Minimal Residual Disease in Acute Myeloid Leukemia: Old and New Concepts
Abstract
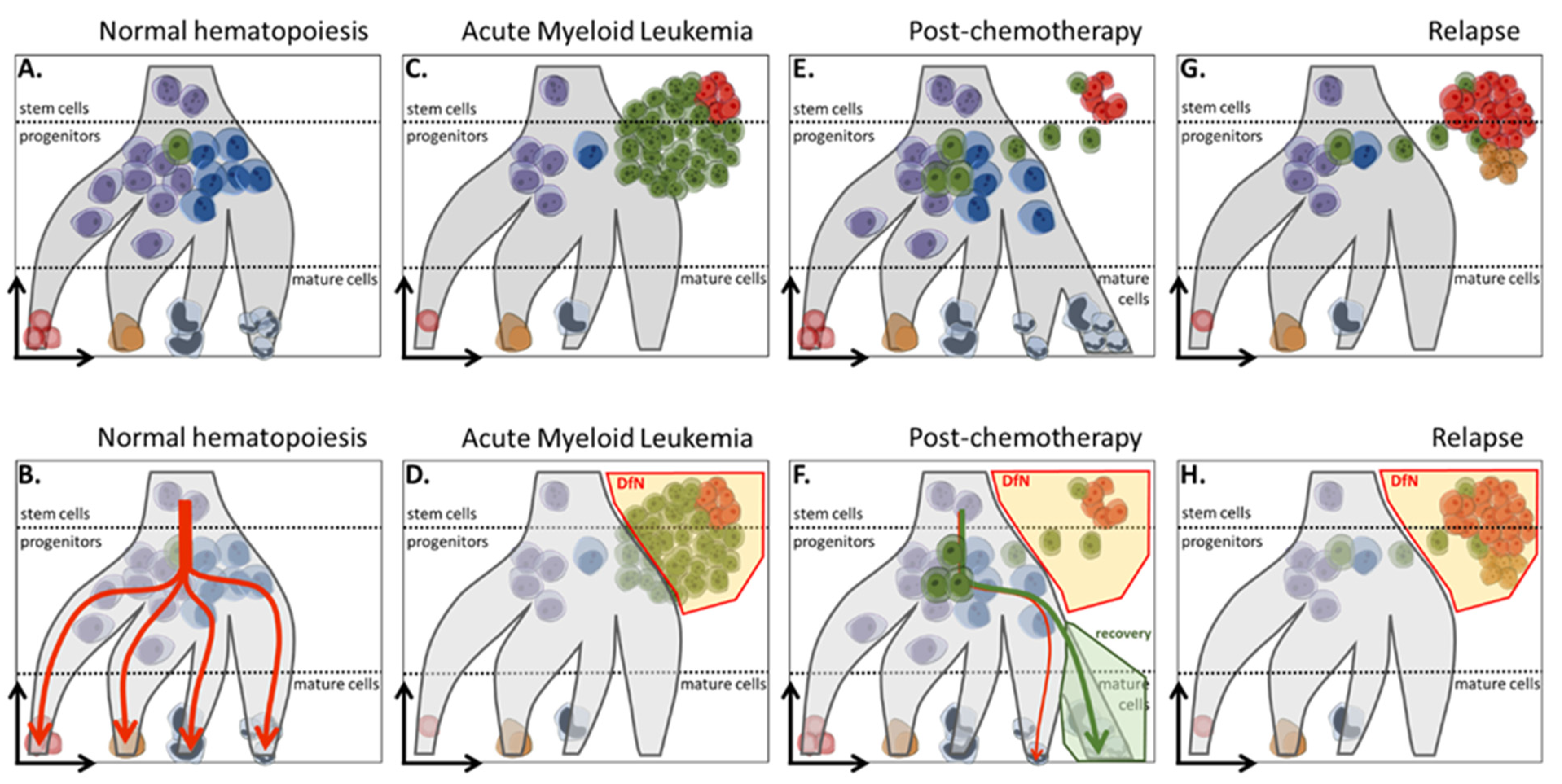
1. MRD Definition
2. One MRD, Different Techniques
2.1. Phenotypic Approaches
2.1.1. Multicolor Flow Cytometry (MFC)
2.1.2. LAIP and DfN Approaches
2.1.3. Current Technical Aspects
2.1.4. Strengths and Weaknesses
2.2. Molecular Approaches
Quantitative PCR (qPCR)
3. Current Knowledge about MRD in AML
3.1. Clinical Trials and Clinical Role
- (1)
- As a predictive biomarker for relapse risk, The HOVON/SAKK AML 42A study [30] involving 517 AML patients under 60 demonstrated that MRD was an independent prognostic factor, distinguishing high-risk relapse patients from those with better overall survival. This was later confirmed by the meta-analysis by Short et al. [11]. Yuan et al. [71] showed that pre-transplant MRD positivity, high white blood cell counts, resistance to chemotherapy, or a positive DNMT3A mutation status were associated with an increased risk of relapse.
- (2)
- As a prognostic tool for survival, Zhang et al. [72] confirmed, through a retrospective analysis of three clinical trials, that positive MRD is consistently associated with a bleak prognosis regardless of the ELN risk group. Specifically, favorable or intermediate-risk patients with positive MRD after one or two cycles of chemotherapy had a higher risk of relapse and lower overall survival than patients with negative MRD.
- (3)
- As a marker of treatment effectiveness, the phase II SORMAIN trial [73] demonstrated the utility of MRD as a predictive criterion for the effectiveness of sorafenib, an FLT3 inhibitor. Patients with negative MRD pre-transplant or positive MRD post-transplant had better relapse-free survival under sorafenib compared to patients with undetectable MRD pre-transplant or detectable MRD post-transplant under placebo. The ARTEMIS trial [74], evaluating the effectiveness of Zedenoleucel (MT-401), an allogeneic multi-tumor-associated antigen-specific T cell therapy, also showed promising results in terms of efficacy and tolerance by converting post-allograft positive MRD to negative.
- (4)
- As a surrogate endpoint in clinical trials, using the terms “AML” and “MRD” on the ClinicalTrials.gov website, approximately 324 clinical trials have been conducted, with 143 currently recruiting patients. In 2022, ELN recognized the use of MRD as a surrogate biomarker, and Walter et al. defined the optimization criteria for clinical trials. According to Walter et al. [70], each study should be conducted as an intention-to-treat, considering every MRD-positive patient as a non-responder.
3.2. Current Guidelines
3.2.1. MFC and qPCR Are the Gold Standard for MRD Monitoring
3.2.2. Novel Concept of Remission and Relapse
4. New Perspectives
4.1. Next-Generation Sequencing
4.2. Digital PCR
4.3. Unsupervised Technique in Cytometry as a Tool for MRD Monitoring
4.4. Leukemic Stem Cell: A Promising Target for MRD Tracking
4.5. Circulating Leukemic Cells and Microfluidics as a Novel Target and Method for MRD Assessment
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Martens, A.C.; Van Bekkum, D.W.; Hagenbeek, A. The BN acute myelocytic leukemia (BNML) (a rat model for studying human acute myelocytic leukemia (AML)). Leukemia 1990, 4, 241–257. [Google Scholar]
- Martens, A.C.; van Bekkum, D.W.; Hagenbeek, A. Minimal residual disease in leukemia: Studies in an animal model for acute myelocytic leukemia (BNML). Int. J. Cell Cloning 1990, 8, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Buccisano, F.; Maurillo, L.; Schuurhuis, G.J.; Del Principe, M.I.; Di Veroli, A.; Gurnari, C.; Venditti, A. The emerging role of measurable residual disease detection in AML in morphologic remission. Semin. Hematol. 2019, 56, 125–130. [Google Scholar] [CrossRef]
- Dekker, S.E.; Rea, D.; Cayuela, J.-M.; Arnhardt, I.; Leonard, J.; Heuser, M. Using Measurable Residual Disease to Optimize Management of AML, ALL, and Chronic Myeloid Leukemia. Am. Soc. Clin. Oncol. Educ. Book 2023, 43, e390010. [Google Scholar] [CrossRef]
- Kumar, S.; Paiva, B.; Anderson, K.C.; Durie, B.; Landgren, O.; Moreau, P.; Munshi, N.; Lonial, S.; Bladé, J.; Mateos, M.-V.; et al. International Myeloma Working Group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol. 2016, 17, e328–e346. [Google Scholar] [CrossRef]
- Saygin, C.; Cannova, J.; Stock, W.; Muffly, L. Measurable residual disease in acute lymphoblastic leukemia: Methods and clinical context in adult patients. Haematologica 2022, 107, 2783–2793. [Google Scholar] [CrossRef] [PubMed]
- Berry, D.A.; Zhou, S.; Higley, H.; Mukundan, L.; Fu, S.; Reaman, G.H.; Wood, B.L.; Kelloff, G.J.; Jessup, J.M.; Radich, J.P. Association of Minimal Residual Disease With Clinical Outcome in Pediatric and Adult Acute Lymphoblastic Leukemia: A Meta-analysis. JAMA Oncol. 2017, 3, e170580. [Google Scholar] [CrossRef]
- Heuser, M.; Freeman, S.D.; Ossenkoppele, G.J.; Buccisano, F.; Hourigan, C.S.; Ngai, L.L.; Tettero, J.M.; Bachas, C.; Baer, C.; Béné, M.-C.; et al. 2021 Update on MRD in acute myeloid leukemia: A consensus document from the European LeukemiaNet MRD Working Party. Blood 2021, 138, 2753–2767. [Google Scholar] [PubMed]
- Döhner, H.; Wei, A.H.; Appelbaum, F.R.; Craddock, C.; DiNardo, C.D.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Godley, L.A.; Hasserjian, R.P.; et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood 2022, 140, 1345–1377. [Google Scholar] [CrossRef]
- Short, N.J.; Fu, C.; Berry, D.A.; Walter, R.B.; Freeman, S.D.; Hourigan, C.S.; Huang, X.; Gonzalez, G.N.; Hwang, H.; Qi, X.; et al. Association of hematologic response and assay sensitivity on the prognostic impact of measurable residual disease in acute myeloid leukemia: A systematic review and meta-analysis. Leukemia 2022, 36, 2817–2826. [Google Scholar] [CrossRef]
- Short, N.J.; Zhou, S.; Fu, C.; Berry, D.A.; Walter, R.B.; Freeman, S.D.; Hourigan, C.S.; Huang, X.; Nogueras Gonzalez, G.; Hwang, H.; et al. Association of Measurable Residual Disease with Survival Outcomes in Patients With Acute Myeloid Leukemia: A Systematic Review and Meta-analysis. JAMA Oncol. 2020, 6, 1890–1899. [Google Scholar] [CrossRef]
- Paietta, E. Consensus on MRD in AML? Blood 2018, 131, 1265–1266. [Google Scholar] [CrossRef] [PubMed]
- Cluzeau, T.; Lemoli, R.M.; McCloskey, J.; Cooper, T. Measurable Residual Disease in High-Risk Acute Myeloid Leukemia. Cancers 2022, 14, 1278. [Google Scholar] [CrossRef] [PubMed]
- Hourigan, C.S. Achieving MRD negativity in AML: How important is this and how do we get there? Hematol. Am. Soc. Hematol. Educ. Program 2022, 2022, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Ivey, A.; Hills, R.K.; Simpson, M.A.; Jovanovic, J.V.; Gilkes, A.; Grech, A.; Patel, Y.; Bhudia, N.; Farah, H.; Mason, J.; et al. Assessment of Minimal Residual Disease in Standard-Risk AML. N. Engl. J. Med. 2016, 374, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Buckley, S.A.; Wood, B.L.; Othus, M.; Hourigan, C.S.; Ustun, C.; Linden, M.A.; DeFor, T.E.; Malagola, M.; Anthias, C.; Valkova, V.; et al. Minimal residual disease prior to allogeneic hematopoietic cell transplantation in acute myeloid leukemia: A meta-analysis. Haematologica 2017, 102, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Schuurhuis, G.J.; Heuser, M.; Freeman, S.; Béné, M.-C.; Buccisano, F.; Cloos, J.; Grimwade, D.; Haferlach, T.; Hills, R.K.; Hourigan, C.S.; et al. Minimal/measurable residual disease in AML: A consensus document from the European LeukemiaNet MRD Working Party. Blood 2018, 131, 1275–1291. [Google Scholar] [CrossRef]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef]
- Döhner, H.; Estey, E.H.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Burnett, A.K.; Dombret, H.; Fenaux, P.; Grimwade, D.; Larson, R.A.; et al. Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood 2010, 115, 453–474. [Google Scholar] [CrossRef]
- Tettero, J.M.; Freeman, S.; Buecklein, V.; Venditti, A.; Maurillo, L.; Kern, W.; Walter, R.B.; Wood, B.L.; Roumier, C.; Philippé, J.; et al. Technical Aspects of Flow Cytometry-based Measurable Residual Disease Quantification in Acute Myeloid Leukemia: Experience of the European LeukemiaNet MRD Working Party. HemaSphere 2022, 6, e676. [Google Scholar] [CrossRef]
- Dillon, R.; Potter, N.; Freeman, S.; Russell, N. How we use molecular minimal residual disease (MRD) testing in acute myeloid leukaemia (AML). Br. J. Haematol. 2021, 193, 231–244. [Google Scholar] [CrossRef]
- Ghannam, J.; Dillon, L.W.; Hourigan, C.S. Next-generation sequencing for measurable residual disease detection in acute myeloid leukaemia. Br. J. Haematol. 2020, 188, 77–85. [Google Scholar] [CrossRef]
- Vonk, C.M.; Al Hinai, A.S.A.; Hanekamp, D.; Valk, P.J.M. Molecular Minimal Residual Disease Detection in Acute Myeloid Leukemia. Cancers 2021, 13, 5431. [Google Scholar] [CrossRef]
- Vassault, A. Interpretation and analysis of the requirements of the standard EN ISO 15189: 2012. Ann. Biol. Clin. 2013, 71, 19–27. [Google Scholar] [CrossRef]
- Getta, B.M.; Devlin, S.M.; Levine, R.L.; Arcila, M.E.; Mohanty, A.S.; Zehir, A.; Tallman, M.S.; Giralt, S.A.; Roshal, M. Multicolor Flow Cytometry and Multigene Next-Generation Sequencing Are Complementary and Highly Predictive for Relapse in Acute Myeloid Leukemia after Allogeneic Transplantation. Biol. Blood Marrow Transplant. 2017, 23, 1064–1071. [Google Scholar] [CrossRef]
- Jongen-Lavrencic, M.; Grob, T.; Hanekamp, D.; Kavelaars, F.G.; Al Hinai, A.; Zeilemaker, A.; Erpelinck-Verschueren, C.A.J.; Gradowska, P.L.; Meijer, R.; Cloos, J.; et al. Molecular Minimal Residual Disease in Acute Myeloid Leukemia. N. Engl. J. Med. 2018, 378, 1189–1199. [Google Scholar] [CrossRef]
- Röhnert, M.A.; Kramer, M.; Schadt, J.; Ensel, P.; Thiede, C.; Krause, S.W.; Bücklein, V.; Hoffmann, J.; Jaramillo, S.; Schlenk, R.F.; et al. Reproducible measurable residual disease detection by multiparametric flow cytometry in acute myeloid leukemia. Leukemia 2022, 36, 2208–2217. [Google Scholar] [PubMed]
- Buccisano, F.; Maurillo, L.; Spagnoli, A.; Del Principe, M.I.; Fraboni, D.; Panetta, P.; Ottone, T.; Consalvo, M.I.; Lavorgna, S.; Bulian, P.; et al. Cytogenetic and molecular diagnostic characterization combined to postconsolidation minimal residual disease assessment by flow cytometry improves risk stratification in adult acute myeloid leukemia. Blood 2010, 116, 2295–2303. [Google Scholar] [CrossRef] [PubMed]
- Loken, M.R.; Alonzo, T.A.; Pardo, L.; Gerbing, R.B.; Raimondi, S.C.; Hirsch, B.A.; Ho, P.A.; Franklin, J.; Cooper, T.M.; Gamis, A.S.; et al. Residual disease detected by multidimensional flow cytometry signifies high relapse risk in patients with de novo acute myeloid leukemia: A report from Children’s Oncology Group. Blood 2012, 120, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
- Terwijn, M.; van Putten, W.L.J.; Kelder, A.; van der Velden, V.H.J.; Brooimans, R.A.; Pabst, T.; Maertens, J.; Boeckx, N.; de Greef, G.E.; Valk, P.J.M.; et al. High prognostic impact of flow cytometric minimal residual disease detection in acute myeloid leukemia: Data from the HOVON/SAKK AML 42A study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2013, 31, 3889–3897. [Google Scholar] [CrossRef] [PubMed]
- Walter, R.B.; Gooley, T.A.; Wood, B.L.; Milano, F.; Fang, M.; Sorror, M.L.; Estey, E.H.; Salter, A.I.; Lansverk, E.; Chien, J.W.; et al. Impact of pretransplantation minimal residual disease, as detected by multiparametric flow cytometry, on outcome of myeloablative hematopoietic cell transplantation for acute myeloid leukemia. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2011, 29, 1190–1197. [Google Scholar] [CrossRef]
- Wood, B.L. Acute Myeloid Leukemia Minimal Residual Disease Detection: The Difference from Normal Approach. Curr. Protoc. Cytom. 2020, 93, e73. [Google Scholar] [CrossRef]
- Buldini, B.; Maurer-Granofszky, M.; Varotto, E.; Dworzak, M.N. Flow-Cytometric Monitoring of Minimal Residual Disease in Pediatric Patients With Acute Myeloid Leukemia: Recent Advances and Future Strategies. Front. Pediatr. 2019, 7, 412. [Google Scholar] [CrossRef]
- Wood, B.L. Flow cytometry in the diagnosis and monitoring of acute leukemia in children. J. Hematop. 2015, 8, 191–199. [Google Scholar] [CrossRef]
- Baer, M.R.; Stewart, C.C.; Dodge, R.K.; Leget, G.; Sulé, N.; Mrózek, K.; Schiffer, C.A.; Powell, B.L.; Kolitz, J.E.; Moore, J.O.; et al. High frequency of immunophenotype changes in acute myeloid leukemia at relapse: Implications for residual disease detection (Cancer and Leukemia Group B Study 8361). Blood 2001, 97, 3574–3580. [Google Scholar] [CrossRef]
- Voskova, D.; Schoch, C.; Schnittger, S.; Hiddemann, W.; Haferlach, T.; Kern, W. Stability of leukemia-associated aberrant immunophenotypes in patients with acute myeloid leukemia between diagnosis and relapse: Comparison with cytomorphologic, cytogenetic, and molecular genetic findings. Cytom. B Clin. Cytom. 2004, 62, 25–38. [Google Scholar] [CrossRef]
- Lee, D.; Grigoriadis, G.; Westerman, D. The role of multiparametric flow cytometry in the detection of minimal residual disease in acute leukaemia. Pathology 2015, 47, 609–621. [Google Scholar] [CrossRef] [PubMed]
- Zeijlemaker, W.; Kelder, A.; Cloos, J.; Schuurhuis, G.J. Immunophenotypic Detection of Measurable Residual (Stem Cell) Disease Using LAIP Approach in Acute Myeloid Leukemia. Curr. Protoc. Cytom. 2019, 91, e66. [Google Scholar] [CrossRef]
- Lacombe, F.; Bernal, E.; Bloxham, D.; Couzens, S.; Porta, M.G.D.; Johansson, U.; Kern, W.; Macey, M.; Matthes, T.; Morilla, R.; et al. Harmonemia: A universal strategy for flow cytometry immunophenotyping-A European LeukemiaNet WP10 study. Leukemia 2016, 30, 1769–1772. [Google Scholar] [CrossRef] [PubMed]
- Matarraz, S.; Almeida, J.; Flores-Montero, J.; Lécrevisse, Q.; Guerri, V.; López, A.; Bárrena, S.; Van Der Velden, V.H.J.; Te Marvelde, J.G.; Van Dongen, J.J.M.; et al. Introduction to the diagnosis and classification of monocytic-lineage leukemias by flow cytometry. Cytom. B Clin. Cytom. 2017, 92, 218–227. [Google Scholar] [CrossRef]
- Dix, C.; Lo, T.-H.; Clark, G.; Abadir, E. Measurable Residual Disease in Acute Myeloid Leukemia Using Flow Cytometry: A Review of Where We Are and Where We Are Going. J. Clin. Med. 2020, 9, 1714. [Google Scholar] [CrossRef]
- Hanekamp, D.; Bachas, C.; van de Loosdrecht, A.; Ossenkoppele, G.; Cloos, J. Re: Myeloblasts in normal bone marrows expressing leukaemia-associated immunophenotypes. Pathology 2020, 52, 289–291. [Google Scholar] [CrossRef]
- Riva, G.; Nasillo, V.; Ottomano, A.M.; Bergonzini, G.; Paolini, A.; Forghieri, F.; Lusenti, B.; Barozzi, P.; Lagreca, I.; Fiorcari, S.; et al. Multiparametric Flow Cytometry for MRD Monitoring in Hematologic Malignancies: Clinical Applications and New Challenges. Cancers 2021, 13, 4582. [Google Scholar] [CrossRef]
- Freeman, S.D.; Virgo, P.; Couzens, S.; Grimwade, D.; Russell, N.; Hills, R.K.; Burnett, A.K. Prognostic relevance of treatment response measured by flow cytometric residual disease detection in older patients with acute myeloid leukemia. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2013, 31, 4123–4131. [Google Scholar] [CrossRef]
- Freeman, S.D.; Hills, R.K.; Virgo, P.; Khan, N.; Couzens, S.; Dillon, R.; Gilkes, A.; Upton, L.; Nielsen, O.J.; Cavenagh, J.D.; et al. Measurable Residual Disease at Induction Redefines Partial Response in Acute Myeloid Leukemia and Stratifies Outcomes in Patients at Standard Risk Without NPM1 Mutations. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2018, 36, 1486–1497. [Google Scholar] [CrossRef] [PubMed]
- Venditti, A.; Piciocchi, A.; Candoni, A.; Melillo, L.; Calafiore, V.; Cairoli, R.; de Fabritiis, P.; Storti, G.; Salutari, P.; Lanza, F.; et al. GIMEMA AML1310 trial of risk-adapted, MRD-directed therapy for young adults with newly diagnosed acute myeloid leukemia. Blood 2019, 134, 935–945. [Google Scholar] [CrossRef]
- Chen, X.; Wood, B.L. Monitoring minimal residual disease in acute leukemia: Technical challenges and interpretive complexities. Blood Rev. 2017, 31, 63–75. [Google Scholar] [CrossRef]
- Lo Coco, F.; Diverio, D.; Pandolfi, P.P.; Biondi, A.; Rossi, V.; Avvisati, G.; Rambaldi, A.; Arcese, W.; Petti, M.C.; Meloni, G. Molecular evaluation of residual disease as a predictor of relapse in acute promyelocytic leukaemia. Lancet 1992, 340, 1437–1438. [Google Scholar] [CrossRef] [PubMed]
- Biondi, A.; Rambaldi, A.; Pandolfi, P.P.; Rossi, V.; Giudici, G.; Alcalay, M.; Lo Coco, F.; Diverio, D.; Pogliani, E.M.; Lanzi, E.M. Molecular monitoring of the myl/retinoic acid receptor-alpha fusion gene in acute promyelocytic leukemia by polymerase chain reaction. Blood 1992, 80, 492–497. [Google Scholar] [CrossRef] [PubMed]
- van der Velden, V.H.J.; Hochhaus, A.; Cazzaniga, G.; Szczepanski, T.; Gabert, J.; van Dongen, J.J.M. Detection of minimal residual disease in hematologic malignancies by real-time quantitative PCR: Principles, approaches, and laboratory aspects. Leukemia 2003, 17, 1013–1034. [Google Scholar] [CrossRef] [PubMed]
- van Dongen, J.J.; Macintyre, E.A.; Gabert, J.A.; Delabesse, E.; Rossi, V.; Saglio, G.; Gottardi, E.; Rambaldi, A.; Dotti, G.; Griesinger, F.; et al. Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease. Report of the BIOMED-1 Concerted Action: Investigation of minimal residual disease in acute leukemia. Leukemia 1999, 13, 1901–1928. [Google Scholar] [CrossRef]
- Gabert, J.; Beillard, E.; van der Velden, V.H.J.; Bi, W.; Grimwade, D.; Pallisgaard, N.; Barbany, G.; Cazzaniga, G.; Cayuela, J.M.; Cavé, H.; et al. Standardization and quality control studies of “real-time” quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia—A Europe Against Cancer program. Leukemia 2003, 17, 2318–2357. [Google Scholar] [CrossRef]
- Beillard, E.; Pallisgaard, N.; van der Velden, V.H.J.; Bi, W.; Dee, R.; van der Schoot, E.; Delabesse, E.; Macintyre, E.; Gottardi, E.; Saglio, G.; et al. Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using “real-time” quantitative reverse-transcriptase polymerase chain reaction (RQ-PCR)—A Europe against cancer program. Leukemia 2003, 17, 2474–2486. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, A.; Carreira, L.; Abalde-Cela, S.; Sampaio-Marques, B.; Areias, A.C.; Ludovico, P.; Diéguez, L. Current and Emerging Techniques for Diagnosis and MRD Detection in AML: A Comprehensive Narrative Review. Cancers 2023, 15, 1362. [Google Scholar] [CrossRef]
- Grimwade, D.; Freeman, S.D. Defining minimal residual disease in acute myeloid leukemia: Which platforms are ready for “prime time”? Blood 2014, 124, 3345–3355. [Google Scholar] [CrossRef] [PubMed]
- Sangle, N.A.; Perkins, S.L. Core-binding factor acute myeloid leukemia. Arch. Pathol. Lab. Med. 2011, 135, 1504–1509. [Google Scholar] [CrossRef]
- Cruz, N.M.; Mencia-Trinchant, N.; Hassane, D.C.; Guzman, M.L. Minimal residual disease in acute myelogenous leukemia. Int. J. Lab. Hematol. 2017, 39, 53–60. [Google Scholar] [CrossRef]
- Kristensen, T.; Møller, M.B.; Friis, L.; Bergmann, O.J.; Preiss, B. NPM1 mutation is a stable marker for minimal residual disease monitoring in acute myeloid leukaemia patients with increased sensitivity compared to WT1 expression. Eur. J. Haematol. 2011, 87, 400–408. [Google Scholar] [CrossRef]
- Forghieri, F.; Comoli, P.; Marasca, R.; Potenza, L.; Luppi, M. Minimal/Measurable Residual Disease Monitoring in NPM1-Mutated Acute Myeloid Leukemia: A Clinical Viewpoint and Perspectives. Int. J. Mol. Sci. 2018, 19, 3492. [Google Scholar] [CrossRef]
- Ossenkoppele, G.; Schuurhuis, G.J. MRD in AML: Does it already guide therapy decision-making? Hematology 2016, 2016, 356–365. [Google Scholar] [CrossRef]
- Hourigan, C.S.; Gale, R.P.; Gormley, N.J.; Ossenkoppele, G.J.; Walter, R.B. Measurable residual disease testing in acute myeloid leukaemia. Leukemia 2017, 31, 1482–1490. [Google Scholar] [CrossRef] [PubMed]
- Gorello, P.; Cazzaniga, G.; Alberti, F.; Dell’Oro, M.G.; Gottardi, E.; Specchia, G.; Roti, G.; Rosati, R.; Martelli, M.F.; Diverio, D.; et al. Quantitative assessment of minimal residual disease in acute myeloid leukemia carrying nucleophosmin (NPM1) gene mutations. Leukemia 2006, 20, 1103–1108. [Google Scholar] [CrossRef]
- Bacher, U.; Dicker, F.; Haferlach, C.; Alpermann, T.; Rose, D.; Kern, W.; Haferlach, T.; Schnittger, S. Quantification of rare NPM1 mutation subtypes by digital PCR. Br. J. Haematol. 2014, 167, 710–714. [Google Scholar] [CrossRef]
- Mencia-Trinchant, N.; Hu, Y.; Alas, M.A.; Ali, F.; Wouters, B.J.; Lee, S.; Ritchie, E.K.; Desai, P.; Guzman, M.L.; Roboz, G.J.; et al. Minimal Residual Disease Monitoring of Acute Myeloid Leukemia by Massively Multiplex Digital PCR in Patients with NPM1 Mutations. J. Mol. Diagn. JMD 2017, 19, 537–548. [Google Scholar] [CrossRef]
- Papadaki, C.; Dufour, A.; Seibl, M.; Schneider, S.; Bohlander, S.K.; Zellmeier, E.; Mellert, G.; Hiddemann, W.; Spiekermann, K. Monitoring minimal residual disease in acute myeloid leukaemia with NPM1 mutations by quantitative PCR: Clonal evolution is a limiting factor. Br. J. Haematol. 2009, 144, 517–523. [Google Scholar] [CrossRef]
- Madaci, L.; Farnault, L.; Abbou, N.; Gabert, J.; Venton, G.; Costello, R. Impact of Next-Generation Sequencing in Diagnosis, Prognosis and Therapeutic Management of Acute Myeloid Leukemia/Myelodysplastic Neoplasms. Cancers 2023, 15, 3280. [Google Scholar] [CrossRef] [PubMed]
- Luo, P.; Jing, W.; Yi, K.; Wu, S.; Zhou, F. Wilms’ tumor 1 gene in hematopoietic malignancies: Clinical implications and future directions. Leuk. Lymphoma 2020, 61, 2059–2067. [Google Scholar] [CrossRef]
- Levis, M.J.; Perl, A.E.; Altman, J.K.; Gocke, C.D.; Bahceci, E.; Hill, J.; Liu, C.; Xie, Z.; Carson, A.R.; McClain, V.; et al. A next-generation sequencing-based assay for minimal residual disease assessment in AML patients with FLT3-ITD mutations. Blood Adv. 2018, 2, 825–831. [Google Scholar] [CrossRef]
- Blätte, T.J.; Schmalbrock, L.K.; Skambraks, S.; Lux, S.; Cocciardi, S.; Dolnik, A.; Döhner, H.; Döhner, K.; Bullinger, L. getITD for FLT3-ITD-based MRD monitoring in AML. Leukemia 2019, 33, 2535–2539. [Google Scholar] [CrossRef]
- Walter, R.B.; Ofran, Y.; Wierzbowska, A.; Ravandi, F.; Hourigan, C.S.; Ngai, L.L.; Venditti, A.; Buccisano, F.; Ossenkoppele, G.J.; Roboz, G.J. Measurable residual disease as a biomarker in acute myeloid leukemia: Theoretical and practical considerations. Leukemia 2021, 35, 1529–1538. [Google Scholar] [CrossRef]
- Yuan, X.; Lai, X.; Wu, Y.; Yang, L.; Shi, J.; Liu, L.; Zhao, Y.; Yu, J.; Huang, H.; Luo, Y. Risk Factors and Prediction Model of Early Relapse in Acute Myeloid Leukemia after Allogeneic Hematopoietic Cell Transplantation. Blood 2022, 140, 12915. [Google Scholar] [CrossRef]
- Zhang, C.; Gu, R.; Zhou, C.; Li, Y.; Liu, Y.; Wei, S.; Lin, D.; Gong, B.; Liu, K.; Fang, Q.; et al. Prognostic Effect and Clinical Application of Early Measurable Residual Disease (MRD) By Flow Cytometry on De Novo Acute Myeloid Leukemia (AML). Blood 2022, 140, 2030–2032. [Google Scholar] [CrossRef]
- Burchert, A.; Bug, G.; Fritz, L.V.; Finke, J.; Stelljes, M.; Röllig, C.; Wollmer, E.; Wäsch, R.; Bornhäuser, M.; Berg, T.; et al. Sorafenib Maintenance After Allogeneic Hematopoietic Stem Cell Transplantation for Acute Myeloid Leukemia With FLT3–Internal Tandem Duplication Mutation (SORMAIN). J. Clin. Oncol. 2020, 38, 2993–3002. [Google Scholar] [CrossRef]
- Koneru, M.; Arslan, S.; Magalhaes-Silverman, M.M.; Liu, H.; Bejanyan, N.; Di Stasi, A.; McCallum, R.; Quintero, S.; Garrett, G.; Wang, K.; et al. Early Results of MT-401 (Zedenoleucel) in Post-Transplant MRD+ aml Patients. Blood 2022, 140 (Suppl. S1), 12718–12719. [Google Scholar] [CrossRef]
- Othman, J.; Potter, N.; Mokretar, K.; Taussig, D.; Khan, A.; Krishnamurthy, P.; Latif, A.-L.; Cahalin, P.; Aries, J.; Amer, M.; et al. FLT3 inhibitors as MRD-guided salvage treatment for molecular failure in FLT3 mutated AML. Leukemia 2023, 37, 2066–2072. [Google Scholar] [CrossRef] [PubMed]
- Scott, B.L.; Pasquini, M.C.; Fei, M.; Fraser, R.; Wu, J.; Devine, S.M.; Porter, D.L.; Maziarz, R.T.; Warlick, E.; Fernandez, H.F.; et al. Myeloablative versus Reduced-Intensity Conditioning for Hematopoietic Cell Transplantation in Acute Myelogenous Leukemia and Myelodysplastic Syndromes-Long-Term Follow-Up of the BMT CTN 0901 Clinical Trial. Transplant. Cell. Ther. 2021, 27, e1–e483. [Google Scholar] [CrossRef]
- Craddock, C.; Jackson, A.; Loke, J.; Siddique, S.; Hodgkinson, A.; Mason, J.; Andrew, G.; Nagra, S.; Malladi, R.; Peniket, A.; et al. Augmented Reduced-Intensity Regimen Does Not Improve Postallogeneic Transplant Outcomes in Acute Myeloid Leukemia. J. Clin. Oncol. 2021, 39, 768–778. [Google Scholar] [CrossRef]
- Hourigan, C.S.; Dillon, L.W.; Gui, G.; Logan, B.R.; Fei, M.; Ghannam, J.; Li, Y.; Licon, A.; Alyea, E.P.; Bashey, A.; et al. Impact of Conditioning Intensity of Allogeneic Transplantation for Acute Myeloid Leukemia With Genomic Evidence of Residual Disease. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2020, 38, 1273–1283. [Google Scholar] [CrossRef] [PubMed]
- Hasserjian, R.P.; Steensma, D.P.; Graubert, T.A.; Ebert, B.L. Clonal hematopoiesis and measurable residual disease assessment in acute myeloid leukemia. Blood 2020, 135, 1729–1738. [Google Scholar] [CrossRef]
- Cappelli, L.V.; Meggendorfer, M.; Baer, C.; Nadarajah, N.; Hutter, S.; Jeromin, S.; Dicker, F.; Kern, W.; Haferlach, T.; Haferlach, C.; et al. Indeterminate and oncogenic potential: CHIP vs CHOP mutations in AML with NPM1 alteration. Leukemia 2022, 36, 394–402. [Google Scholar] [CrossRef]
- Heuser, M.; Heida, B.; Büttner, K.; Wienecke, C.P.; Teich, K.; Funke, C.; Brandes, M.; Klement, P.; Liebich, A.; Wichmann, M.; et al. Posttransplantation MRD monitoring in patients with AML by next-generation sequencing using DTA and non-DTA mutations. Blood Adv. 2021, 5, 2294–2304. [Google Scholar] [CrossRef]
- Maiti, A.; DiNardo, C.D.; Wang, S.A.; Jorgensen, J.; Kadia, T.M.; Daver, N.G.; Short, N.J.; Yilmaz, M.; Pemmaraju, N.; Borthakur, G.; et al. Prognostic value of measurable residual disease after venetoclax and decitabine in acute myeloid leukemia. Blood Adv. 2021, 5, 1876–1883. [Google Scholar] [CrossRef]
- Pratz, K.W.; Jonas, B.A.; Pullarkat, V.; Recher, C.; Schuh, A.C.; Thirman, M.J.; Garcia, J.S.; DiNardo, C.D.; Vorobyev, V.; Fracchiolla, N.S.; et al. Measurable Residual Disease Response and Prognosis in Treatment-Naïve Acute Myeloid Leukemia With Venetoclax and Azacitidine. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2022, 40, 855–865. [Google Scholar] [CrossRef]
- Willekens, C.; Blanchet, O.; Renneville, A.; Cornillet-Lefebvre, P.; Pautas, C.; Guieze, R.; Ifrah, N.; Dombret, H.; Jourdan, E.; Preudhomme, C.; et al. Prospective long-term minimal residual disease monitoring using RQ-PCR in RUNX1-RUNX1T1-positive acute myeloid leukemia: Results of the French CBF-2006 trial. Haematologica 2016, 101, 328–335. [Google Scholar] [CrossRef]
- Rücker, F.G.; Agrawal, M.; Corbacioglu, A.; Weber, D.; Kapp-Schwoerer, S.; Gaidzik, V.I.; Jahn, N.; Schroeder, T.; Wattad, M.; Lübbert, M.; et al. Measurable residual disease monitoring in acute myeloid leukemia with t(8;21)(q22;q22.1): Results from the AML Study Group. Blood 2019, 134, 1608–1618. [Google Scholar] [CrossRef]
- Ding, L.; Ley, T.J.; Larson, D.E.; Miller, C.A.; Koboldt, D.C.; Welch, J.S.; Ritchey, J.K.; Young, M.A.; Lamprecht, T.; McLellan, M.D.; et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 2012, 481, 506–510. [Google Scholar] [CrossRef] [PubMed]
- Voso, M.T.; Ottone, T.; Lavorgna, S.; Venditti, A.; Maurillo, L.; Lo-Coco, F.; Buccisano, F. MRD in AML: The Role of New Techniques. Front. Oncol. 2019, 9, 655. [Google Scholar] [CrossRef] [PubMed]
- Welch, J.S.; Ley, T.J.; Link, D.C.; Miller, C.A.; Larson, D.E.; Koboldt, D.C.; Wartman, L.D.; Lamprecht, T.L.; Liu, F.; Xia, J.; et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 2012, 150, 264–278. [Google Scholar] [CrossRef]
- Yoest, J.M.; Shirai, C.L.; Duncavage, E.J. Sequencing-Based Measurable Residual Disease Testing in Acute Myeloid Leukemia. Front. Cell Dev. Biol. 2020, 8, 249. [Google Scholar] [CrossRef] [PubMed]
- Klco, J.M.; Miller, C.A.; Griffith, M.; Petti, A.; Spencer, D.H.; Ketkar-Kulkarni, S.; Wartman, L.D.; Christopher, M.; Lamprecht, T.L.; Helton, N.M.; et al. Association Between Mutation Clearance After Induction Therapy and Outcomes in Acute Myeloid Leukemia. JAMA 2015, 314, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Moon, J.H.; Ahn, J.-S.; Kim, Y.-K.; Lee, S.-S.; Ahn, S.-Y.; Jung, S.-H.; Yang, D.-H.; Lee, J.-J.; Choi, S.H.; et al. Next-generation sequencing–based posttransplant monitoring of acute myeloid leukemia identifies patients at high risk of relapse. Blood 2018, 132, 1604–1613. [Google Scholar] [CrossRef]
- Thol, F.; Kölking, B.; Damm, F.; Reinhardt, K.; Klusmann, J.-H.; Reinhardt, D.; von Neuhoff, N.; Brugman, M.H.; Schlegelberger, B.; Suerbaum, S.; et al. Next-generation sequencing for minimal residual disease monitoring in acute myeloid leukemia patients with FLT3-ITD or NPM1 mutations. Genes. Chromosomes Cancer 2012, 51, 689–695. [Google Scholar] [CrossRef] [PubMed]
- Patkar, N.; Kakirde, C.; Shaikh, A.F.; Salve, R.; Bhanshe, P.; Chatterjee, G.; Rajpal, S.; Joshi, S.; Chaudhary, S.; Kodgule, R.; et al. Clinical impact of panel-based error-corrected next generation sequencing versus flow cytometry to detect measurable residual disease (MRD) in acute myeloid leukemia (AML). Leukemia 2021, 35, 1392–1404. [Google Scholar] [CrossRef]
- Press, R.D.; Eickelberg, G.; Froman, A.; Yang, F.; Stentz, A.; Flatley, E.M.; Fan, G.; Lim, J.Y.; Meyers, G.; Maziarz, R.T.; et al. Next-generation sequencing-defined minimal residual disease before stem cell transplantation predicts acute myeloid leukemia relapse. Am. J. Hematol. 2019, 94, 902–912. [Google Scholar] [CrossRef]
- Li, Y.; Solis-Ruiz, J.; Yang, F.; Long, N.; Tong, C.H.; Lacbawan, F.L.; Racke, F.K.; Press, R.D. NGS-defined measurable residual disease (MRD) after initial chemotherapy as a prognostic biomarker for acute myeloid leukemia. Blood Cancer J. 2023, 13, 59. [Google Scholar] [CrossRef]
- Thol, F.; Gabdoulline, R.; Liebich, A.; Klement, P.; Schiller, J.; Kandziora, C.; Hambach, L.; Stadler, M.; Koenecke, C.; Flintrop, M.; et al. Measurable residual disease monitoring by NGS before allogeneic hematopoietic cell transplantation in AML. Blood 2018, 132, 1703–1713. [Google Scholar] [CrossRef]
- Young, A.L.; Wong, T.N.; Hughes, A.E.O.; Heath, S.E.; Ley, T.J.; Link, D.C.; Druley, T.E. Quantifying ultra-rare pre-leukemic clones via targeted error-corrected sequencing. Leukemia 2015, 29, 1608–1611. [Google Scholar] [CrossRef] [PubMed]
- Salk, J.J.; Schmitt, M.W.; Loeb, L.A. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat. Rev. Genet. 2018, 19, 269–285. [Google Scholar] [CrossRef]
- Vogelstein, B.; Kinzler, K.W. Digital PCR. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef]
- Colmenares, R.; Álvarez, N.; Barrio, S.; Martínez-López, J.; Ayala, R. The Minimal Residual Disease Using Liquid Biopsies in Hematological Malignancies. Cancers 2022, 14, 1310. [Google Scholar] [CrossRef]
- Dube, S.; Qin, J.; Ramakrishnan, R. Mathematical analysis of copy number variation in a DNA sample using digital PCR on a nanofluidic device. PLoS ONE 2008, 3, e2876. [Google Scholar] [CrossRef]
- Quan, P.-L.; Sauzade, M.; Brouzes, E. dPCR: A Technology Review. Sensors 2018, 18, 1271. [Google Scholar] [CrossRef]
- Pettersson, L.; Johansson Alm, S.; Almstedt, A.; Chen, Y.; Orrsjö, G.; Shah-Barkhordar, G.; Zhou, L.; Kotarsky, H.; Vidovic, K.; Asp, J.; et al. Comparison of RNA- and DNA-based methods for measurable residual disease analysis in NPM1-mutated acute myeloid leukemia. Int. J. Lab. Hematol. 2021, 43, 664–674. [Google Scholar] [CrossRef] [PubMed]
- Bill, M.; Grimm, J.; Jentzsch, M.; Kloss, L.; Goldmann, K.; Schulz, J.; Beinicke, S.; Häntschel, J.; Cross, M.; Vucinic, V.; et al. Digital droplet PCR-based absolute quantification of pre-transplant NPM1 mutation burden predicts relapse in acute myeloid leukemia patients. Ann. Hematol. 2018, 97, 1757–1765. [Google Scholar] [CrossRef] [PubMed]
- Ferret, Y.; Boissel, N.; Helevaut, N.; Madic, J.; Nibourel, O.; Marceau-Renaut, A.; Bucci, M.; Geffroy, S.; Celli-Lebras, K.; Castaigne, S.; et al. Clinical relevance of IDH1/2 mutant allele burden during follow-up in acute myeloid leukemia. A study by the French ALFA group. Haematologica 2018, 103, 822–829. [Google Scholar] [CrossRef] [PubMed]
- Grassi, S.; Guerrini, F.; Ciabatti, E.; Puccetti, R.; Salehzadeh, S.; Metelli, M.R.; Di Vita, A.; Domenichini, C.; Caracciolo, F.; Orciuolo, E.; et al. Digital Droplet PCR is a Specific and Sensitive Tool for Detecting IDH2 Mutations in Acute Myeloid LeuKemia Patients. Cancers 2020, 12, 1738. [Google Scholar] [CrossRef] [PubMed]
- Winters, A.; Goosman, M.; Stevens, B.M.; Purev, E.; Smith, C.; Pollyea, D.A.; Jordan, C.T.; Gutman, J.A. Tracking of AML-Associated Mutations Via Droplet Digital PCR Is Predictive of Outcomes Post-Transplant. Blood 2018, 132, 2138. [Google Scholar] [CrossRef]
- Parkin, B.; Londoño-Joshi, A.; Kang, Q.; Tewari, M.; Rhim, A.D.; Malek, S.N. Ultrasensitive mutation detection identifies rare residual cells causing acute myelogenous leukemia relapse. J. Clin. Investig. 2017, 127, 3484–3495. [Google Scholar] [CrossRef]
- Brunetti, C.; Anelli, L.; Zagaria, A.; Minervini, A.; Minervini, C.F.; Casieri, P.; Coccaro, N.; Cumbo, C.; Tota, G.; Impera, L.; et al. Droplet Digital PCR Is a Reliable Tool for Monitoring Minimal Residual Disease in Acute Promyelocytic Leukemia. J. Mol. Diagn. JMD 2017, 19, 437–444. [Google Scholar] [CrossRef]
- Yuan, D.; Cui, M.; Yu, S.; Wang, H.; Jing, R. Droplet digital PCR for quantification of PML-RARα in acute promyelocytic leukemia: A comprehensive comparison with real-time PCR. Anal. Bioanal. Chem. 2019, 411, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Lesieur, A.; Thomas, X.; Nibourel, O.; Boissel, N.; Fenwarth, L.; De Botton, S.; Fournier, E.; Celli-Lebras, K.; Raffoux, E.; Recher, C.; et al. Minimal residual disease monitoring in acute myeloid leukemia with non-A/B/D-NPM1 mutations by digital polymerase chain reaction: Feasibility and clinical use. Haematologica 2021, 106, 1767–1769. [Google Scholar] [CrossRef] [PubMed]
- Spitzer, M.H.; Nolan, G.P. Mass Cytometry: Single Cells, Many Features. Cell 2016, 165, 780–791. [Google Scholar] [CrossRef]
- Bjornson, Z.B.; Nolan, G.P.; Fantl, W.J. Single-cell mass cytometry for analysis of immune system functional states. Curr. Opin. Immunol. 2013, 25, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Qiu, P.; Zeng, Z.; Jorgensen, J.L.; Mak, D.H.; Burks, J.K.; Schober, W.; McQueen, T.J.; Cortes, J.; Tanner, S.D.; et al. Single-cell mass cytometry reveals intracellular survival/proliferative signaling in FLT3-ITD-mutated AML stem/progenitor cells. Cytom. Part J. Int. Soc. Anal. Cytol. 2015, 87, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Gjertsen, B.T.; Tislevoll, B.S.; Fagerholt, O.H.E.; Hellesøy, M. Early response evaluation in AML using mass cytometry. HemaSphere 2019, 3, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Amir, E.D.; Davis, K.L.; Tadmor, M.D.; Simonds, E.F.; Levine, J.H.; Bendall, S.C.; Shenfeld, D.K.; Krishnaswamy, S.; Nolan, G.P.; Pe’er, D. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 2013, 31, 545–552. [Google Scholar] [CrossRef]
- Qiu, P.; Simonds, E.F.; Bendall, S.C.; Gibbs, K.D.; Bruggner, R.V.; Linderman, M.D.; Sachs, K.; Nolan, G.P.; Plevritis, S.K. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat. Biotechnol. 2011, 29, 886–891. [Google Scholar] [CrossRef]
- Polikowsky, H.G.; Drake, K.A. Supervised Machine Learning with CITRUS for Single Cell Biomarker Discovery. Mass Cytom. Methods Protoc. 2019, 1989, 309–332. [Google Scholar]
- Weber, L.M.; Robinson, M.D. Comparison of clustering methods for high-dimensional single-cell flow and mass cytometry data. Cytom. Part J. Int. Soc. Anal. Cytol. 2016, 89, 1084–1096. [Google Scholar] [CrossRef]
- Lacombe, F.; Lechevalier, N.; Vial, J.P.; Béné, M.C. An R-Derived FlowSOM Process to Analyze Unsupervised Clustering of Normal and Malignant Human Bone Marrow Classical Flow Cytometry Data. Cytom. A 2019, 95, 1191–1197. [Google Scholar] [CrossRef]
- Lacombe, F.; Dupont, B.; Lechevalier, N.; Vial, J.P.; Béné, M.C. Innovation in Flow Cytometry Analysis: A New Paradigm Delineating Normal or Diseased Bone Marrow Subsets Through Machine Learning. HemaSphere 2019, 3, e173. [Google Scholar] [CrossRef] [PubMed]
- Van Gassen, S.; Callebaut, B.; Van Helden, M.J.; Lambrecht, B.N.; Demeester, P.; Dhaene, T.; Saeys, Y. FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytom. Part J. Int. Soc. Anal. Cytol. 2015, 87, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Vial, J.P.; Lechevalier, N.; Lacombe, F.; Dumas, P.-Y.; Bidet, A.; Leguay, T.; Vergez, F.; Pigneux, A.; Béné, M.C. Unsupervised Flow Cytometry Analysis Allows for an Accurate Identification of Minimal Residual Disease Assessment in Acute Myeloid Leukemia. Cancers 2021, 13, 629. [Google Scholar] [CrossRef]
- Rossi, G.; Carella, A.M.; Minervini, M.M.; Savino, L.; Fontana, A.; Pellegrini, F.; Greco, M.M.; Merla, E.; Quarta, G.; Loseto, G.; et al. Minimal residual disease after allogeneic stem cell transplant: A comparison among multiparametric flow cytometry, Wilms tumor 1 expression and chimerism status (Complete chimerism versus Low Level Mixed Chimerism) in acute leukemia. Leuk. Lymphoma 2013, 54, 2660–2666. [Google Scholar] [CrossRef] [PubMed]
- de Moraes, A.C.R.; Maranho, C.K.; Rauber, G.S.; Santos-Silva, M.C. Importance of Detecting Multidrug Resistance Proteins in Acute Leukemia Prognosis and Therapy. J. Clin. Lab. Anal. 2013, 27, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Essers, M.A.G.; Trumpp, A. Targeting leukemic stem cells by breaking their dormancy. Mol. Oncol. 2010, 4, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Uchida, N.; Tanaka, S.; Suzuki, N.; Tomizawa-Murasawa, M.; Sone, A.; Najima, Y.; Takagi, S.; Aoki, Y.; Wake, A.; et al. Induction of cell cycle entry eliminates human leukemia stem cells in a mouse model of AML. Nat. Biotechnol. 2010, 28, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Costello, R.T.; Mallet, F.; Gaugler, B.; Sainty, D.; Arnoulet, C.; Gastaut, J.A.; Olive, D. Human acute myeloid leukemia CD34+/CD38- progenitor cells have decreased sensitivity to chemotherapy and Fas-induced apoptosis, reduced immunogenicity, and impaired dendritic cell transformation capacities. Cancer Res. 2000, 60, 4403–4411. [Google Scholar]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef]
- Laverdière, I.; Boileau, M.; Neumann, A.L.; Frison, H.; Mitchell, A.; Ng, S.W.K.; Wang, J.C.Y.; Minden, M.D.; Eppert, K. Leukemic stem cell signatures identify novel therapeutics targeting acute myeloid leukemia. Blood Cancer J. 2018, 8, 52. [Google Scholar] [CrossRef]
- Hanekamp, D.; Cloos, J.; Schuurhuis, G.J. Leukemic stem cells: Identification and clinical application. Int. J. Hematol. 2017, 105, 549–557. [Google Scholar] [CrossRef]
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef]
- Stelmach, P.; Trumpp, A. Leukemic stem cells and therapy resistance in acute myeloid leukemia. Haematologica 2023, 108, 353–366. [Google Scholar] [CrossRef]
- Heo, S.-K.; Noh, E.-K.; Ju, L.J.; Sung, J.Y.; Jeong, Y.K.; Cheon, J.; Koh, S.J.; Min, Y.J.; Choi, Y.; Jo, J.-C. CD45dimCD34+CD38-CD133+ cells have the potential as leukemic stem cells in acute myeloid leukemia. BMC Cancer 2020, 20, 285. [Google Scholar] [CrossRef]
- Al-Mawali, A.; Pinto, A.D.; Al-Zadjali, S. CD34+CD38-CD123+ Cells Are Present in Virtually All Acute Myeloid Leukaemia Blasts: A Promising Single Unique Phenotype for Minimal Residual Disease Detection. Acta Haematol. 2017, 138, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Hauswirth, A.W.; Florian, S.; Printz, D.; Sotlar, K.; Krauth, M.-T.; Fritsch, G.; Schernthaner, G.-H.; Wacheck, V.; Selzer, E.; Sperr, W.R.; et al. Expression of the target receptor CD33 in CD34+/CD38-/CD123+ AML stem cells. Eur. J. Clin. Investig. 2007, 37, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Zeijlemaker, W.; Grob, T.; Meijer, R.; Hanekamp, D.; Kelder, A.; Carbaat-Ham, J.C.; Oussoren-Brockhoff, Y.J.M.; Snel, A.N.; Veldhuizen, D.; Scholten, W.J.; et al. CD34+CD38- leukemic stem cell frequency to predict outcome in acute myeloid leukemia. Leukemia 2019, 33, 1102–1112. [Google Scholar] [CrossRef] [PubMed]
- Vergez, F.; Green, A.S.; Tamburini, J.; Sarry, J.-E.; Gaillard, B.; Cornillet-Lefebvre, P.; Pannetier, M.; Neyret, A.; Chapuis, N.; Ifrah, N.; et al. High levels of CD34+CD38low/-CD123+ blasts are predictive of an adverse outcome in acute myeloid leukemia: A Groupe Ouest-Est des Leucemies Aigues et Maladies du Sang (GOELAMS) study. Haematologica 2011, 96, 1792–1798. [Google Scholar] [CrossRef] [PubMed]
- Vergez, F.; Nicolau-Travers, M.-L.; Bertoli, S.; Rieu, J.-B.; Tavitian, S.; Bories, P.; Luquet, I.; Mas, V.D.; Largeaud, L.; Sarry, A.; et al. CD34+CD38-CD123+ Leukemic Stem Cell Frequency Predicts Outcome in Older Acute Myeloid Leukemia Patients Treated by Intensive Chemotherapy but Not Hypomethylating Agents. Cancers 2020, 12, 1174. [Google Scholar] [CrossRef] [PubMed]
- van Rhenen, A.; van Dongen, G.A.M.S.; Kelder, A.; Rombouts, E.J.; Feller, N.; Moshaver, B.; Stigter-van Walsum, M.; Zweegman, S.; Ossenkoppele, G.J.; Jan Schuurhuis, G. The novel AML stem cell associated antigen CLL-1 aids in discrimination between normal and leukemic stem cells. Blood 2007, 110, 2659–2666. [Google Scholar] [CrossRef]
- Canali, A.; Vergnolle, I.; Bertoli, S.; Largeaud, L.; Nicolau, M.-L.; Rieu, J.-B.; Tavitian, S.; Huguet, F.; Picard, M.; Bories, P.; et al. Prognostic Impact of Unsupervised Early Assessment of Bulk and Leukemic Stem Cell Measurable Residual Disease in Acute Myeloid Leukemia. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2023, 29, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Zou, D.; Cui, D. Advances in isolation and detection of circulating tumor cells based on microfluidics. Cancer Biol. Med. 2018, 15, 335–353. [Google Scholar]
- Khoo, B.L.; Shang, M.; Ng, C.H.; Lim, C.T.; Chng, W.J.; Han, J. Liquid biopsy for minimal residual disease detection in leukemia using a portable blast cell biochip. NPJ Precis. Oncol. 2019, 3, 30. [Google Scholar] [CrossRef]
- Jackson, J.M.; Taylor, J.B.; Witek, M.A.; Hunsucker, S.A.; Waugh, J.P.; Fedoriw, Y.; Shea, T.C.; Soper, S.A.; Armistead, P.M. Microfluidics for the detection of minimal residual disease in acute myeloid leukemia patients using circulating leukemic cells selected from blood. Analyst 2016, 141, 640–651. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chea, M.; Rigolot, L.; Canali, A.; Vergez, F. Minimal Residual Disease in Acute Myeloid Leukemia: Old and New Concepts. Int. J. Mol. Sci. 2024, 25, 2150. https://doi.org/10.3390/ijms25042150
Chea M, Rigolot L, Canali A, Vergez F. Minimal Residual Disease in Acute Myeloid Leukemia: Old and New Concepts. International Journal of Molecular Sciences. 2024; 25(4):2150. https://doi.org/10.3390/ijms25042150
Chicago/Turabian StyleChea, Mathias, Lucie Rigolot, Alban Canali, and Francois Vergez. 2024. "Minimal Residual Disease in Acute Myeloid Leukemia: Old and New Concepts" International Journal of Molecular Sciences 25, no. 4: 2150. https://doi.org/10.3390/ijms25042150
APA StyleChea, M., Rigolot, L., Canali, A., & Vergez, F. (2024). Minimal Residual Disease in Acute Myeloid Leukemia: Old and New Concepts. International Journal of Molecular Sciences, 25(4), 2150. https://doi.org/10.3390/ijms25042150






