GLN147C | 2.71 |
| GLU335A | ASN146C | 3.03 |
| GLU335A | GLN147C | 2.75 |
| PRO336A | GLN147C | 2.71 |
| PRO336A | PHE148C | 3.48 |
| GLU319B | VAL67A | 3.08 |
| GLU320B | THR66A | 3.33 |
| LYS326B | VAL68D | 2.56 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yepes-Molina, L.; Teruel, J.A.; Johanson, U.; Carvajal, M. Brassica oleracea L. var. italica Aquaporin Reconstituted Proteoliposomes as Nanosystems for Resveratrol Encapsulation. Int. J. Mol. Sci. 2024, 25, 1987. https://doi.org/10.3390/ijms25041987
Yepes-Molina L, Teruel JA, Johanson U, Carvajal M. Brassica oleracea L. var. italica Aquaporin Reconstituted Proteoliposomes as Nanosystems for Resveratrol Encapsulation. International Journal of Molecular Sciences. 2024; 25(4):1987. https://doi.org/10.3390/ijms25041987
Chicago/Turabian StyleYepes-Molina, Lucia, José A. Teruel, Urban Johanson, and Micaela Carvajal. 2024. "Brassica oleracea L. var. italica Aquaporin Reconstituted Proteoliposomes as Nanosystems for Resveratrol Encapsulation" International Journal of Molecular Sciences 25, no. 4: 1987. https://doi.org/10.3390/ijms25041987
APA StyleYepes-Molina, L., Teruel, J. A., Johanson, U., & Carvajal, M. (2024). Brassica oleracea L. var. italica Aquaporin Reconstituted Proteoliposomes as Nanosystems for Resveratrol Encapsulation. International Journal of Molecular Sciences, 25(4), 1987. https://doi.org/10.3390/ijms25041987