Unraveling the Genetic Landscape of Neurological Disorders: Insights into Pathogenesis, Techniques for Variant Identification, and Therapeutic Approaches
Abstract
1. Introduction
2. Genetic Techniques in Addressing Neurodegenerative Disorders
3. Genetic Mutations and Corresponding Cellular Alterations in Neurodegenerative Disorders
3.1. Amyotrophic Lateral Sclerosis (ALS)
3.1.1. Genetic and Pathological Overlap between ALS and FTD
3.1.2. Epidemiology
3.1.3. Genetic Causes and Risk Factor
- Superoxide dismutase (SOD1)
- Chromosome 9 open reading frame 72 (C9ORF72)
- TARDBP
- FUS
3.1.4. Additional Risk Loci from Genome-Wide Association Studies
3.2. Alzheimer’s Disease (AD)
3.2.1. Epidemiology
3.2.2. Genetic Causes and Risk Factors
- APOE
- SORL1
- MAPT
- TREM2
- ABCA7
3.2.3. Additional Risk Loci from Genome-Wide Association Studies (GWASs)
3.3. Parkinson’s Disease (PD)
3.3.1. Epidemiology
3.3.2. Genetic Causes and Risk Factors
- SNCA
- LRRK2
- VPS35
- GBA
3.3.3. Additional Risk Loci from Genome-Wide Association Studies
3.3.4. Other Neurodegenerative Disorders
4. Gene Therapy for Neurodegenerative Diseases
5. Gene Expression
5.1. The Exogenous Introduction of Genes into the CNS
5.2. DNA Editing
5.3. CRISPR-Mediated Base Editing and Prime Editing
6. Genetic Therapy for AD
6.1. Targeting MAPT
6.2. Targeting APOE
6.3. Targeting APP
7. Gene Therapy for PD
7.1. Modulating Neuronal Signaling
7.2. Targeting Disease Genes—SNCA, GBA, and LRRK2
8. Gene Therapy for ALS
Targeting SOD1
9. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nichols, E.; Steinmetz, J.D.; Vollset, S.E.; Fukutaki, K.; Chalek, J.; Abd-Allah, F.; Abdoli, A.; Abualhasan, A.; Abu-Gharbieh, E.; Akram, T.T. Estimation of the global prevalence of dementia in 2019 and forecasted prevalence in 2050: An analysis for the Global Burden of Disease Study 2019. Lancet Public Health 2022, 7, e105–e125. [Google Scholar] [CrossRef] [PubMed]
- Blackhurst, B.M.; Funk, K.E. Viral pathogens increase risk of neurodegenerative disease. Nat. Rev. Neurol. 2023, 19, 259–260. [Google Scholar] [CrossRef]
- Cerejeira, J.; Lagarto, L.; Mukaetova-Ladinska, E.B. Behavioral and psychological symptoms of dementia. Front. Neurol. 2012, 3, 73. [Google Scholar] [CrossRef] [PubMed]
- Calderón-Garcidueñas, L. Common Fatal Neurodegenerative Diseases Revisited: Beyond Age, Comorbidities, and Devastating Terminal Neuropathology There Is Hope with Prevention. Front. Neurol. 2022, 13, 901447. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, G.G.; Botond, G.; Budka, H. Protein coding of neurodegenerative dementias: The neuropathological basis of biomarker diagnostics. Acta Neuropathol. 2010, 119, 389–408. [Google Scholar] [CrossRef] [PubMed]
- Goutman, S.A.; Hardiman, O.; Al-Chalabi, A.; Chió, A.; Savelieff, M.G.; Kiernan, M.C.; Feldman, E.L. Recent advances in the diagnosis and prognosis of amyotrophic lateral sclerosis. Lancet Neurol. 2022, 21, 480–493. [Google Scholar] [CrossRef] [PubMed]
- Spires-Jones, T.L.; Attems, J.; Thal, D.R. Interactions of pathological proteins in neurodegenerative diseases. Acta Neuropathol. 2017, 134, 187–205. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.A.; Kammenga, J.E.; Harvey, S.C. Genetic variation in neurodegenerative diseases and its accessibility in the model organism Caenorhabditis elegans. Hum. Genom. 2017, 11, 12. [Google Scholar] [CrossRef]
- Bocchetta, M.; Mega, A.; Bernardi, L.; Di Maria, E.; Benussi, L.; Binetti, G.; Borroni, B.; Colao, R.; Di Fede, G.; Fostinelli, S.; et al. Genetic counseling and testing for alzheimer’s disease and frontotemporal lobar degeneration: An Italian consensus protocol. J. Alzheimer’s Dis. 2016, 51, 277–291. [Google Scholar] [CrossRef]
- Fernández, M.V.; Kim, J.H.; Budde, J.P.; Black, K.; Medvedeva, A.; Saef, B.; Deming, Y.; Del-Aguila, J.; Ibañez, L.; Dube, U.; et al. Analysis of neurodegenerative Mendelian genes in clinically diagnosed Alzheimer Disease. PLoS Genet. 2017, 13, e1007045. [Google Scholar] [CrossRef]
- Woollacott, I.O.C.; Rohrer, J.D. The clinical spectrum of sporadic and familial forms of frontotemporal dementia. J. Neurochem. 2016, 138, 6–31. [Google Scholar] [CrossRef]
- Faghihi, M.A.; Mottagui-Tabar, S.; Wahlestedt, C. Genetics of neurological disorders. Expert Rev. Mol. Diagn. 2004, 4, 317–332. [Google Scholar] [CrossRef]
- Koretsky, M.J.; Alvarado, C.; Makarious, M.B.; Vitale, D.; Levine, K.; Bandres-Ciga, S.; Dadu, A.; Scholz, S.W.; Sargent, L.; Faghri, F.; et al. Genetic risk factor clustering within and across neurodegenerative diseases. Brain 2023, 146, 4486–4494. [Google Scholar] [CrossRef]
- Jiang, T.; Tan, M.-S.; Tan, L.; Yu, J.-T. Application of next-generation sequencing technologies in Neurology. Ann. Transl. Med. 2014, 2, 125. [Google Scholar] [CrossRef]
- Guo, M.H.; Hirschhorn, J.N.; Dauber, A. Insights and implications of genome-wide association studies of height. J. Clin. Endocrinol. Metab. 2018, 103, 3155–3168. [Google Scholar] [CrossRef]
- Craig, J. Complex Diseases: Research and Applications. Nat. Educ. 2008, 1, 184. [Google Scholar]
- Tan, M.-S.; Jiang, T.; Tan, L.; Yu, J.-T. Genome-wide association studies in neurology. Ann. Transl. Med. 2014, 2, 125. [Google Scholar] [CrossRef]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef] [PubMed]
- Young, A.I. Solving the missing heritability problem. PLoS Genet. 2019, 15, e1008222. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, S. Genetics of neurodegenerative diseases: Insights from high-throughput resequencing. Hum. Mol. Genet. 2010, 19, R65–R70. [Google Scholar] [CrossRef] [PubMed]
- Dilliott, A.A.; Abdelhady, A.; Sunderland, K.M.; Farhan, S.M.K.; Abrahao, A.; Binns, M.A.; Black, S.E.; Borrie, M.; Casaubon, L.K.; Dowlatshahi, D.; et al. Contribution of rare variant associations to neurodegenerative disease presentation. npj Genom. Med. 2021, 6, 80. [Google Scholar] [CrossRef]
- Goswami, C.; Chattopadhyay, A.; Chuang, E.Y. Rare variants: Data types and analysis strategies. Ann. Transl. Med. 2021, 9, 961. [Google Scholar] [CrossRef]
- Zhao, Y.; Fang, L.T.; Shen, T.-W.; Choudhari, S.; Talsania, K.; Chen, X.; Shetty, J.; Kriga, Y.; Tran, B.; Zhu, B.; et al. Whole genome and exome sequencing reference datasets from a multi-center and cross-platform benchmark study. Sci. Data 2021, 8, 296. [Google Scholar] [CrossRef] [PubMed]
- Pervez, M.T.; Hasnain, M.J.U.; Abbas, S.H.; Moustafa, M.F.; Aslam, N.; Shah, S.S.M. A comprehensive review of performance of next-generation sequencing platforms. BioMed Res. Int. 2022, 2022, 3457806. [Google Scholar] [CrossRef]
- Chen, Y.-C.; Liu, T.; Yu, C.-H.; Chiang, T.-Y.; Hwang, C.-C. Effects of GC bias in next-generation-sequencing data on de novo genome assembly. PLoS ONE 2013, 8, e62856. [Google Scholar] [CrossRef]
- Eid, J.; Fehr, A.; Gray, J.; Luong, K.; Lyle, J.; Otto, G.; Peluso, P.; Rank, D.; Baybayan, P.; Bettman, B.; et al. Real-time DNA sequencing from single polymerase molecules. Science 2009, 323, 133–138. [Google Scholar] [CrossRef]
- Clarke, J.; Wu, H.-C.; Jayasinghe, L.; Patel, A.; Reid, S.; Bayley, H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 2009, 4, 265–270. [Google Scholar] [CrossRef]
- Su, Y.; Fan, L.; Shi, C.; Wang, T.; Zheng, H.; Luo, H.; Zhang, S.; Hu, Z.; Fan, Y.; Dong, Y.; et al. Deciphering neurodegenerative diseases using long-read sequencing. Neurology 2021, 97, 423–433. [Google Scholar] [CrossRef]
- Oehler, J.B.; Wright, H.; Stark, Z.; Mallett, A.J.; Schmitz, U. The application of long-read sequencing in clinical settings. Hum. Genom. 2023, 17, 73. [Google Scholar] [CrossRef] [PubMed]
- Hafford-Tear, N.J.; Tsai, Y.-C.; Sadan, A.N.; Sanchez-Pintado, B.; Zarouchlioti, C.; Maher, G.J.; Liskova, P.; Tuft, S.J.; Hardcastle, A.J.; Clark, T.A.; et al. CRISPR/Cas9-targeted enrichment and long-read sequencing of the Fuchs endothelial corneal dystrophy–associated TCF4 triplet repeat. Genet. Med. 2019, 21, 2092–2102. [Google Scholar] [CrossRef] [PubMed]
- Miga, K.H.; Koren, S.; Rhie, A.; Vollger, M.R.; Gershman, A.; Bzikadze, A.; Brooks, S.; Howe, E.; Porubsky, D.; Logsdon, G.A.; et al. Telomere-to-telomere assembly of a complete human X chromosome. Nature 2020, 585, 79–84. [Google Scholar] [CrossRef]
- Nurk, S.; Koren, S.; Rhie, A.; Rautiainen, M.; Bzikadze, A.V.; Mikheenko, A.; Vollger, M.R.; Altemose, N.; Uralsky, L.; Gershman, A.; et al. The complete sequence of a human genome. Science 2022, 376, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Chaisson, M.J.P.; Huddleston, J.; Dennis, M.Y.; Sudmant, P.H.; Malig, M.; Hormozdiari, F.; Antonacci, F.; Surti, U.; Sandstrom, R.; Boitano, M.; et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature 2015, 517, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Ebert, P.; Audano, P.A.; Zhu, Q.; Rodriguez-Martin, B.; Porubsky, D.; Bonder, M.J.; Sulovari, A.; Ebler, J.; Zhou, W.; Mari, R.S.; et al. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science 2021, 372, 48. [Google Scholar] [CrossRef]
- Theunissen, F.; Flynn, L.L.; Anderton, R.S.; Mastaglia, F.; Pytte, J.; Jiang, L.; Hodgetts, S.; Burns, D.K.; Saunders, A.; Fletcher, S.; et al. Structural variants may be a source of missing heritability in sALS. Front. Neurosci. 2020, 14, 47. [Google Scholar] [CrossRef]
- Parobkova, E.; Matej, R. Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degenerations: Similarities in Genetic Background. Diagnostics 2021, 11, 509. [Google Scholar] [CrossRef] [PubMed]
- Perrone, F.; Nguyen, H.P.; Van Mossevelde, S.; Moisse, M.; Sieben, A.; Santens, P.; De Bleecker, J.; Vandenbulcke, M.; Engelborghs, S.; Baets, J.; et al. Investigating the role of ALS genes CHCHD10 and TUBA4A in Belgian FTD-ALS spectrum patients. Neurobiol. Aging 2017, 51, 177.e9–177.e16. [Google Scholar] [CrossRef]
- Ahmed, R.M.; Devenney, E.M.; Strikwerda-Brown, C.; Hodges, J.R.; Piguet, O.; Kiernan, M.C. Phenotypic variability in ALS-FTD and effect on survival. Neurology 2020, 94, e2005–e2013. [Google Scholar] [CrossRef]
- Abramzon, Y.A.; Fratta, P.; Traynor, B.J.; Chia, R. The Overlapping Genetics of Amyotrophic Lateral Sclerosis and Frontotemporal Dementia. Front. Neurosci. 2020, 14, 42. [Google Scholar] [CrossRef]
- Olney, N.T.; Spina, S.; Miller, B.L. Frontotemporal Dementia. Neurol. Clin. 2017, 35, 339–374. [Google Scholar] [CrossRef]
- Broce, I.J.; Castruita, P.A.; Yokoyama, J.S. Moving Toward Patient-Tailored Treatment in ALS and FTD: The Potential of Genomic Assessment as a Tool for Biological Discovery and Trial Recruitment. Front. Neurosci. 2021, 15, 639078. [Google Scholar] [CrossRef]
- Marin, B.; Boumédiene, F.; Logroscino, G.; Couratier, P.; Babron, M.-C.; Leutenegger, A.-L.; Copetti, M.; Preux, P.-M.; Beghi, E. Variation in worldwide incidence of amyotrophic lateral sclerosis: A meta-analysis. Int. J. Epidemiol. 2017, 46, 57–74. [Google Scholar] [CrossRef]
- Feldman, E.L.; Goutman, S.A.; Petri, S.; Mazzini, L.; Savelieff, M.G.; Shaw, P.J.; Sobue, G. Amyotrophic lateral sclerosis. Lancet 2022, 400, 1363–1380. [Google Scholar] [CrossRef]
- Chia, R.; Chiò, A.; Traynor, B.J. Novel genes associated with amyotrophic lateral sclerosis: Diagnostic and clinical implications. Lancet Neurol. 2018, 17, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Genç, B.; Gautam, M.; Helmold, B.R.; Koçak, N.; Günay, A.; Goshu, G.M.; Silverman, R.B.; Ozdinler, P.H. NU-9 improves health of hSOD1G93A mouse upper motor neurons in vitro, especially in combination with riluzole or edaravone. Sci. Rep. 2022, 12, 5383. [Google Scholar] [CrossRef] [PubMed]
- Merjane, J.; Chung, R.; Patani, R.; Lisowski, L. Molecular mechanisms of amyotrophic lateral sclerosis as broad therapeutic targets for gene therapy applications utilizing adeno-associated viral vectors. Med. Res. Rev. 2023, 43, 829–854. [Google Scholar] [CrossRef]
- Aschenbrenner, D.S. New Drug Approved For ALS. AJN Am. J. Nurs. 2023, 123, 22–23. [Google Scholar] [CrossRef]
- Goutman, S.A.; Hardiman, O.; Al-Chalabi, A.; Chió, A.; Savelieff, M.G.; Kiernan, M.C.; Feldman, E.L. Emerging insights into the complex genetics and pathophysiology of amyotrophic lateral sclerosis. Lancet Neurol. 2022, 21, 465–479. [Google Scholar] [CrossRef] [PubMed]
- Ryan, M.; Heverin, M.; McLaughlin, R.L.; Hardiman, O. Lifetime risk and heritability of amyotrophic lateral sclerosis. JAMA Neurol. 2019, 76, 1367–1374. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, A.; Kenna, K.P.; Renton, A.E.; Ticozzi, N.; Faghri, F.; Chia, R.; Dominov, J.A.; Kenna, B.J.; Nalls, M.A.; Keagle, P.; et al. Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron 2018, 97, 1268–1283.e6. [Google Scholar] [CrossRef]
- Shahrizaila, N.; Sobue, G.; Kuwabara, S.; Kim, S.H.; Birks, C.; Fan, D.S.; Bae, J.S.; Hu, C.J.; Gourie-Devi, M.; Noto, Y.; et al. Amyotrophic lateral sclerosis and motor neuron syndromes in Asia. J. Neurol. Neurosurg. Psychiatry 2016, 87, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.-Y.; Zhou, Z.-R.; Che, C.-H.; Liu, C.-Y.; He, R.-L.; Huang, H.-P. Genetic epidemiology of amyotrophic lateral sclerosis: A systematic review and meta-analysis. J. Neurol. Neurosurg. Psychiatry 2017, 88, 540–549. [Google Scholar] [CrossRef] [PubMed]
- Rosen, D.R.; Siddique, T.; Patterson, D.; Figlewicz, D.A.; Sapp, P.; Hentati, A.; Donaldson, D.; Goto, J.; O’Regan, J.P.; Deng, H.-X.; et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 1993, 362, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Valentine, J.S.; Doucette, P.A.; Potter, S.Z. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu. Rev. Biochem. 2005, 74, 563–593. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Nishiyama, A.; Warita, H.; Aoki, M. Genetics of amyotrophic lateral sclerosis: Seeking therapeutic targets in the era of gene therapy. J. Hum. Genet. 2023, 68, 131–152. [Google Scholar] [CrossRef]
- Azadmanesh, J.; Borgstahl, G.E.O. A Review of the catalytic mechanism of human manganese superoxide dismutase. Antioxidants 2018, 7, 25. [Google Scholar] [CrossRef]
- Corcia, P.; Valdmanis, P.; Millecamps, S.; Lionnet, C.; Blasco, H.; Mouzat, K.; Daoud, H.; Belzil, V.; Morales, R.; Pageot, N.; et al. Phenotype and genotype analysis in amyotrophic lateral sclerosis with TARDBP gene mutations. Neurology 2012, 78, 1519–1526. [Google Scholar] [CrossRef]
- Hayashi, Y.; Homma, K.; Ichijo, H. SOD1 in neurotoxicity and its controversial roles in SOD1 mutation-negative ALS. Adv. Biol. Regul. 2016, 60, 95–104. [Google Scholar] [CrossRef]
- Fujisawa, T.; Homma, K.; Yamaguchi, N.; Kadowaki, H.; Tsuburaya, N.; Naguro, I.; Matsuzawa, A.; Takeda, K.; Takahashi, Y.; Goto, J.; et al. A novel monoclonal antibody reveals a conformational alteration shared by amyotrophic lateral sclerosis-linked SOD1 mutants. Ann. Neurol. 2012, 72, 739–749. [Google Scholar] [CrossRef]
- Mathis, S.; Couratier, P.; Julian, A.; Vallat, J.-M.; Corcia, P.; Le Masson, G. Management and therapeutic perspectives in amyotrophic lateral sclerosis. Expert Rev. Neurother. 2017, 17, 263–276. [Google Scholar] [CrossRef]
- Li, K.; Hala, T.J.; Seetharam, S.; Poulsen, D.J.; Wright, M.C.; Lepore, A.C. GLT1 overexpression in SOD1G93A mouse cervical spinal cord does not preserve diaphragm function or extend disease. Neurobiol. Dis. 2015, 78, 12–23. [Google Scholar] [CrossRef]
- Szelechowski, M.; Amoedo, N.; Obre, E.; Léger, C.; Allard, L.; Bonneu, M.; Claverol, S.; Lacombe, D.; Oliet, S.; Chevallier, S.; et al. Metabolic reprogramming in amyotrophic lateral sclerosis. Sci. Rep. 2018, 8, 3953. [Google Scholar] [CrossRef]
- Le Masson, G.; Przedborski, S.; Abbott, L. A computational model of motor neuron degeneration. Neuron 2014, 83, 975–988. [Google Scholar] [CrossRef]
- Lamanauskas, N.; Nistri, A. Riluzole blocks persistent Na+ and Ca2+ currents and modulates release of glutamate via presynaptic NMDA receptors on neonatal rat hypoglossal motoneurons in vitro. Eur. J. Neurosci. 2008, 27, 2501–2514. [Google Scholar] [CrossRef]
- Ayers, J.I.; Fromholt, S.E.; O’neal, V.M.; Diamond, J.H.; Borchelt, D.R. Prion-like propagation of mutant SOD1 misfolding and motor neuron disease spread along neuroanatomical pathways. Acta Neuropathol. 2016, 131, 103–114. [Google Scholar] [CrossRef]
- Polymenidou, M.; Cleveland, D.W. The seeds of neurodegeneration: Prion-like spreading in ALS. Cell 2011, 147, 498–508. [Google Scholar] [CrossRef]
- Saeed, M.; Yang, Y.; Deng, H.-X.; Hung, W.-Y.; Siddique, N.; Dellefave, L.; Gellera, C.; Andersen, P.M.; Siddique, T. Age and founder effect of SOD1 A4V mutation causing ALS. Neurology 2009, 72, 1634–1639. [Google Scholar] [CrossRef]
- Zou, Z.-Y.; Liu, M.-S.; Li, X.-G.; Cui, L.-Y. H46R SOD1 mutation is consistently associated with a relatively benign form of amyotrophic lateral sclerosis with slow progression. Amyotroph. Lateral Scler. Front. Degener. 2016, 17, 610–613. [Google Scholar] [CrossRef]
- Kato, M.; Aoki, M.; Ohta, M.; Nagai, M.; Ishizaki, F.; Nakamura, S.; Itoyama, Y. Marked reduction of the Cu/Zn superoxide dismutase polypeptide in a case of familial amyotrophic lateral sclerosis with the homozygous mutation. Neurosci. Lett. 2001, 312, 165–168. [Google Scholar] [CrossRef]
- Hideshima, M.; Beck, G.; Yamadera, M.; Motoyama, Y.; Ikenaka, K.; Kakuda, K.; Tsuda, H.; Nagano, S.; Fujimura, H.; Morii, E.; et al. A clinicopathological study of ALS with L126S mutation in the SOD1 gene presenting with isolated inferior olivary hypertrophy. Neuropathology 2020, 40, 191–195. [Google Scholar] [CrossRef]
- Gagliardi, D.; Ahmadinejad, M.; Del Bo, R.; Meneri, M.; Comi, G.P.; Corti, S.; Ronchi, D. Homozygous SOD1 variation L144S produces a severe form of amyotrophic lateral sclerosis in an Iranian family. Neurol. Genet. 2021, 8, e645. [Google Scholar] [CrossRef] [PubMed]
- Berdyński, M.; Miszta, P.; Safranow, K.; Andersen, P.M.; Morita, M.; Filipek, S.; Żekanowski, C.; Kuźma-Kozakiewicz, M. SOD1 mutations associated with amyotrophic lateral sclerosis analysis of variant severity. Sci. Rep. 2022, 12, 103. [Google Scholar] [CrossRef]
- DeJesus-Hernandez, M.; Mackenzie, I.R.; Boeve, B.F.; Boxer, A.L.; Baker, M.; Rutherford, N.J.; Nicholson, A.M.; Finch, N.A.; Flynn, H.; Adamson, J.; et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011, 72, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, A.; Wen, J.; Rinaldi, D.; Houot, M.; Sayah, S.; Camuzat, A.; Fournier, C.; Fontanella, S.; Routier, A.; Couratier, P.; et al. Early cognitive, structural, and microstructural changes in presymptomatic C9orf72 carriers younger than 40 years. JAMA Neurol. 2018, 75, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Koppers, M.; Blokhuis, A.M.; Westeneng, H.; Terpstra, M.L.; Zundel, C.A.C.; de Sá, R.V.; Schellevis, R.D.; Waite, A.J.; Blake, D.J.; Veldink, J.H.; et al. C9orf72 ablation in mice does not cause motor neuron degeneration or motor deficits. Ann. Neurol. 2015, 78, 426–438. [Google Scholar] [CrossRef]
- Lee, Y.-B.; Chen, H.-J.; Peres, J.N.; Gomez-Deza, J.; Attig, J.; Štalekar, M.; Troakes, C.; Nishimura, A.L.; Scotter, E.L.; Vance, C.; et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013, 5, 1178–1186. [Google Scholar] [CrossRef]
- Lee, Y.-B.; Baskaran, P.; Gomez-Deza, J.; Chen, H.-J.; Nishimura, A.L.; Smith, B.N.; Troakes, C.; Adachi, Y.; Stepto, A.; Petrucelli, L.; et al. C9orf72 poly GA RAN-translated protein plays a key role in amyotrophic lateral sclerosis via aggregation and toxicity. Hum. Mol. Genet. 2017, 26, 4765–4777. [Google Scholar] [CrossRef]
- Selvaraj, B.T.; Livesey, M.R.; Zhao, C.; Gregory, J.M.; James, O.T.; Cleary, E.M.; Chouhan, A.K.; Gane, A.B.; Perkins, E.M.; Dando, O.; et al. C9ORF72 repeat expansion causes vulnerability of motor neurons to Ca2+-permeable AMPA receptor-mediated excitotoxicity. Nat. Commun. 2018, 9, 347. [Google Scholar] [CrossRef]
- Mitra, J.; Guerrero, E.N.; Hegde, P.M.; Liachko, N.F.; Wang, H.; Vasquez, V.; Gao, J.; Pandey, A.; Taylor, J.P.; Kraemer, B.C.; et al. Motor neuron disease-associated loss of nuclear TDP-43 is linked to DNA double-strand break repair defects. Proc. Natl. Acad. Sci. USA 2019, 116, 4696–4705. [Google Scholar] [CrossRef]
- Kabashi, E.; Valdmanis, P.N.; Dion, P.; Spiegelman, D.; McConkey, B.J.; Velde, C.V.; Bouchard, J.-P.; Lacomblez, L.; Pochigaeva, K.; Salachas, F.; et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 2008, 40, 572–574. [Google Scholar] [CrossRef]
- Braak, H.; Brettschneider, J.; Ludolph, A.C.; Lee, V.M.; Trojanowski, J.Q.; Del Tredici, K. Amyotrophic lateral sclerosis—A model of corticofugal axonal spread. Nat. Rev. Neurol. 2013, 9, 708–714. [Google Scholar] [CrossRef]
- Mitsuzawa, S.; Akiyama, T.; Nishiyama, A.; Suzuki, N.; Kato, M.; Warita, H.; Izumi, R.; Osana, S.; Koyama, S.; Kato, T.; et al. TARDBP p.G376D mutation, found in rapid progressive familial ALS, induces mislocalization of TDP-43. eNeurologicalSci 2018, 11, 20–22. [Google Scholar] [CrossRef]
- Kametani, F.; Obi, T.; Shishido, T.; Akatsu, H.; Murayama, S.; Saito, Y.; Yoshida, M.; Hasegawa, M. Mass spectrometric analysis of accumulated TDP-43 in amyotrophic lateral sclerosis brains. Sci. Rep. 2016, 6, 23281. [Google Scholar] [CrossRef]
- Donde, A.; Sun, M.; Ling, J.P.; Braunstein, K.E.; Pang, B.; Wen, X.; Cheng, X.; Chen, L.; Wong, P.C. Splicing repression is a major function of TDP-43 in motor neurons. Acta Neuropathol. 2019, 138, 813–826. [Google Scholar] [CrossRef]
- Tank, E.M.; Figueroa-Romero, C.; Hinder, L.M.; Bedi, K.; Archbold, H.C.; Li, X.; Weskamp, K.; Safren, N.; Paez-Colasante, X.; Pacut, C.; et al. Abnormal RNA stability in amyotrophic lateral sclerosis. Nat. Commun. 2018, 9, 2845. [Google Scholar] [CrossRef]
- Wood, A.; Gurfinkel, Y.; Polain, N.; Lamont, W.; Rea, S.L. Molecular mechanisms underlying TDP-43 pathology in cellular and animal models of ALS and FTLD. Int. J. Mol. Sci. 2021, 22, 4705. [Google Scholar] [CrossRef]
- Kwiatkowski, T.J., Jr.; Bosco, D.A.; Leclerc, A.L.; Tamrazian, E.; Vanderburg, C.R.; Russ, C.; Davis, A.; Gilchrist, J.; Kasarskis, E.J.; Munsat, T.; et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 2009, 323, 1205–1208. [Google Scholar] [CrossRef] [PubMed]
- Huebers, A.; Just, W.; Rosenbohm, A.; Mueller, K.; Marroquin, N.; Goebel, I.; Hoegel, J.; Thiele, H.; Altmueller, J.; Nuernberg, P.; et al. De novo FUS mutations are the most frequent genetic cause in early-onset German ALS patients. Neurobiol. Aging 2015, 36, 3117.e1–3117.e6. [Google Scholar]
- Akiyama, T.; Warita, H.; Kato, M.; Nishiyama, A.; Izumi, R.; Ikeda, C.; Kamada, M.; Suzuki, N.; Aoki, M. Genotype–phenotype relationships in familial amyotrophic lateral sclerosis with FUS/TLS mutations in Japan. Muscle Nerve 2016, 54, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Neumann, M.; Rademakers, R.; Roeber, S.; Baker, M.; Kretzschmar, H.A.; Mackenzie, I.R.A. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain 2009, 132, 2922–2931. [Google Scholar] [CrossRef] [PubMed]
- Salam, S.; Tacconelli, S.; Smith, B.N.; Mitchell, J.C.; Glennon, E.; Nikolaou, N.; Houart, C.; Vance, C. Identification of a novel interaction of FUS and syntaphilin may explain synaptic and mitochondrial abnormalities caused by ALS mutations. Sci. Rep. 2021, 11, 13613. [Google Scholar] [CrossRef]
- Li, Y.R.; King, O.D.; Shorter, J.; Gitler, A.D. Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 2013, 201, 361–372. [Google Scholar] [CrossRef]
- van Rheenen, W.; van der Spek, R.A.A.; Bakker, M.K.; van Vugt, J.J.F.A.; Hop, P.J.; Zwamborn, R.A.J.; de Klein, N.; Westra, H.-J.; Bakker, O.B.; Deelen, P.; et al. Common and rare variant association analyses in amyotrophic lateral sclerosis identify 15 risk loci with distinct genetic architectures and neuron-specific biology. Nat. Genet. 2021, 53, 1636–1648. [Google Scholar] [CrossRef]
- Nguyen, H.P.; Van Broeckhoven, C.; van der Zee, J. ALS genes in the genomic era and their implications for FTD. Trends Genet. 2018, 34, 404–423. [Google Scholar] [CrossRef]
- Lipstein, N.; Verhoeven-Duif, N.M.; Michelassi, F.E.; Calloway, N.; van Hasselt, P.M.; Pienkowska, K.; van Haaften, G.; van Haelst, M.M.; van Empelen, R.; Cuppen, I.; et al. Synaptic UNC13A protein variant causes increased neurotransmission and dyskinetic movement disorder. J. Clin. Investig. 2017, 127, 1005–1018. [Google Scholar] [CrossRef]
- Borg, R.; Purkiss, A.; Cacciottolo, R.; Herrera, P.; Cauchi, R.J. Loss of amyotrophic lateral sclerosis risk factor SCFD1 causes motor dysfunction in Drosophila. Neurobiol. Aging 2023, 126, 67–76. [Google Scholar] [CrossRef]
- Liampas, I.; Siokas, V.; Aloizou, A.-M.; Bakirtzis, C.; Tsouris, Z.; Nousia, A.; Nasios, G.; Papadimitriou, D.; Liakos, P.; Bogdanos, D.P.; et al. MOBP rs616147 Polymorphism and Risk of Amyotrophic Lateral Sclerosis in a Greek Population: A Case-Control Study. Medicina 2021, 57, 1337. [Google Scholar] [CrossRef]
- Zhang, Z.; Sun, S. Linking ALS mutations to the disruption of the MHC-II pathway. Proc. Natl. Acad. Sci. USA 2023, 120, e2315240120. [Google Scholar] [CrossRef]
- Soustelle, L.; Aimond, F.; López-Andrés, C.; Brugioti, V.; Raoul, C.; Layalle, S. ALS-Associated KIF5A Mutation Causes Locomotor Deficits Associated with Cytoplasmic Inclusions, Alterations of Neuromuscular Junctions, and Motor Neuron Loss. J. Neurosci. 2023, 43, 8058–8072. [Google Scholar] [CrossRef]
- De Decker, M.; Zelina, P.; Moens, T.G.; Eggermont, K.; Moisse, M.; Veldink, J.H.; Van Den Bosch, L.; Pasterkamp, R.J.; Van Damme, P. C21orf2 mutations found in ALS disrupt primary cilia function. bioRxiv 2022. [Google Scholar] [CrossRef]
- Restuadi, R.; Steyn, F.J.; Kabashi, E.; Ngo, S.T.; Cheng, F.-F.; Nabais, M.F.; Thompson, M.J.; Qi, T.; Wu, Y.; Henders, A.K.; et al. Functional characterisation of the amyotrophic lateral sclerosis risk locus GPX3/TNIP1. Genome Med. 2022, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; Yang, T.M.; Ou, R.W.; Wei, Q.Q.; Shang, H.F. Genome-wide genetic links between amyotrophic lateral sclerosis and autoimmune diseases. BMC Med. 2021, 19, 27. [Google Scholar] [CrossRef] [PubMed]
- Oakes, J.A.; Davies, M.C.; Collins, M.O. TBK1: A new player in ALS linking autophagy and neuroinflammation. Mol. Brain 2017, 10, 5. [Google Scholar] [CrossRef] [PubMed]
- Karch, C.M.; Wen, N.; Fan, C.C.; Yokoyama, J.S.; Kouri, N.; Ross, O.A.; Höglinger, G.; Müller, U.; Ferrari, R.; Hardy, J.; et al. Selective Genetic Overlap Between Amyotrophic Lateral Sclerosis and Diseases of the Frontotemporal Dementia Spectrum. JAMA Neurol. 2018, 75, 860–875. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; He, X.; Cui, B.; Zhao, F.; Zhou, C. NEK1 mutations and the risk of amyotrophic lateral sclerosis (ALS): A meta-analysis. Neurol. Sci. 2021, 42, 1277–1285. [Google Scholar] [CrossRef]
- Laszlo, Z.I.; Hindley, N.; Avila, A.S.; Kline, R.A.; Eaton, S.L.; Lamont, D.J.; Smith, C.; Spires-Jones, T.L.; Wishart, T.M.; Henstridge, C.M. Synaptic proteomics reveal distinct molecular signatures of cognitive change and C9ORF72 repeat expansion in the human ALS cortex. Acta Neuropathol. Commun. 2022, 10, 156. [Google Scholar] [CrossRef]
- Hendriks, W.J.A.J.; van Cruchten, R.T.P.; Pulido, R. Hereditable variants of classical protein tyrosine phosphatase genes: Will they prove innocent or guilty? Front. Cell Dev. Biol. 2023, 10, 1051311. [Google Scholar] [CrossRef]
- Andrés-Benito, P.; Moreno, J.; Aso, E.; Povedano, M.; Ferrer, I. Amyotrophic lateral sclerosis, gene deregulation in the anterior horn of the spinal cord and frontal cortex area 8: Implications in frontotemporal lobar degeneration. Aging 2017, 9, 823–851. [Google Scholar] [CrossRef]
- Alzheimer, A. Uber eigenartige Erkrankung der Hirnrinde. All Z Psychiatr. 1907, 64, 146–148. [Google Scholar]
- Maurer, K.; Volk, S.; Gerbaldo, H. Auguste D and Alzheimer’s disease. Lancet 1997, 349, 1546–1549. [Google Scholar] [CrossRef]
- Montine, T.J.; Phelps, C.H.; Beach, T.G.; Bigio, E.H.; Cairns, N.J.; Dickson, D.W.; Duyckaerts, C.; Frosch, M.P.; Masliah, E.; Mirra, S.S.; et al. National Institute on Aging–Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease: A practical approach. Acta Neuropathol. 2012, 123, 1–11. [Google Scholar] [CrossRef]
- Roth, M. The natural history of mental disorder in old age. J. Ment. Sci. 1955, 101, 281–301. [Google Scholar] [CrossRef]
- Sirkis, D.W.; Bonham, L.W.; Johnson, T.P.; La Joie, R.; Yokoyama, J.S. Dissecting the clinical heterogeneity of early-onset Alzheimer’s disease. Mol. Psychiatry 2022, 27, 2674–2688. [Google Scholar] [CrossRef]
- Reitz, C.; Rogaeva, E.; Beecham, G.W. Late-onset vs nonmendelian early-onset Alzheimer disease: A distinction without a difference? Neurol. Genet. 2020, 6. [Google Scholar] [CrossRef]
- Dementia, E.O. A National Challenge, a Future Crisis; Alzheimers Association: Washington, DC, USA, 2006. [Google Scholar]
- Alzheimer’s Association. 2019 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2019, 15, 321–387. [Google Scholar] [CrossRef]
- Goate, A.; Chartier-Harlin, M.C.; Mullan, M.; Brown, J.; Crawford, F.; Fidani, L.; Giuffra, L.; Haynes, A.; Irving, N.; James, L.; et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature 1991, 349, 704–706. [Google Scholar] [CrossRef] [PubMed]
- Chartier-Harlin, M.C.; Crawford, F.; Houlden, H.; Warren, A.; Hughes, D.; Fidani, L.; Goate, A.; Rossor, M.; Roques, P.; Hardy, J.; et al. Early-onset Alzheimer’s disease caused by mutations at codon 717 of the β-amyloid precursor protein gene. Nature 1991, 353, 844–846. [Google Scholar] [CrossRef] [PubMed]
- Murrell, J.; Farlow, M.; Ghetti, B.; Benson, M.D. A mutation in the amyloid precursor protein associated with hereditary Alzheimer’s disease. Science 1991, 254, 97–99. [Google Scholar] [CrossRef]
- Mullan, M.; Tsuji, S.; Miki, T.; Katsuya, T.; Naruse, S.; Kaneko, K.; Shimizu, T.; Kojima, T.; Nakano, I.; Ogihara, T.; et al. Clinical comparison of Alzheimer’s disease in pedigrees with the codon 717 Val→ Ile mutation in the amyloid precursor protein gene. Neurobiol. Aging 1993, 14, 407–419. [Google Scholar] [CrossRef]
- Hardy, J. The discovery of Alzheimer-causing mutations in the APP gene and the formulation of the “amyloid cascade hypothesis. FEBS J. 2017, 284, 1040–1044. [Google Scholar] [CrossRef]
- Sherrington, R.; Rogaev, E.I.; Liang, Y.A.; Rogaeva, E.A.; Levesque, G.; Ikeda, M.; Chi, H.; Lin, C.; Li, G.; Holman, K.; et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature 1995, 375, 754–760. [Google Scholar] [CrossRef]
- Campion, D.; Flaman, J.M.; Brice, A.; Hannequin, D.; Dubois, B.; Martin, C.; Moreau, V.; Charbonnier, F.; Didierjean, O.; Tardieu, S.; et al. Mutations of the presenilin I gene in families with early-onset Alzheimer’s disease. Hum. Mol. Genet. 1995, 4, 2373–2377. [Google Scholar] [CrossRef]
- Cacace, R.; Sleegers, K.; Van Broeckhoven, C. Molecular genetics of early-onset Alzheimer’s disease revisited. Alzheimer’s Dement. 2016, 12, 733–748. [Google Scholar] [CrossRef]
- Shim, K.H.; Kang, S.; An, S.S.A.; Kang, M.J. Identification of the Third Case of PSEN1 Tyr389His Variant in Early-Onset Alzheimer’s Disease in Korea. Int. J. Mol. Sci. 2022, 23, 16192. [Google Scholar] [CrossRef]
- Rogaev, E. Familial Alzheimer’s disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer’s disease type 3 gene. Nature 1995, 376, 775–778. [Google Scholar] [CrossRef]
- Levy-Lahad, E.; Wasco, W.; Poorkaj, P.; Romano, D.M.; Oshima, J.; Pettingell, W.H.; Yu, C.E.; Jondro, P.D.; Schmidt, S.D.; Wang, K.; et al. Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science 1995, 269, 973–977. [Google Scholar] [CrossRef]
- Finckh, U.; Müller-Thomsen, T.; Mann, U.; Eggers, C.; Marksteiner, J.; Meins, W.; Binetti, G.; Alberici, A.; Hock, C.; Nitsch, R.M.; et al. High prevalence of pathogenic mutations in patients with early-onset dementia detected by sequence analyses of four different genes. Am. J. Hum. Genet. 2000, 66, 110–117. [Google Scholar] [CrossRef]
- Van Cauwenberghe, C.; Van Broeckhoven, C.; Sleegers, K. The genetic landscape of Alzheimer disease: Clinical implications and perspectives. Genet. Med. 2016, 18, 421–430. [Google Scholar] [CrossRef]
- Pericak-Vance, A.M.; Bebout, J.L.; Gaskell, P.C.; Yamaoka, L.H.; Hung, W.Y.; Alberts, M.J.; Walker, A.P.; Bartlett, R.J.; A Haynes, C.; A Welsh, K. Linkage studies in familial Alzheimer disease: Evidence for chromosome 19 linkage. Am. J. Hum. Genet. 1991, 48, 1034–1050. [Google Scholar]
- Namba, Y.; Tomonaga, M.; Kawasaki, H.; Otomo, E.; Ikeda, K. Apolipoprotein E immunoreactivity in cerebral amyloid deposits and neurofibrillary tangles in Alzheimer’s disease and kuru plaque amyloid in Creutzfeldt-Jakob disease. Brain Res. 1991, 541, 163–166. [Google Scholar] [CrossRef] [PubMed]
- Strittmatter, W.J.; Saunders, A.M.; Schmechel, D.; Pericak-Vance, M.; Enghild, J.; Salvesen, G.S.; Roses, A.D. Apolipoprotein E: High-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. USA 1993, 90, 1977–1981. [Google Scholar] [CrossRef] [PubMed]
- Corder, E.H. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 1993, 261, 921–923. [Google Scholar] [CrossRef] [PubMed]
- Arboleda-Velasquez, J.F.; Lopera, F.; O’hare, M.; Delgado-Tirado, S.; Marino, C.; Chmielewska, N.; Saez-Torres, K.L.; Amarnani, D.; Schultz, A.P.; Sperling, R.A.; et al. Resistance to autosomal dominant Alzheimer’s disease in an APOE3 Christchurch homozygote: A case report. Nat. Med. 2019, 25, 1680–1683. [Google Scholar] [CrossRef]
- Scherzer, C.R.; Offe, K.; Gearing, M.; Rees, H.D.; Fang, G.; Heilman, C.J.; Schaller, C.; Bujo, H.; Levey, A.I.; Lah, J.J. Loss of apolipoprotein E receptor LR11 in Alzheimer disease. Arch. Neurol. 2004, 61, 1200–1205. [Google Scholar] [CrossRef]
- Rogaeva, E.; Meng, Y.; Lee, J.H.; Gu, Y.; Kawarai, T.; Zou, F.; Katayama, T.; Baldwin, C.T.; Cheng, R.; Hasegawa, H.; et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat. Genet. 2007, 39, 168–177. [Google Scholar] [CrossRef]
- Lee, J.H.; Barral, S.; Reitz, C. The neuronal sortilin-related receptor gene SORL1 and late-onset Alzheimer’s disease. Curr. Neurol. Neurosci. Rep. 2008, 8, 384–391. [Google Scholar] [CrossRef]
- Strang, K.H.; Golde, T.E.; Giasson, B.I. MAPT mutations, tauopathy, and mechanisms of neurodegeneration. Lab. Investig. 2019, 99, 912–928. [Google Scholar] [CrossRef]
- Roks, G.; Dermaut, B.; Heutink, P.; Julliams, A.; Backhovens, H.; Van de Broeck, M.; Serneels, S.; Hofman, A.; Van Broeckhoven, C.; van Duijn, C.M. Mutation screening of the tau gene in patients with early-onset Alzheimer’s disease. Neurosci. Lett. 1999, 277, 137–139. [Google Scholar] [CrossRef]
- Baker, M.; Litvan, I.; Houlden, H.; Adamson, J.; Dickson, D.; Perez-Tur, J.; Hardy, J.; Lynch, T.; Bigio, E.; Hutton, M. Association of an extended haplotype in the tau gene with progressive supranuclear palsy. Hum. Mol. Genet. 1999, 8, 711–715. [Google Scholar] [CrossRef]
- Caffrey, T.M.; Wade-Martins, R. Functional MAPT haplotypes: Bridging the gap between genotype and neuropathology. Neurobiol. Dis. 2007, 27, 1–10. [Google Scholar] [CrossRef]
- Kalinderi, K.; Fidani, L.; Bostantjopoulou, S. From 1997 to 2007: A decade journey through the H1 haplotype on 17q21 chromosome. Park. Relat. Disord. 2009, 15, 2–5. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Jia, J. The +347 C promoter allele up-regulates MAPT expression and is associated with Alzheimer’s disease among the Chinese Han. Neurosci. Lett. 2009, 450, 340–343. [Google Scholar] [CrossRef]
- Filipello, F.; Morini, R.; Corradini, I.; Zerbi, V.; Canzi, A.; Michalski, B.; Erreni, M.; Markicevic, M.; Starvaggi-Cucuzza, C.; Otero, K.; et al. The microglial innate immune receptor TREM2 is required for synapse elimination and normal brain connectivity. Immunity 2018, 48, 979–991.e8. [Google Scholar] [CrossRef]
- Colonna, M. TREMs in the immune system and beyond. Nat. Rev. Immunol. 2003, 3, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Chouery, E.; Delague, V.; Bergougnoux, A.; Koussa, S.; Serre, J.-L.; Mégarbané, A. Mutations in TREM2 lead to pure early-onset dementia without bone cysts. Hum. Mutat. 2008, 29, E194–E204. [Google Scholar] [CrossRef]
- Guerreiro, R.; Wojtas, A.; Bras, J.; Carrasquillo, M.; Rogaeva, E.; Majounie, E.; Cruchaga, C.; Sassi, C.; Kauwe, J.S.; Younkin, S.; et al. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 2013, 368, 117–127. [Google Scholar] [CrossRef]
- Carmona, S.; Zahs, K.; Wu, E.; Dakin, K.; Bras, J.; Guerreiro, R. The role of TREM2 in Alzheimer’s disease and other neurodegenerative disorders. Lancet Neurol. 2018, 17, 721–730. [Google Scholar] [CrossRef]
- Colonna, M.; Wang, Y. TREM2 variants: New keys to decipher Alzheimer disease pathogenesis. Nat. Rev. Neurosci. 2016, 17, 201–207. [Google Scholar] [CrossRef]
- Sakae, N.; Liu, C.-C.; Shinohara, M.; Frisch-Daiello, J.; Ma, L.; Yamazaki, Y.; Tachibana, M.; Younkin, L.; Kurti, A.; Carrasquillo, M.M.; et al. ABCA7 Deficiency Accelerates Amyloid-β Generation and Alzheimer’s Neuronal Pathology. J. Neurosci. 2016, 36, 3848–3859. [Google Scholar] [CrossRef]
- Aikawa, T.; Ren, Y.; Yamazaki, Y.; Tachibana, M.; Johnson, M.R.; Anderson, C.T.; Martens, Y.A.; Holm, M.-L.; Asmann, Y.W.; Saito, T.; et al. ABCA7 haplodeficiency disturbs microglial immune responses in the mouse brain. Proc. Natl. Acad. Sci. USA 2019, 116, 23790–23796. [Google Scholar] [CrossRef]
- Lyssenko, N.N.; Praticò, D. ABCA7 and the altered lipidostasis hypothesis of Alzheimer’s disease. Alzheimer’s Dement. 2021, 17, 164–174. [Google Scholar] [CrossRef]
- Stepler, K.E.; Gillyard, T.R.; Reed, C.B.; Avery, T.M.; Davis, J.S.; Robinson, R.A. ABCA7, a Genetic Risk Factor Associated with Alzheimer’s Disease Risk in African Americans. J. Alzheimer’s Dis. 2022, 86, 5–19. [Google Scholar] [CrossRef] [PubMed]
- Sinha, N.; Reagh, Z.M.; Tustison, N.J.; Berg, C.N.; Shaw, A.; Myers, C.E.; Hill, D.; Yassa, M.A.; Gluck, M.A. ABCA7 risk variant in healthy older African Americans is associated with a functionally isolated entorhinal cortex mediating deficient generalization of prior discrimination training. Hippocampus 2019, 29, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Bertram, L.; McQueen, M.B.; Mullin, K.; Blacker, D.; E Tanzi, R. Systematic meta-analyses of Alzheimer disease genetic association studies: The AlzGene database. Nat. Genet. 2007, 39, 17–23. [Google Scholar] [CrossRef]
- Bellenguez, C.; Küçükali, F.; Jansen, I.E.; Kleineidam, L.; Moreno-Grau, S.; Amin, N.; Naj, A.C.; Campos-Martin, R.; Grenier-Boley, B.; Andrade, V.; et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet. 2022, 54, 412–436. [Google Scholar] [CrossRef] [PubMed]
- Gustafsen, C.; Glerup, S.; Pallesen, L.T.; Olsen, D.; Andersen, O.M.; Nykjær, A.; Madsen, P.; Petersen, C.M. Sortilin and SorLA Display Distinct Roles in Processing and Trafficking of Amyloid Precursor Protein. J. Neurosci. 2013, 33, 64–71. [Google Scholar] [CrossRef]
- Brouwers, N.; Van Cauwenberghe, C.; Engelborghs, S.; Lambert, J.-C.; Bettens, K.; Le Bastard, N.; Pasquier, F.; Gil Montoya, A.; Peeters, K.; Mattheijssens, M.; et al. Alzheimer risk associated with a copy number variation in the complement receptor 1 increasing C3b/C4b binding sites. Mol. Psychiatry 2011, 17, 223. [Google Scholar] [CrossRef]
- Hartl, D.; Aesg; May, P.; Gu, W.; Mayhaus, M.; Pichler, S.; Spaniol, C.; Glaab, E.; Bobbili, D.R.; Antony, P.; et al. A rare loss-of-function variant of ADAM17 is associated with late-onset familial Alzheimer disease. Mol. Psychiatry 2020, 25, 629–639. [Google Scholar] [CrossRef]
- McQuade, A.; Blurton-Jones, M. Microglia in Alzheimer’s Disease: Exploring How Genetics and Phenotype Influence Risk. J. Mol. Biol. 2019, 431, 1805–1817. [Google Scholar] [CrossRef]
- Chang, C.-J.; Chang, M.-Y.; Lee, Y.-C.; Chen, K.-Y.; Hsu, T.-I.; Wu, Y.-H.; Chuang, J.-Y.; Kao, T.-J. Nck2 is essential for limb trajectory selection by spinal motor axons. Dev. Dyn. 2018, 247, 1043–1056. [Google Scholar] [CrossRef]
- Tesi, N.; van der Lee, S.; Hulsman, M.; van Schoor, N.M.; Huisman, M.; Pijnenburg, Y.; van der Flier, W.M.; Reinders, M.; Holstege, H. Cognitively Healthy Centenarians are genetically protected against Alzheimer’s disease specifically in immune and endo-lysosomal systems. medRxiv 2023. [Google Scholar] [CrossRef]
- Lambert, E.; Saha, O.; Landeira, B.S.; de Farias, A.R.M.; Hermant, X.; Carrier, A.; Pelletier, A.; Gadaut, J.; Davoine, L.; Dupont, C.; et al. The Alzheimer susceptibility gene BIN1 induces isoform-dependent neurotoxicity through early endosome defects. Acta Neuropathol. Commun. 2022, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Chou, V.; Pearse, R.V.; Aylward, A.J.; Ashour, N.; Taga, M.; Terzioglu, G.; Fujita, M.; Fancher, S.B.; Sigalov, A.; Benoit, C.R.; et al. INPP5D regulates inflammasome activation in human microglia. Nat. Commun. 2023, 14, 7552. [Google Scholar] [CrossRef] [PubMed]
- Toca, A.B.; Arroyo, A.M.; Sainz, A.R.; Martínez, J.A.; Zárraga, I.S.; Calle, I.A.; Portillo, J.B.; Picon, S.V.; Catalli, C.; Gómez-Beldarrain, M. Mutations in MME gene causing distal hereditary motor neuropathy and MCI-AD. J. Neurol. Sci. 2021, 429, 118990. [Google Scholar] [CrossRef]
- Wang, D.; El-Amouri, S.S.; Dai, M.; Kuan, C.-Y.; Hui, D.Y.; Brady, R.O.; Pan, D. Engineering a lysosomal enzyme with a derivative of receptor-binding domain of apoE enables delivery across the blood–brain barrier. Proc. Natl. Acad. Sci. USA 2013, 110, 2999–3004. [Google Scholar] [CrossRef]
- Lefort, R. Reversing Synapse Loss in Alzheimer’s Disease: Rho-Guanosine Triphosphatases and Insights from Other Brain Disorders. Neurotherapeutics 2015, 12, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Jorfi, M.; Maaser-Hecker, A.; Tanzi, R.E. The neuroimmune axis of Alzheimer’s disease. Genome Med. 2023, 15, 6. [Google Scholar] [CrossRef]
- Bano, D.; Ehninger, D.; Bagetta, G. Decoding metabolic signatures in Alzheimer’s disease: A mitochondrial perspective. Cell Death Discov. 2023, 9, 432. [Google Scholar] [CrossRef]
- Yan, H.; Liu, M.; Gao, Y.; Yuan, Y.; Liu, X.; Wang, Y.; Li, L.; Wang, Q.; Wang, Y.; Shi, C.; et al. Assessing the impact of novel risk loci on Alzheimer’s and Parkinson’s diseases in a Chinese Han cohort. Front. Neurol. 2024, 15, 1326692. [Google Scholar] [CrossRef]
- Nott, A.; Holtman, I.R. Genetic insights into immune mechanisms of Alzheimer’s and Parkinson’s disease. Front. Immunol. 2023, 14, 1168539. [Google Scholar] [CrossRef]
- Ferreira, A.; Royaux, I.; Liu, J.; Wang, Z.; Su, G.; Moechars, D.; Callewaert, N.; De Muynck, L. The 3-O sulfation of heparan sulfate proteoglycans contributes to the cellular internalization of tau aggregates. BMC Cell Biol. 2022, 23, 61. [Google Scholar] [CrossRef]
- Zhang, X.; Zou, M.; Wu, Y.; Jiang, D.; Wu, T.; Zhao, Y.; Wu, D.; Cui, J.; Li, G. Regulation of the Late Onset alzheimer’s Disease Associated HLA-DQA1/DRB1 Expression. Am. J. Alzheimer’s Dis. Other Dementiasr 2022, 37, 15333175221085066. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, H.; Wang, H.; Yang, K.; Luan, J.; Wang, S. TREM2: Potential therapeutic targeting of microglia for Alzheimer’s disease. Biomed. Pharmacother. 2023, 165, 115218. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.-N.; Li, J.-Q.; Tan, C.-C.; Wang, H.-F.; Tan, M.-S.; Cao, X.-P.; Yu, J.-T.; Tan, L.; Initiative, T.A.D.N. TREML2 Mutation Mediate Alzheimer’s Disease Risk by Altering Neuronal Degeneration. Front. Neurosci. 2019, 13, 455. [Google Scholar] [CrossRef] [PubMed]
- Tao, Q.-Q.; Chen, Y.-C.; Wu, Z.-Y. The role of CD2AP in the Pathogenesis of Alzheimer’s Disease. Aging Dis. 2019, 10, 901–907. [Google Scholar] [CrossRef] [PubMed]
- Stefani, F.; Zhang, L.; Taylor, S.; Donovan, J.; Rollinson, S.; Doyotte, A.; Brownhill, K.; Bennion, J.; Pickering-Brown, S.; Woodman, P. UBAP1 Is a Component of an Endosome-Specific ESCRT-I Complex that Is Essential for MVB Sorting. Curr. Biol. 2011, 21, 1245–1250. [Google Scholar] [CrossRef] [PubMed]
- Talwar, P.; Silla, Y.; Grover, S.; Gupta, M.; Agarwal, R.; Kushwaha, S.; Kukreti, R. Genomic convergence and network analysis approach to identify candidate genes in Alzheimer’s disease. BMC Genom. 2014, 15, 199. [Google Scholar] [CrossRef]
- Lee, J.Y.; Harney, D.J.; Teo, J.D.; Kwok, J.B.; Sutherland, G.T.; Larance, M.; Don, A.S. The major TMEM106B dementia risk allele affects TMEM106B protein levels, fibril formation, and myelin lipid homeostasis in the ageing human hippocampus. Mol. Neurodegener. 2023, 18, 63. [Google Scholar] [CrossRef]
- Andrews, S.J.; Renton, A.E.; Fulton-Howard, B.; Podlesny-Drabiniok, A.; Marcora, E.; Goate, A.M. The complex genetic architecture of Alzheimer’s disease: Novel insights and future directions. EBioMedicine 2023, 90, 104511. [Google Scholar] [CrossRef]
- Lang, S.; Pfeffer, S.; Lee, P.-H.; Cavalié, A.; Helms, V.; Förster, F.; Zimmermann, R. An Update on Sec61 Channel Functions, Mechanisms, and Related Diseases. Front. Physiol. 2017, 8, 887. [Google Scholar] [CrossRef]
- Gastwirt, R.F.; McAndrew, C.W.; Donoghue, D.J. Speedy/RINGO Regulation of CDKs in Cell Cycle, Checkpoint Activation and Apoptosis. Cell Cycle 2007, 6, 1188–1193. [Google Scholar] [CrossRef]
- Kim, Y.B.; Komor, A.C.; Levy, J.M.; Packer, M.S.; Zhao, K.T.; Liu, D.R. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 2017, 35, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Hook, V.; Yoon, M.; Mosier, C.; Ito, G.; Podvin, S.; Head, B.P.; Rissman, R.; O’Donoghue, A.J.; Hook, G. Cathepsin B in neurodegeneration of Alzheimer’s disease, traumatic brain injury, and related brain disorders. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2020, 1868, 140428. [Google Scholar] [CrossRef] [PubMed]
- Asanomi, Y.; Shigemizu, D.; Akiyama, S.; Miyashita, A.; Mitsumori, R.; Hara, N.; Ikeuchi, T.; Niida, S.; Ozaki, K. A functional variant of SHARPIN confers increased risk of late-onset Alzheimer’s disease. J. Hum. Genet. 2022, 67, 203–208. [Google Scholar] [CrossRef]
- De Farias, A.-R.M.; Saha, O.; Lambert, J.-C.; Costa, M.R. Role of the late-onset Alzheimer’s disease risk genes bin1 and ptk2b in the hyperexcitability of hiPSC-derived neurons. Alzheimer’s Dement. 2021, 17, e053632. [Google Scholar] [CrossRef]
- Nelson, A.R.; Sagare, A.P.; Zlokovic, B.V. Role of clusterin in the brain vascular clearance of amyloid-β. Proc. Natl. Acad. Sci. USA 2017, 114, 8681–8682. [Google Scholar] [CrossRef] [PubMed]
- Lewandowski, C.T.; Laham, M.S.; Thatcher, G.R. Remembering your A, B, C’s: Alzheimer’s disease and ABCA1. Acta Pharm. Sin. B 2022, 12, 995–1018. [Google Scholar] [CrossRef]
- Becic, A.; Leifeld, J.; Shaukat, J.; Hollmann, M. Tetraspanins as Potential Modulators of Glutamatergic Synaptic Function. Front. Mol. Neurosci. 2022, 14, 801882. [Google Scholar] [CrossRef]
- Xiao, B.; Li, J.; Zhou, M.; Li, X.; Huang, X.; Hang, J.; Sun, Z.; Li, L. Structure and function of B-cell linker and its role in the development of B cell-related diseases. Nan Fang Yi Ke Da Xue Xue Bao= J. South. Med. Univ. 2019, 39, 253–256. [Google Scholar] [CrossRef]
- Matveeva, N.; Kiselev, I.; Baulina, N.; Semina, E.; Kakotkin, V.; Agapov, M.; Kulakova, O.; Favorova, O. Shared genetic architecture of COVID-19 and Alzheimer’s disease. Front. Aging Neurosci. 2023, 15, 1287322. [Google Scholar] [CrossRef]
- Raj, T.; Li, Y.I.; Wong, G.; Humphrey, J.; Wang, M.; Ramdhani, S.; Wang, Y.-C.; Ng, B.; Gupta, I.; Haroutunian, V.; et al. Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer’s disease susceptibility. Nat. Genet. 2018, 50, 1584–1592. [Google Scholar] [CrossRef] [PubMed]
- Rustenhoven, J.; Smith, A.M.; Smyth, L.C.; Jansson, D.; Scotter, E.L.; Swanson, M.E.V.; Aalderink, M.; Coppieters, N.; Narayan, P.; Handley, R.; et al. PU.1 regulates Alzheimer’s disease-associated genes in primary human microglia. Mol. Neurodegener. 2018, 13, 44. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-Y.; Deng, Y.-S.; Dai, S.-K.; Mi, T.-W.; Li, R.-Y.; Liu, P.-P.; Liu, C.; He, B.-D.; He, X.-C.; Du, H.-Z.; et al. Loss of microglial EED impairs synapse density, learning, and memory. Mol. Psychiatry 2022, 27, 2999–3009. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Knupp, A.; Szabo, M.P.; Williams, C.A.; Kinoshita, C.; Hailey, D.W.; Wang, Y.; Andersen, O.M.; Young, J.E. The Alzheimer’s gene SORL1 is a regulator of endosomal traffic and recycling in human neurons. Cell. Mol. Life Sci. 2022, 79, 162. [Google Scholar] [CrossRef] [PubMed]
- Foster, W.J.; Taylor, H.B.C.; Padamsey, Z.; Jeans, A.F.; Galione, A.; Emptage, N.J. Hippocampal mGluR1-dependent long-term potentiation requires NAADP-mediated acidic store Ca 2+ signaling. Sci. Signal. 2018, 11, eaat9093. [Google Scholar] [CrossRef] [PubMed]
- Dalmasso, M.C.; de Rojas, I.; Olivar, N.; Muchnik, C.; Angel, B.; Gloger, S.; Abalos, M.S.S.; Chacón, M.V.; Aránguiz, R.; Orellana, P.; et al. The first genome-wide association study in the Argentinian and Chilean populations identifies shared genetics with Europeans in Alzheimer’s disease. Alzheimer’s Dement. Early View. [CrossRef]
- Eysert, F.; Coulon, A.; Boscher, E.; Vreulx, A.-C.; Flaig, A.; Mendes, T.; Hughes, S.; Grenier-Boley, B.; Hanoulle, X.; Demiautte, F.; et al. Alzheimer’s genetic risk factor FERMT2 (Kindlin-2) controls axonal growth and synaptic plasticity in an APP-dependent manner. Mol. Psychiatry 2021, 26, 5592–5607. [Google Scholar] [CrossRef]
- Tan, M.-S.; Initiative, A.D.N.; Yang, Y.-X.; Xu, W.; Wang, H.-F.; Tan, L.; Zuo, C.-T.; Dong, Q.; Tan, L.; Suckling, J.; et al. Associations of Alzheimer’s disease risk variants with gene expression, amyloidosis, tauopathy, and neurodegeneration. Alzheimer’s Res. Ther. 2021, 13, 15. [Google Scholar] [CrossRef]
- Park, Y.H.; Pyun, J.-M.; Hodges, A.; Jang, J.-W.; Bice, P.J.; Kim, S.; Saykin, A.J.; Kwangsik Nho for the AddNeuroMed consortium and the Alzheimer’s Disease Neuroimaging Initiative. Dysregulated expression levels of APH1B in peripheral blood are associated with brain atrophy and amyloid-β deposition in Alzheimer’s disease. Alzheimer’s Res. Ther. 2021, 13, 183. [Google Scholar] [CrossRef]
- Kim, N.-Y.; Cho, M.-H.; Won, S.-H.; Kang, H.-J.; Yoon, S.-Y.; Kim, D.-H. Sorting nexin-4 regulates β-amyloid production by modulating β-site-activating cleavage enzyme-1. Alzheimer’s Res. Ther. 2017, 9, 4. [Google Scholar] [CrossRef]
- Li, Y.; Xu, M.; Xiang, B.-L.; Li, X.; Zhang, D.-F.; Zhao, H.; Bi, R.; Yao, Y.-G. Functional genomics identify causal variant underlying the protective CTSH locus for Alzheimer’s disease. Neuropsychopharmacology 2023, 48, 1555–1566. [Google Scholar] [CrossRef]
- Yoo, H.-S.; Shanmugalingam, U.; Smith, P.D. Potential roles of branched-chain amino acids in neurodegeneration. Nutrition 2022, 103-104, 111762. [Google Scholar] [CrossRef]
- Ho, A.; Ngala, B.; Yamada, C.; Garcia, C.; Duarte, C.; Akkaoui, J.; Ciolac, D.; Nusbaum, A.; Kochen, W.; Efremova, D.; et al. IL-34 exacerbates pathogenic features of Alzheimer’s disease and calvaria osteolysis in triple transgenic (3x-Tg) female mice. Biomed. Pharmacother. 2023, 166, 115435. [Google Scholar] [CrossRef]
- Li, K.; Ran, B.; Wang, Y.; Liu, L.; Li, W. PLCγ2 impacts microglia-related effectors revealing variants and pathways important in Alzheimer’s disease. Front. Cell Dev. Biol. 2022, 10, 999061. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.-W.; Qin, J.; Chen, Y.-F.; Tu, Y.; Xing, Y.-Y.; Wang, Y.; Yang, L.-Y.; Lu, S.-Y.; Geng, L.; Shi, W.; et al. 16p11.2 CNV gene Doc2α functions in neurodevelopment and social behaviors through interaction with Secretagogin. Cell Rep. 2023, 42, 112691. [Google Scholar] [CrossRef] [PubMed]
- Dugan, A.J.; Nelson, P.T.; Katsumata, Y.; Shade, L.M.; Teylan, M.A.; Boehme, K.L.; Mukherjee, S.; Kauwe, J.S.; Hohman, T.J.; Schneider, J.A.; et al. Association between WWOX/MAF variants and dementia-related neuropathologic endophenotypes. Neurobiol. Aging 2022, 111, 95–106. [Google Scholar] [CrossRef]
- Oli, V.; Gupta, R.; Kumar, P. FOXO and related transcription factors binding elements in the regulation of neurodegenerative disorders. J. Chem. Neuroanat. 2021, 116, 102012. [Google Scholar] [CrossRef]
- Rasmi, Y.; Shokati, A.; Hassan, A.; Aziz, S.G.-G.; Bastani, S.; Jalali, L.; Moradi, F.; Alipour, S. The role of DNA methylation in progression of neurological disorders and neurodegenerative diseases as well as the prospect of using DNA methylation inhibitors as therapeutic agents for such disorders. IBRO Neurosci. Rep. 2023, 14, 28–37. [Google Scholar] [CrossRef]
- Liu, X.; Yin, L.; Li, T.; Lin, L.; Zhang, J.; Li, Y. Reduction of WDR81 impairs autophagic clearance of aggregated proteins and cell viability in neurodegenerative phenotypes. PLoS Genet. 2021, 17, e1009415. [Google Scholar] [CrossRef]
- Xicota, L.; Lagarde, J.; Eysert, F.; Grenier-Boley, B.; Rivals, I.; Botté, A.; Forlani, S.; Landron, S.; Gautier, C.; Gabriel, C.; et al. Modifications of the endosomal compartment in fibroblasts from sporadic Alzheimer’s disease patients are associated with cognitive impairment. Transl. Psychiatry 2023, 13, 54. [Google Scholar] [CrossRef]
- Lona-Durazo, F.; Reynolds, R.H.; Scholz, S.W.; Ryten, M.; Taliun, S.A.G. Regional genetic correlations highlight relationships between neurodegenerative disease loci and the immune system. Commun. Biol. 2023, 6, 729. [Google Scholar] [CrossRef]
- Aghaizu, N.D.; Jin, H.; Whiting, P.J. Dysregulated Wnt Signalling in the Alzheimer’s Brain. Brain Sci. 2020, 10, 902. [Google Scholar] [CrossRef]
- Cao, M.; Liu, J.; Zhang, X.; Yang, T.; Wang, Y.; Hou, Y.; Song, Q.; Cui, Y.; Wang, Y.; Wang, P. ABI3 Is a Novel Early Biomarker of Alzheimer’s Disease. J. Alzheimer’s Dis. 2022, 87, 335–344. [Google Scholar] [CrossRef]
- Wainberg, M.; Andrews, S.J.; Tripathy, S.J. Shared genetic risk loci between Alzheimer’s disease and related dementias, Parkinson’s disease, and amyotrophic lateral sclerosis. Alzheimer’s Res. Ther. 2023, 15, 113. [Google Scholar] [CrossRef]
- Miners, S.; Ashby, E.; Baig, S.; Harrison, R.; Tayler, H.; Speedy, E.; Prince, J.A.; Love, S.; Kehoe, P.G. Angiotensin-converting enzyme levels and activity in Alzheimer’s disease: Differences in brain and CSF ACE and association with ACE1 genotypes. Am. J. Transl. Res. 2009, 1, 163–177. [Google Scholar]
- Breen, C.; Papale, L.A.; Clark, L.R.; Bergmann, P.E.; Madrid, A.; Asthana, S.; Johnson, S.C.; Keleş, S.; Alisch, R.S.; Hogan, K.J. Whole genome methylation sequencing in blood identifies extensive differential DNA methylation in late-onset dementia due to Alzheimer’s disease. Alzheimer’s Dement. Early View. [CrossRef]
- Siew, J.J.; Chern, Y.; Khoo, K.-H.; Angata, T. Roles of Siglecs in neurodegenerative diseases. Mol. Asp. Med. 2023, 90, 101141. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Vidal, G.S.; Djurisic, M.; William, C.M.; Birnbaum, M.E.; Garcia, K.C.; Hyman, B.T.; Shatz, C.J. Human LilrB2 Is a β-Amyloid Receptor and Its Murine Homolog PirB Regulates Synaptic Plasticity in an Alzheimer’s Model. Science 2013, 341, 1399–1404. [Google Scholar] [CrossRef]
- Zhao, Y.; Yun, D.; Zou, X.; Jiang, T.; Li, G.; Hu, L.; Chen, J.; Xu, J.; Mao, Y.; Chen, H.; et al. Whole exome-wide association study identifies a missense variant in SLC2A4RG associated with glioblastoma risk. Am. J. Cancer Res. 2017, 7, 1937–1947. [Google Scholar]
- Giri, M.; Zhang, M.; Lü, Y. Genes associated with Alzheimer’s disease: An overview and current status. Clin. Interv. Aging 2016, 11, 665–681. [Google Scholar] [CrossRef]
- Hampel, H.; Hardy, J.; Blennow, K.; Chen, C.; Perry, G.; Kim, S.H.; Villemagne, V.L.; Aisen, P.; Vendruscolo, M.; Iwatsubo, T.; et al. The Amyloid-beta Pathway in Alzheimer’s Disease. Mol. Psychiatry 2021, 26, 5481–5503. [Google Scholar] [CrossRef]
- Qiu, Y.; Sha, L.; Zhang, X.; Li, G.; Zhu, W.; Xu, Q. Induction of A Disintegrin and Metalloproteinase with Thrombospondin motifs 1 by a rare variant or cognitive activities reduces hippocampal amyloid-β and consequent Alzheimer’s disease risk. Front. Aging Neurosci. 2022, 14, 896522. [Google Scholar] [CrossRef]
- Jun, G.; Naj, A.C.; Beecham, G.W.; Wang, L.S.; Buros, J.; Gallins, P.J.; Buxbaum, J.D.; Ertekin-Taner, N.; Fallin, M.D.; Friedland, R.; et al. Meta-analysis confirms CR1, CLU, and PICALM as Alzheimer disease risk loci and reveals interactions with APOE genotypes. Arch. Neurol. 2010, 67, 1473–1484. [Google Scholar] [CrossRef]
- Kunkle, B.W.; Grenier-Boley, B.; Sims, R.; Bis, J.C.; Damotte, V.; Naj, A.C.; Boland, A.; Vronskaya, M.; Van Der Lee, S.J.; Amlie-Wolf, A.; et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019, 51, 414–430. [Google Scholar] [CrossRef]
- Marioni, R.E.; Harris, S.E.; Zhang, Q.; McRae, A.F.; Hagenaars, S.P.; Hill, W.D.; Davies, G.; Ritchie, C.W.; Gale, C.R.; Starr, J.M.; et al. GWAS on family history of Alzheimer’s disease. Transl. Psychiatry 2018, 8, 99. [Google Scholar] [CrossRef]
- Parkinson, J. An essay on the shaking palsy. J. Neuropsychiatry Clin. Neurosci. 2002, 14, 223–236. [Google Scholar] [CrossRef]
- Poewe, W.; Seppi, K.; Tanner, C.M.; Halliday, G.M.; Brundin, P.; Volkmann, J.; Schrag, A.E.; Lang, A.E. Parkinson disease. Nat. Rev. Dis. Primers 2017, 3, 17013. [Google Scholar] [CrossRef]
- Marras, C.; Beck, J.C.; Bower, J.H.; Roberts, E.; Ritz, B.; Ross, G.W.; Abbott, R.D.; Savica, R.; Van Den Eeden, S.K.; Willis, A.W.; et al. Prevalence of Parkinson’s disease across North America. NPJ Parkinson’s Dis. 2018, 4, 21. [Google Scholar] [CrossRef]
- Aguirre-Mardones, C.; Iranzo, A.; Vilas, D.; Serradell, M.; Gaig, C.; Santamaría, J.; Tolosa, E. Prevalence and timeline of nonmotor symptoms in idiopathic rapid eye movement sleep behavior disorder. J. Neurol. 2015, 262, 1568–1578. [Google Scholar] [CrossRef]
- Twelves, D.; Perkins, K.S.; Counsell, C. Systematic review of incidence studies of Parkinson’s disease. Mov. Disord. 2003, 18, 19–31. [Google Scholar] [CrossRef]
- Dorsey, E.R.; Elbaz, A.; Nichols, E.; Abbasi, N.; Abd-Allah, F.; Abdelalim, A.; Adsuar, J.C.; Ansha, M.G.; Brayne, C.; Choi, J.Y.J.; et al. Global, regional, and national burden of Parkinson’s disease, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2018, 17, 939–953. [Google Scholar] [CrossRef]
- Simon, D.K.; Tanner, C.M.; Brundin, P. Parkinson disease epidemiology, pathology, genetics, and pathophysiology. Clin. Geriatr. Med. 2020, 36, 1–12. [Google Scholar] [CrossRef]
- Burré, J.; Sharma, M.; Südhof, T.C. Cell biology and pathophysiology of α-synuclein. Cold Spring Harb. Perspect. Med. 2018, 8, a024091. [Google Scholar] [CrossRef] [PubMed]
- Beyer, K.; Munoz-Marmol, A.M.; Sanz, C.; Marginet-Flinch, R.; Ferrer, I.; Ariza, A. New brain-specific beta-synuclein isoforms show expression ratio changes in Lewy body diseases. Neurogenetics 2012, 13, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Polymeropoulos, M.H.; Lavedan, C.; Leroy, E.; Ide, S.E.; Dehejia, A.; Dutra, A.; Pike, B.; Root, H.; Rubenstein, J.; Boyer, R. Mutation in the α-synuclein gene identified in families with Parkinson’s disease. Science 1997, 276, 2045–2047. [Google Scholar] [CrossRef]
- Spillantini, M.G.; Crowther, R.A.; Jakes, R.; Hasegawa, M.; Goedert, M. α-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with Lewy bodies. Proc. Natl. Acad. Sci. USA 1998, 95, 6469–6473. [Google Scholar] [CrossRef]
- Hernandez, D.G.; Reed, X.; Singleton, A.B. Genetics in Parkinson disease: Mendelian versus non-Mendelian inheritance. J. Neurochem. 2016, 139, 59–74. [Google Scholar] [CrossRef]
- Vázquez-Vélez, G.E.; Zoghbi, H.Y. Parkinson’s disease genetics and pathophysiology. Annu. Rev. Neurosci. 2021, 44, 87–108. [Google Scholar] [CrossRef]
- Singleton, A.B.; Farrer, M.; Johnson, J.; Singleton, A.; Hague, S.; Kachergus, J.; Hulihan, M.; Peuralinna, T.; Dutra, A.; Nussbaum, R.; et al. α-Synuclein locus triplication causes Parkinson’s disease. Science 2003, 302, 841. [Google Scholar] [CrossRef]
- Konno, T.; Ross, O.A.; Puschmann, A.; Dickson, D.W.; Wszolek, Z.K. Autosomal dominant Parkinson’s disease caused by SNCA duplications. Park. Relat. Disord. 2016, 22, S1–S6. [Google Scholar] [CrossRef]
- Trinh, J.; Guella, I.; Farrer, M.J. Disease penetrance of late-onset parkinsonism: A meta-analysis. JAMA Neurol. 2014, 71, 1535–1539. [Google Scholar] [CrossRef]
- Soldner, F.; Stelzer, Y.; Shivalila, C.S.; Abraham, B.J.; Latourelle, J.C.; Barrasa, M.I.; Goldmann, J.; Myers, R.H.; Young, R.A.; Jaenisch, R. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature 2016, 533, 95–99. [Google Scholar] [CrossRef]
- Funayama, M.; Hasegawa, K.; Kowa, H.; Saito, M.; Tsuji, S.; Obata, F. A new locus for Parkinson’s disease (PARK8) maps to chromosome 12p11. 2–q13. 1. Ann. Neurol. Off. J. Am. Neurol. Assoc. Child Neurol. Soc. 2002, 51, 296–301. [Google Scholar]
- Araki, M.; Ito, G.; Tomita, T. Physiological and pathological functions of LRRK2: Implications from substrate proteins. Neuronal Signal. 2018, 2, NS20180005. [Google Scholar] [CrossRef]
- Thaler, A.; Ash, E.; Gan-Or, Z.; Orr-Urtreger, A.; Giladi, N. The LRRK2 G2019S mutation as the cause of Parkinson’s disease in Ashkenazi Jews. J. Neural Transm. 2009, 116, 1473–1482. [Google Scholar] [CrossRef]
- Healy, D.G.; Falchi, M.; O’Sullivan, S.S.; Bonifati, V.; Durr, A.; Bressman, S.; Brice, A.; Aasly, J.; Zabetian, C.P.; Goldwurm, S.; et al. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson’s disease: A case-control study. Lancet Neurol. 2008, 7, 583–590. [Google Scholar] [CrossRef]
- Di Maio, R.; Hoffman, E.K.; Rocha, E.M.; Keeney, M.T.; Sanders, L.H.; De Miranda, B.R.; Zharikov, A.; Van Laar, A.; Stepan, A.F.; Lanz, T.A.; et al. LRRK2 activation in idiopathic Parkinson’s disease. Sci. Transl. Med. 2018, 10, eaar5429. [Google Scholar] [CrossRef]
- Herzig, M.C.; Kolly, C.; Persohn, E.; Theil, D.; Schweizer, T.; Hafner, T.; Stemmelen, C.; Troxler, T.J.; Schmid, P.; Danner, S.; et al. LRRK2 protein levels are determined by kinase function and are crucial for kidney and lung homeostasis in mice. Hum. Mol. Genet. 2011, 20, 4209–4223. [Google Scholar] [CrossRef]
- Vilas, D.; Gelpi, E.; Aldecoa, I.; Grau, O.; Rodriguez-Diehl, R.; Jaumà, S.; Martí, M.J.; Tolosa, E. Lack of central and peripheral nervous system synuclein pathology in R1441G LRRK2-associated Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry 2019, 90, 832–833. [Google Scholar] [CrossRef] [PubMed]
- Williams, E.T.; Chen, X.; Moore, D.J. VPS35, the retromer complex and Parkinson’s disease. J. Park. Dis. 2017, 7, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Zimprich, A.; Benet-Pagès, A.; Struhal, W.; Graf, E.; Eck, S.H.; Offman, M.N.; Haubenberger, D.; Spielberger, S.; Schulte, E.C.; Lichtner, P.; et al. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am. J. Hum. Genet. 2011, 89, 168–175. [Google Scholar] [CrossRef]
- Sidransky, E.; Lopez, G. The link between the GBA gene and parkinsonism. Lancet Neurol. 2012, 11, 986–998. [Google Scholar] [CrossRef]
- Sidransky, E.; Samaddar, T.; Tayebi, N. Mutations in GBA are associated with familial Parkinson disease susceptibility and age at onset. Neurology 2009, 73, 1424–1426. [Google Scholar] [CrossRef]
- Duran, R.; Mencacci, N.E.; Angeli, A.V.; Shoai, M.; Deas, E.; Houlden, H.; Mehta, A.; Hughes, D.; Cox, T.M.; Deegan, P.; et al. The glucocerobrosidase E326K variant predisposes to Parkinson’s disease, but does not cause Gaucher’s disease. Mov. Disord. 2013, 28, 232–236. [Google Scholar] [CrossRef]
- Mallett, V.; Ross, J.P.; Alcalay, R.N.; Ambalavanan, A.; Sidransky, E.; Dion, P.A.; Rouleau, G.A.; Gan-Or, Z. GBA p.T369M substitution in Parkinson disease: Polymorphism or association? A meta-analysis. Neurol. Genet. 2016, 2, e104. [Google Scholar] [CrossRef]
- Riboldi, G.M.; Di Fonzo, A.B. GBA, Gaucher disease, and Parkinson’s disease: From genetic to clinic to new therapeutic approaches. Cells 2019, 8, 364. [Google Scholar] [CrossRef]
- Simon-Sanchez, J.; Schulte, C.; Bras, J.M.; Sharma, M.; Gibbs, J.R.; Berg, D.; Paisan-Ruiz, C.; Lichtner, P.; Scholz, S.W.; Hernandez, D.G.; et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat. Genet. 2009, 41, 1308–1312. [Google Scholar] [CrossRef] [PubMed]
- Nalls, M.A.; Blauwendraat, C.; Vallerga, C.L.; Heilbron, K.; Bandres-Ciga, S.; Chang, D.; Tan, M.; Kia, D.A.; Noyce, A.J.; Xue, A. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 2019, 18, 1091–1102. [Google Scholar] [CrossRef] [PubMed]
- Buenaventura, R.G.; Harvey, A.C.; Burns, M.P.; Main, B.S. Traumatic brain injury induces an adaptive immune response in the meningeal transcriptome that is amplified by aging. Front. Neurosci. 2023, 17, 1210175. [Google Scholar] [CrossRef]
- Iwaki, H.; Blauwendraat, C.; Leonard, H.L.; Liu, G.; Maple-Grødem, J.; Corvol, J.-C.; Pihlstrøm, L.; van Nimwegen, M.; Hutten, S.J.; Nguyen, K.-D.H.; et al. Genetic risk of Parkinson disease and progression: An analysis of 13 longitudinal cohorts. Neurol. Genet. 2019, 5, e348. [Google Scholar] [CrossRef]
- Straniero, L.; Rimoldi, V.; Samarani, M.; Goldwurm, S.; Di Fonzo, A.; Krüger, R.; Deleidi, M.; Aureli, M.; Soldà, G.; Duga, S.; et al. The GBAP1 pseudogene acts as a ceRNA for the glucocerebrosidase gene GBA by sponging miR-22-3p. Sci. Rep. 2017, 7, 12702. [Google Scholar] [CrossRef]
- Kaiser, S.; Zhang, L.; Mollenhauer, B.; Jacob, J.; Longerich, S.; Del-Aguila, J.; Marcus, J.; Raghavan, N.; Stone, D.; Fagboyegun, O.; et al. A proteogenomic view of Parkinson’s disease causality and heterogeneity. npj Park. Dis. 2023, 9, 24. [Google Scholar] [CrossRef]
- Ivanova, D.; Dobson, K.L.; Gajbhiye, A.; Davenport, E.C.; Hacker, D.; Ultanir, S.K.; Trost, M.; Cousin, M.A. Control of synaptic vesicle release probability via VAMP4 targeting to endolysosomes. Sci. Adv. 2021, 7, eabf3873. [Google Scholar] [CrossRef]
- Singh, S.; Park, S.-G.J.B. Functional association between NUCKS1 gene and Parkinson disease: A potential susceptibility biomarker. Bioinformation 2019, 15, 548–556. [Google Scholar] [CrossRef]
- Purlyte, E.; Dhekne, H.S.; Sarhan, A.R.; Gomez, R.; Lis, P.; Wightman, M.; Martinez, T.N.; Tonelli, F.; Pfeffer, S.R.; Alessi, D.R. Rab29 activation of the Parkinson’s disease-associated LRRK2 kinase. EMBO J. 2017, 37, 1–18. [Google Scholar] [CrossRef]
- Apicco, D.J.; Shlevkov, E.; Nezich, C.L.; Tran, D.T.; Guilmette, E.; Nicholatos, J.W.; Bantle, C.M.; Chen, Y.; Glajch, K.E.; Abraham, N.A.; et al. The Parkinson’s disease-associated gene ITPKB protects against α-synuclein aggregation by regulating ER-to-mitochondria calcium release. Proc. Natl. Acad. Sci. USA 2020, 118, e2006476118. [Google Scholar] [CrossRef]
- Andres-Alonso, M.; Ammar, M.R.; Butnaru, I.; Gomes, G.M.; Sanhueza, G.A.; Raman, R.; Yuanxiang, P.; Borgmeyer, M.; Lopez-Rojas, J.; Raza, S.A.; et al. SIPA1L2 controls trafficking and local signaling of TrkB-containing amphisomes at presynaptic terminals. Nat. Commun. 2019, 10, 5448. [Google Scholar] [CrossRef] [PubMed]
- Pike, A.F.; Szabò, I.; Veerhuis, R.; Bubacco, L. The potential convergence of NLRP3 inflammasome, potassium, and dopamine mechanisms in Parkinson’s disease. Npj Park. Dis. 2022, 8, 32. [Google Scholar] [CrossRef] [PubMed]
- Yu, E.; Larivière, R.; Thomas, R.A.; Liu, L.; Senkevich, K.; Rahayel, S.; Trempe, J.-F.; Fon, E.A.; Gan-Or, Z. Identification of novel variants, genes and pathways potentially linked to Parkinson’s disease using machine learning. medRxiv 2023. [Google Scholar] [CrossRef]
- Cappelletti, C.; Henriksen, S.P.; Geut, H.; Rozemuller, A.J.M.; van de Berg, W.D.J.; Pihlstrøm, L.; Toft, M. Transcriptomic profiling of Parkinson’s disease brains reveals disease stage specific gene expression changes. Acta Neuropathol. 2023, 146, 227–244. [Google Scholar] [CrossRef] [PubMed]
- Styrpejko, D.J.; Cuajungco, M.P. Transmembrane 163 (TMEM163) Protein: A New Member of the Zinc Efflux Transporter Family. Biomedicines 2021, 9, 220. [Google Scholar] [CrossRef]
- Chang, K.-H.; Wu, Y.-R.; Chen, Y.-C.; Fung, H.-C.; Lee-Chen, G.-J.; Chen, C.-M. STK39, But Not BST1, HLA-DQB1, and SPPL2B Polymorphism, Is Associated With Han-Chinese Parkinson’s Disease in Taiwan. Medicine 2015, 94, e1690. [Google Scholar] [CrossRef]
- Riessland, M.; Kolisnyk, B.; Kim, T.W.; Cheng, J.; Ni, J.; Pearson, J.A.; Park, E.J.; Dam, K.; Acehan, D.; Ramos-Espiritu, L.S.; et al. Loss of SATB1 Induces p21-Dependent Cellular Senescence in Post-mitotic Dopaminergic Neurons. Cell Stem Cell 2019, 25, 514–530.e8. [Google Scholar] [CrossRef] [PubMed]
- Parikshak, N.N.; Swarup, V.; Belgard, T.G.; Irimia, M.; Ramaswami, G.; Gandal, M.J.; Hartl, C.; Leppa, V.; Ubieta, L.d.l.T.; Huang, J.; et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 2016, 540, 423–427. [Google Scholar] [CrossRef]
- Nagpal, L.; Kornberg, M.D.; Snyder, S.H. Inositol hexakisphosphate kinase-2 non-catalytically regulates mitophagy by attenuating PINK1 signaling. Proc. Natl. Acad. Sci. USA 2022, 119, e2121946119. [Google Scholar] [CrossRef] [PubMed]
- Markopoulou, K.; Chase, B.A.; Premkumar, A.P.; Schoneburg, B.; Kartha, N.; Wei, J.; Yu, H.; Epshteyn, A.; Garduno, L.; Pham, A.; et al. Variable Effects of PD-Risk Associated SNPs and Variants in Parkinsonism-Associated Genes on Disease Phenotype in a Community-Based Cohort. Front. Neurol. 2021, 12, 662278. [Google Scholar] [CrossRef] [PubMed]
- Nizon, M.; Laugel, V.; Flanigan, K.M.; Pastore, M.; Waldrop, M.A.; Rosenfeld, J.A.; Marom, R.; Xiao, R.; Gerard, A.; Pichon, O.; et al. Variants in MED12L, encoding a subunit of the mediator kinase module, are responsible for intellectual disability associated with transcriptional defect. Anesthesia Analg. 2019, 21, 2713–2722. [Google Scholar] [CrossRef]
- Custodia, A.; Aramburu-Núñez, M.; Correa-Paz, C.; Posado-Fernández, A.; Gómez-Larrauri, A.; Castillo, J.; Gómez-Muñoz, A.; Sobrino, T.; Ouro, A. Ceramide Metabolism and Parkinson’s Disease—Therapeutic Targets. Biomolecules 2021, 11, 945. [Google Scholar] [CrossRef]
- Zhao, A.; Li, Y.; Niu, M.; Li, G.; Luo, N.; Zhou, L.; Kang, W.; Liu, J. SNPs in SNCA, MCCC1, DLG2, GBF1 and MBNL2 are associated with Parkinson’s disease in southern Chinese population. J. Cell. Mol. Med. 2020, 24, 8744–8752. [Google Scholar] [CrossRef]
- Zhang, J.; Zeng, H.; Zhu, L.; Deng, L.; Fang, X.; Deng, X.; Liang, H.; Tang, C.; Cao, X.; Lu, Y.; et al. The Potential Mutation of GAK Gene in the Typical Sporadic Parkinson’s Disease from the Han Population of Chinese Mainland. Mol. Neurobiol. 2016, 53, 7119–7136. [Google Scholar] [CrossRef]
- Hu, M.; Li, P.; Wang, C.; Feng, X.; Geng, Q.; Chen, W.; Marthi, M.; Zhang, W.; Gao, C.; Reid, W.; et al. Parkinson’s disease-risk protein TMEM175 is a proton-activated proton channel in lysosomes. Cell 2022, 185, 2292–2308.e20. [Google Scholar] [CrossRef]
- Mufti, K.; Yu, E.; Rudakou, U.; Krohn, L.; Ruskey, J.A.; Asayesh, F.; Laurent, S.B.; Spiegelman, D.; Arnulf, I.; Hu, M.T.; et al. Novel Associations of BST1 and LAMP3 With REM Sleep Behavior Disorder. Neurology 2021, 96, e1412. [Google Scholar] [CrossRef]
- Alcalay, R.N.; Levy, O.A.; Wolf, P.; Oliva, P.; Zhang, X.K.; Waters, C.H.; Fahn, S.; Kang, U.J.; Liong, C.; Ford, B.; et al. SCARB2 variants and glucocerebrosidase activity in Parkinson’s disease. Npj Park. Dis. 2016, 2, 16004. [Google Scholar] [CrossRef]
- Siddiqui, I.J.; Pervaiz, N.; Abbasi, A.A. The Parkinson Disease gene SNCA: Evolutionary and structural insights with pathological implication. Sci. Rep. 2016, 6, 24475. [Google Scholar] [CrossRef]
- Di Maio, R.; General, I.J.; Furbee, E.; Ayoob, J.C.; Castro, S.L.; Bahar, I.; Greenamyre, J.T.; Pullara, F. Disulfide bridge formation prevents CaMKII/Calmodulin interaction in Parkinson’s disease. bioRxiv 2020. [Google Scholar] [CrossRef]
- Fanning, S.; Cirka, H.; Thies, J.L.; Jeong, J.; Niemi, S.M.; Yoon, J.; Ho, G.P.H.; Pacheco, J.A.; Dettmer, U.; Liu, L.; et al. Lipase regulation of cellular fatty acid homeostasis as a Parkinson’s disease therapeutic strategy. npj Park. Dis. 2022, 8, 74. [Google Scholar] [CrossRef]
- Charvin, D. mGlu4 allosteric modulation for treating Parkinson’s disease. Neuropharmacology 2018, 135, 308–315. [Google Scholar] [CrossRef]
- Fan, L.; Shi, C.; Hu, X.; Zhang, Z.; Zheng, H.; Luo, H.; Fan, Y.; Zhang, S.; Hu, Z.; Yang, J.; et al. Analysis of 12 GWAS-Linked Loci With Parkinson’s Disease in the Chinese Han Population. Front. Neurol. 2021, 12, 623913. [Google Scholar] [CrossRef]
- Zhao, C.; Jia, M.; Song, H.; Yu, Z.; Wang, W.; Li, Q.; Zhang, L.; Zhao, W.; Cao, X. The E3 Ubiquitin Ligase TRIM40 Attenuates Antiviral Immune Responses by Targeting MDA5 and RIG-I. Cell Rep. 2017, 21, 1613–1623. [Google Scholar] [CrossRef]
- Su, W.-M.; Gu, X.-J.; Hou, Y.-B.; Zhang, L.-Y.; Cao, B.; Ou, R.-W.; Wu, Y.; Chen, X.-P.; Song, W.; Zhao, B.; et al. Association Analysis of WNT3, HLA-DRB5 and IL1R2 Polymorphisms in Chinese Patients With Parkinson’s Disease and Multiple System Atrophy. Front. Genet. 2021, 12, 765833. [Google Scholar] [CrossRef]
- Angelopoulou, E.; Paudel, Y.N.; Julian, T.; Shaikh, M.F.; Piperi, C. Pivotal Role of Fyn Kinase in Parkinson’s Disease and Levodopa-Induced Dyskinesia: A Novel Therapeutic Target? Mol. Neurobiol. 2021, 58, 1372–1391. [Google Scholar] [CrossRef]
- Chen, D.; Zhang, R.; Shen, W.; Fu, H.; Liu, S.; Sun, K.; Sun, X. RPS12-specific shRNA inhibits the proliferation, migration of BGC823 gastric cancer cells with S100A4 as a downstream effector. Int. J. Oncol. 2013, 42, 1763–1769. [Google Scholar] [CrossRef]
- Diaz-Ortiz, M.E.; Seo, Y.; Posavi, M.; Cordon, M.C.; Clark, E.; Jain, N.; Charan, R.; Gallagher, M.D.; Unger, T.L.; Amari, N.; et al. GPNMB confers risk for Parkinson’s disease through interaction with α-synuclein. Science 2022, 377, eabk0637. [Google Scholar] [CrossRef]
- Jones-Tabah, J.; He, K.; Senkevich, K.; Karpilovsky, N.; Deyab, G.; Cousineau, Y.; Nikanorova, D.; Goldsmith, T.; del Cid Pellitero, E.; Chen, C.X.-Q.; et al. The Parkinson’s disease risk gene cathepsin B promotes fibrillar alpha-synuclein clearance, lysosomal function and glucocerebrosidase activity in dopaminergic neurons. bioRxiv 2023. [Google Scholar] [CrossRef]
- Fletcher, E.J.R.; Jamieson, A.D.; Williams, G.; Doherty, P.; Duty, S. Targeted repositioning identifies drugs that increase fibroblast growth factor 20 production and protect against 6-hydroxydopamine-induced nigral cell loss in rats. Sci. Rep. 2019, 9, 8336. [Google Scholar] [CrossRef]
- Cosgrove, M.S.; Ding, Y.; Rennie, W.A.; Lane, M.J.; Hanes, S.D. The Bin3 RNA methyltransferase targets 7SK RNA to control transcription and translation. Wiley Interdiscip. Rev. RNA 2012, 3, 633–647. [Google Scholar] [CrossRef] [PubMed]
- Chattaragada, M.S.; Riganti, C.; Sassoe, M.; Principe, M.; Santamorena, M.M.; Roux, C.; Curcio, C.; Evangelista, A.; Allavena, P.; Salvia, R.; et al. FAM49B, a novel regulator of mitochondrial function and integrity that suppresses tumor metastasis. Oncogene 2018, 37, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Decet, M.; Soukup, S.-F. Endophilin-A/SH3GL2 calcium switch for synaptic autophagy induction is impaired by a Parkinson’s risk variant. Autophagy 2023, 1–3. [Google Scholar] [CrossRef]
- Kara, E.; Crimi, A.; Wiedmer, A.; Emmenegger, M.; Manzoni, C.; Bandres-Ciga, S.; D’sa, K.; Reynolds, R.H.; Botía, J.A.; Losa, M.; et al. An integrated genomic approach to dissect the genetic landscape regulating the cell-to-cell transfer of α-synuclein. Cell Rep. 2021, 35, 109189. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Ferreira, N.; Karakaya, M.; Cengiz, N.; Beijer, D.; Brigatti, K.W.; Gonzaga-Jauregui, C.; Fuhrmann, N.; Hölker, I.; Thelen, M.P.; Zetzsche, S.; et al. De Novo and Inherited Variants in GBF1 are Associated with Axonal Neuropathy Caused by Golgi Fragmentation. Am. J. Hum. Genet. 2020, 107, 763–777. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.-L.; Yang, Y.-P.; Mao, C.-J.; Zhang, X.-Q.; Wang, C.-T.; Yang, J.; Lv, D.-J.; Wang, F.; Hu, L.-F.; Liu, C.-F. A role of BAG3 in regulating SNCA/α-synuclein clearance via selective macroautophagy. Neurobiol. Aging 2017, 60, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Park, D.; Wu, Y.; De Camilli, P. Absence of Sac2/INPP5F enhances the phenotype of a Parkinson’s disease mutation of synaptojanin 1. Proc. Natl. Acad. Sci. USA 2020, 117, 12428–12434. [Google Scholar] [CrossRef]
- Clarin, J.D.; Reddy, N.; Alexandropoulos, C.; Gao, W.-J. The role of cell adhesion molecule IgSF9b at the inhibitory synapse and psychiatric disease. Neurosci. Biobehav. Rev. 2024, 156, 105476. [Google Scholar] [CrossRef] [PubMed]
- Rui, Q.; Ni, H.; Li, D.; Gao, R.; Chen, G. The Role of LRRK2 in Neurodegeneration of Parkinson Disease. Curr. Neuropharmacol. 2018, 16, 1348–1357. [Google Scholar] [CrossRef]
- Li, N.-N.; Tan, E.-K.; Chang, X.-L.; Mao, X.-Y.; Zhang, J.-H.; Zhao, D.-M.; Liao, Q.; Yu, W.-J.; Peng, R. Genetic Association Study betweenSTK39 and CCDC62/HIP1R and Parkinson’s Disease. PLoS ONE 2013, 8, e79211. [Google Scholar] [CrossRef]
- Wang, L.-H.; Lin, Y.-M.; Lin, C.-Y.; Chern, Y.; Wang, G.-S. Neurodegeneration induces a developmental RNA processing program by calpain-mediated MBNL2 degradation. bioRxiv 2020. [Google Scholar] [CrossRef]
- Guella, I.; Sherman, H.E.; Appel-Cresswell, S.; Rajput, A.; Rajput, A.H.; Farrer, M.J. Parkinsonism in GTP cyclohydrolase 1 mutation carriers. Brain 2015, 138, e349. [Google Scholar] [CrossRef]
- Li, G.; Cui, S.; Du, J.; Liu, J.; Zhang, P.; Fu, Y.; He, Y.; Zhou, H.; Ma, J.; Chen, S. Association of GALC, ZNF184, IL1R2 and ELOVL7 With Parkinson’s Disease in Southern Chinese. Front. Aging Neurosci. 2018, 10, 402. [Google Scholar] [CrossRef]
- Lesage, S.; Drouet, V.; Majounie, E.; Deramecourt, V.; Jacoupy, M.; Nicolas, A.; Cormier-Dequaire, F.; Hassoun, S.M.; Pujol, C.; Ciura, S.; et al. Loss of VPS13C Function in Autosomal-Recessive Parkinsonism Causes Mitochondrial Dysfunction and Increases PINK1/Parkin-Dependent Mitophagy. Am. J. Hum. Genet. 2016, 98, 500–513. [Google Scholar] [CrossRef]
- Ruhl, D.A.; Bomba-Warczak, E.; Watson, E.T.; Bradberry, M.M.; Peterson, T.A.; Basu, T.; Frelka, A.; Evans, C.S.; Briguglio, J.S.; Basta, T.; et al. Synaptotagmin 17 controls neurite outgrowth and synaptic physiology via distinct cellular pathways. Nat. Commun. 2019, 10, 3532. [Google Scholar] [CrossRef]
- Garfias, S.; Domínguez, B.T.; Rojas, A.T.; Arroyo, M.; Rodríguez, U.; Boll, C.; Sosa, A.; Sciutto, E.; Adalid-Peralta, L.; López, Y.M.; et al. Peripheral blood lymphocyte phenotypes in Alzheimer and Parkinson’s diseases. Neurología 2022, 37, 110–121. [Google Scholar] [CrossRef]
- Wang, S.; Bleeck, A.; Kasri, N.N.; Kleefstra, T.; van Rhijn, J.-R.; Schubert, D. SETD1A Mediated H3K4 Methylation and Its Role in Neurodevelopmental and Neuropsychiatric Disorders. Front. Mol. Neurosci. 2021, 14, 772000. [Google Scholar] [CrossRef]
- Cheng, L.; Chen, L.; Wei, X.; Wang, Y.; Ren, Z.; Zeng, S.; Zhang, X.; Wen, H.; Gao, C.; Liu, H. NOD2 promotes dopaminergic degeneration regulated by NADPH oxidase 2 in 6-hydroxydopamine model of Parkinson’s disease. J. Neuroinflamm. 2018, 15, 243. [Google Scholar] [CrossRef] [PubMed]
- Dharshini, S.A.P.; Jemimah, S.; Taguchi, Y.H.; Gromiha, M.M. Exploring Common Therapeutic Targets for Neurodegenerative Disorders Using Transcriptome Study. Front. Genet. 2021, 12, 639160. [Google Scholar] [CrossRef]
- Di Lorenzo, G.; Iavarone, F.; Maddaluno, M.; Plata-Gómez, A.B.; Aureli, S.; Meza, C.P.Q.; Cinque, L.; Palma, A.; Reggio, A.; Cirillo, C.; et al. Phosphorylation of FAM134C by CK2 controls starvation-induced ER-phagy. Sci. Adv. 2022, 8, eabo1215. [Google Scholar] [CrossRef]
- Hori, R.T.; Khan, M.M.; Xiao, J.; Hargrove, P.W.; Moss, T.; LeDoux, M.S. Behavioral and molecular effects of Ubtf knockout and knockdown in mice. Brain Res. 2022, 1793, 148053. [Google Scholar] [CrossRef]
- Mani, C.; Acharya, G.; Kshirsagar, S.; Vijayan, M.; Khan, H.; Reddy, P.H.; Palle, K. A Novel Role for BRIP1/FANCJ in Neuronal Cells Health and in Resolving Oxidative Stress-Induced DNA Lesions. J. Alzheimer’s Dis. 2022, 85, 207–221. [Google Scholar] [CrossRef]
- Tian, J.; Vemula, S.R.; Xiao, J.; Valente, E.M.; Defazio, G.; Petrucci, S.; Gigante, A.F.; Rudzińska-Bar, M.; Wszolek, Z.K.; Kennelly, K.D.; et al. Whole-exome sequencing for variant discovery in blepharospasm. Mol. Genet. Genom. Med. 2018, 6, 601–626. [Google Scholar] [CrossRef]
- Obergasteiger, J.; Castonguay, A.-M.; Pizzi, S.; Magnabosco, S.; Frapporti, G.; Lobbestael, E.; Baekelandt, V.; Hicks, A.A.; Pramstaller, P.P.; Gravel, C.; et al. The small GTPase Rit2 modulates LRRK2 kinase activity, is required for lysosomal function and protects against alpha-synuclein neuropathology. npj Park. Dis. 2023, 9, 44. [Google Scholar] [CrossRef]
- Cai, C.; Tang, Y.-D.; Zhai, J.; Zheng, C. The RING finger protein family in health and disease. Signal Transduct. Target. Ther. 2022, 7, 300. [Google Scholar] [CrossRef]
- Falabella, M.; Vernon, H.J.; Hanna, M.G.; Claypool, S.M.; Pitceathly, R.D. Cardiolipin, Mitochondria, and Neurological Disease. Trends Endocrinol. Metab. 2021, 32, 224–237. [Google Scholar] [CrossRef]
- Yong, Y.; Wu, Q.; Meng, X.; Lu, R.; Xia, H.; Pei, F.; Yang, X. Dyrk1a Phosphorylation of α-Synuclein Mediating Apoptosis of Dopaminergic Neurons in Parkinson’s Disease. Park. Dis. 2023, 2023, 8848642. [Google Scholar] [CrossRef]
- Xu, W.; Han, S.-D.; Zhang, C.; Li, J.-Q.; Wang, Y.-J.; Tan, C.-C.; Li, H.-Q.; Dong, Q.; Mei, C.; Tan, L.; et al. The FAM171A2 gene is a key regulator of progranulin expression and modifies the risk of multiple neurodegenerative diseases. Sci. Adv. 2020, 6, eabb3063. [Google Scholar] [CrossRef]
- Cheng, W.-W.; Zhu, Q.; Zhang, H.-Y. Identifying Risk Genes and Interpreting Pathogenesis for Parkinson’s Disease by a Multiomics Analysis. Genes 2020, 11, 1100. [Google Scholar] [CrossRef]
- Blauwendraat, C.; Nalls, M.A.; Singleton, A.B. The genetic architecture of Parkinson’s disease. Lancet Neurol. 2020, 19, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Goldman, S.M.; Marek, K.; Ottman, R.; Meng, C.; Comyns, K.; Chan, P.; Ma, J.; Marras, C.; Langston, J.W.; Ross, G.W. Concordance for Parkinson’s disease in twins: A 20-year update. Ann. Neurol. 2019, 85, 600–605. [Google Scholar] [CrossRef]
- Lambert, J.C.; Ibrahim-Verbaas, C.A.; Harold, D.; Naj, A.C.; Sims, R.; Bellenguez, C.; Jun, G.; DeStefano, A.L.; Bis, J.C.; Beecham, G.W.; et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013, 45, 1452–1458. [Google Scholar] [CrossRef]
- Nalls, M.A.; Pankratz, N.; Lill, C.M.; Do, C.B.; Hernandez, D.G.; Saad, M.; DeStefano, A.L.; Kara, E.; Bras, J.; Sharma, M. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 2014, 46, 989–993. [Google Scholar] [CrossRef]
- Ridge, P.G.; Mukherjee, S.; Crane, P.K.; Kauwe, J.S.K. Alzheimer’s disease genetics consortium Alzheimer’s Disease: Analyzing the missing heritability. PLoS ONE 2013, 8, e79771. [Google Scholar] [CrossRef]
- Ciani, M.; Bonvicini, C.; Scassellati, C.; Carrara, M.; Maj, C.; Fostinelli, S.; Binetti, G.; Ghidoni, R.; Benussi, L. the missing heritability of sporadic frontotemporal dementia: New insights from rare variants in neurodegenerative candidate genes. Int. J. Mol. Sci. 2019, 20, 3903. [Google Scholar] [CrossRef]
- Saudou, F.; Humbert, S. The Biology of Huntingtin. Neuron 2016, 89, 910–926. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-M.; Correia, K.; Loupe, J.; Kim, K.-H.; Barker, D.; Hong, E.P.; Chao, M.J.; Long, J.D.; Lucente, D.; Vonsattel, J.P.G.; et al. CAG Repeat Not Polyglutamine Length Determines Timing of Huntington’s Disease Onset. Cell 2019, 178, 887–900.e14. [Google Scholar] [CrossRef]
- Harding, R.J.; Tong, Y.-F. Proteostasis in Huntington’s disease: Disease mechanisms and therapeutic opportunities. Acta Pharmacol. Sin. 2018, 39, 754–769. [Google Scholar] [CrossRef]
- Folger, A.; Wang, Y. The Cytotoxicity and Clearance of Mutant Huntingtin and Other Misfolded Proteins. Cells 2021, 10, 2835. [Google Scholar] [CrossRef]
- Donnelly, K.M.; Coleman, C.M.; Fuller, M.L.; Reed, V.L.; Smerina, D.; Tomlinson, D.S.; Pearce, M.M.P. Hunting for the cause: Evidence for prion-like mechanisms in Huntington’s disease. Front. Neurosci. 2022, 16, 946822. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, D.L.; Barria, M.A. Prion Diseases: A Unique Transmissible Agent or a Model for Neurodegenerative Diseases? Biomolecules 2021, 11, 207. [Google Scholar] [CrossRef] [PubMed]
- Watson, N.; Brandel, J.-P.; Green, A.; Hermann, P.; Ladogana, A.; Lindsay, T.; Mackenzie, J.; Pocchiari, M.; Smith, C.; Zerr, I.; et al. The importance of ongoing international surveillance for Creutzfeldt–Jakob disease. Nat. Rev. Neurol. 2021, 17, 362–379. [Google Scholar] [CrossRef] [PubMed]
- Mey, G.M.; Mahajan, K.R.; DeSilva, T.M. Neurodegeneration in multiple sclerosis. Wiley Interdiscip. Rev. Syst. Biol. Med. 2023, 15, e1583. [Google Scholar] [CrossRef] [PubMed]
- Walton, C.; King, R.; Rechtman, L.; Kaye, W.; Leray, E.; Marrie, R.A.; Robertson, N.; La Rocca, N.; Uitdehaag, B.; Van Der Mei, I.; et al. Rising prevalence of multiple sclerosis worldwide: Insights from the Atlas of MS, third edition. Mult. Scler. J. 2020, 26, 1816–1821. [Google Scholar] [CrossRef] [PubMed]
- International Multiple Sclerosis Genetics Consortium. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 2019, 365, eaav7188. [Google Scholar] [CrossRef]
- Lublin, F.D.; Reingold, S.C.; Cohen, J.A.; Cutter, G.R.; Sørensen, P.S.; Thompson, A.J.; Wolinsky, J.S.; Balcer, L.J.; Banwell, B.; Barkhof, F.; et al. Defining the clinical course of multiple sclerosis: The 2013 revisions. Neurology 2014, 83, 278–286. [Google Scholar] [CrossRef]
- Das, S.K.; Menezes, M.E.; Bhatia, S.; Wang, X.; Emdad, L.; Sarkar, D.; Fisher, P.B. Gene therapies for cancer: Strategies, challenges and successes. J. Cell. Physiol. 2015, 230, 259–271. [Google Scholar] [CrossRef]
- Bulcha, J.T.; Wang, Y.; Ma, H.; Tai, P.W.L.; Gao, G. Viral vector platforms within the gene therapy landscape. Signal Transduct. Target. Ther. 2021, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- O’connor, D.M.; Boulis, N.M. Gene therapy for neurodegenerative diseases. Trends Mol. Med. 2015, 21, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorth, M.; Narvekar, A. Non viral vectors in gene therapy-an overview. J. Clin. Diagn. Res. JCDR 2015, 9, GE01. [Google Scholar] [CrossRef]
- Puhl, D.L.; D’Amato, A.R.; Gilbert, R.J. Challenges of gene delivery to the central nervous system and the growing use of biomaterial vectors. Brain Res. Bull. 2019, 150, 216–230. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhu, M.; Zhang, Y.; Diao, Y. Crossing the blood-brain barrier with AAV vectors. Metab. Brain Dis. 2020, 36, 45–52. [Google Scholar] [CrossRef]
- Kumar, S.R.; Markusic, D.M.; Biswas, M.; A High, K.; Herzog, R.W. Clinical development of gene therapy: Results and lessons from recent successes. Mol. Ther.-Methods Clin. Dev. 2016, 3, 16034. [Google Scholar] [CrossRef]
- Pai, S.-Y.; Logan, B.R.; Griffith, L.M.; Buckley, R.H.; Parrott, R.E.; Dvorak, C.C.; Kapoor, N.; Hanson, I.C.; Filipovich, A.H.; Jyonouchi, S.; et al. Transplantation Outcomes for Severe Combined Immunodeficiency, 2000–2009. N. Engl. J. Med. 2014, 371, 434–446. [Google Scholar] [CrossRef]
- Demirci, S.; Leonard, A.; Haro-Mora, J.J.; Uchida, N.; Tisdale, J.F. CRISPR/Cas9 for Sickle Cell Disease: Applications, Future Possibilities, and Challenges. Adv. Exp. Med. Biol. 2019, 1144, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Mendell, J.R.; Al-Zaidy, S.; Shell, R.; Arnold, W.D.; Rodino-Klapac, L.R.; Prior, T.W.; Lowes, L.; Alfano, L.; Berry, K.; Church, K.; et al. Single-dose gene-replacement therapy for spinal muscular atrophy. N. Engl. J. Med. 2017, 377, 1713–1722. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Roy, S. Gene-based therapies for neurodegenerative diseases. Nat. Neurosci. 2021, 24, 297–311. [Google Scholar] [CrossRef]
- Mullard, A. First in vivo CRISPR candidate enters the clinic. Nat. Rev. Drug Discov. 2019, 18, 656. [Google Scholar] [CrossRef]
- Zhang, H.-X.; Zhang, Y.; Yin, H. Genome editing with mRNA encoding ZFN, TALEN, and Cas9. Mol. Ther. 2019, 27, 735–746. [Google Scholar] [CrossRef]
- Jankele, R.; Svoboda, P. TAL effectors: Tools for DNA targeting. Brief. Funct. Genom. 2014, 13, 409–419. [Google Scholar] [CrossRef]
- Pickar-Oliver, A.; Gersbach, C.A. The next generation of CRISPR–Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019, 20, 490–507. [Google Scholar] [CrossRef]
- Choi, E.H.; Suh, S.; Sears, A.E.; Hołubowicz, R.; Kedhar, S.R.; Browne, A.W.; Palczewski, K. Genome editing in the treatment of ocular diseases. Exp. Mol. Med. 2023, 55, 1678–1690. [Google Scholar] [CrossRef] [PubMed]
- Willems, J.; de Jong, A.P.H.; Scheefhals, N.; Mertens, E.; Catsburg, L.A.E.; Poorthuis, R.B.; de Winter, F.; Verhaagen, J.; Meye, F.J.; MacGillavry, H.D. ORANGE: A CRISPR/Cas9-based genome editing toolbox for epitope tagging of endogenous proteins in neurons. PLoS Biol. 2020, 18, e3000665. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Lee, J.M.; Riley, G.R.; Jang, W.; Rubinstein, W.S.; Church, D.M.; Maglott, D.R. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014, 42, D980–D985. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef]
- Levy, J.M.; Yeh, W.-H.; Pendse, N.; Davis, J.R.; Hennessey, E.; Butcher, R.; Koblan, L.W.; Comander, J.; Liu, Q.; Liu, D.R. Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat. Biomed. Eng. 2020, 4, 97–110. [Google Scholar] [CrossRef]
- Grünewald, J.; Zhou, R.; Garcia, S.P.; Iyer, S.; Lareau, C.A.; Aryee, M.J.; Joung, J.K. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 2019, 569, 433–437. [Google Scholar] [CrossRef]
- Zuo, E.; Sun, Y.; Yuan, T.; He, B.; Zhou, C.; Ying, W.; Liu, J.; Wei, W.; Zeng, R.; Li, Y.; et al. A rationally engineered cytosine base editor retains high on-target activity while reducing both DNA and RNA off-target effects. Nat. Methods 2020, 17, 600–604. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Rafii, M.S.; Baumann, T.L.; Bakay, R.A.; Ostrove, J.M.; Siffert, J.; Fleisher, A.S.; Herzog, C.D.; Barba, D.; Pay, M.; Salmon, D.P.; et al. A phase1 study of stereotactic gene delivery of AAV2-NGF for Alzheimer’s disease. Alzheimer’s Dement. 2014, 10, 571–581. [Google Scholar] [CrossRef]
- Rocco, M.L.; Soligo, M.; Manni, L.; Aloe, L. Nerve growth factor: Early studies and recent clinical trials. Curr. Neuropharmacol. 2018, 16, 1455–1465. [Google Scholar] [CrossRef]
- Tuszynski, M.H.; Yang, J.H.; Barba, D.; Hoi-Sang, U.; Bakay, R.A.; Pay, M.M.; Masliah, E.; Conner, J.M.; Kobalka, P.; Roy, S.; et al. Nerve growth factor gene therapy: Activation of neuronal responses in Alzheimer disease. JAMA Neurol. 2015, 72, 1139–1147. [Google Scholar] [CrossRef]
- Rafii, M.S.; Tuszynski, M.H.; Thomas, R.G.; Barba, D.; Brewer, J.B.; Rissman, R.A.; Siffert, J.; Aisen, P.S. Adeno-associated viral vector (serotype 2)–nerve growth factor for patients with alzheimer disease: A randomized clinical trial. JAMA Neurol. 2018, 75, 834–841. [Google Scholar] [CrossRef] [PubMed]
- Castle, M.J.; Baltanás, F.C.; Kovacs, I.; Nagahara, A.H.; Barba, D.; Tuszynski, M.H. Postmortem analysis in a clinical trial of AAV2-NGF gene therapy for Alzheimer’s disease identifies a need for improved vector delivery. Hum. Gene Ther. 2020, 31, 415–422. [Google Scholar] [CrossRef] [PubMed]
- DeVos, S.L.; Miller, R.L.; Schoch, K.M.; Holmes, B.B.; Kebodeaux, C.S.; Wegener, A.J.; Chen, G.; Shen, T.; Tran, H.; Nichols, B.; et al. Tau reduction prevents neuronal loss and reverses pathological tau deposition and seeding in mice with tauopathy. Sci. Transl. Med. 2017, 9, eaag0481. [Google Scholar] [CrossRef] [PubMed]
- Sud, R.; Geller, E.T.; Schellenberg, G.D. Antisense-mediated exon skipping decreases tau protein expression: A potential therapy for tauopathies. Mol. Ther.-Nucleic Acids 2014, 3, e180. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; McInnes, J.; Wierda, K.; Holt, M.; Herrmann, A.G.; Jackson, R.J.; Wang, Y.-C.; Swerts, J.; Beyens, J.; Miskiewicz, K.; et al. Tau association with synaptic vesicles causes presynaptic dysfunction. Nat. Commun. 2017, 8, 15295. [Google Scholar] [CrossRef]
- Velazquez, R.; Ferreira, E.; Tran, A.; Turner, E.C.; Belfiore, R.; Branca, C.; Oddo, S. Acute tau knockdown in the hippocampus of adult mice causes learning and memory deficits. Aging Cell 2018, 17, e12775. [Google Scholar] [CrossRef] [PubMed]
- Reiman, E.M.; Arboleda-Velasquez, J.F.; Quiroz, Y.T.; Huentelman, M.J.; Beach, T.G.; Caselli, R.J.; Chen, Y.; Su, Y.; Myers, A.J.; Hardy, J.; et al. Exceptionally low likelihood of Alzheimer’s dementia in APOE2 homozygotes from a 5,000-person neuropathological study. Nat. Commun. 2020, 11, 667. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, Y.; Zhao, N.; Caulfield, T.R.; Liu, C.-C.; Bu, G. Apolipoprotein E and Alzheimer disease: Pathobiology and targeting strategies. Nat. Rev. Neurol. 2019, 15, 501–518. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.J.; Keiser, M.S.; Meltzer, J.C.; Fykstra, D.P.; Dierksmeier, S.E.; Melloni, A.; Nakajima, T.; Tecedor, L.; Ranum, P.T.; Carrell, E.; et al. APOE2 gene therapy reduces amyloid deposition, and improves markers of neuroinflammation and neurodegeneration in a mouse model of Alzheimer disease. bioRxiv 2023. [Google Scholar] [CrossRef]
- Yu, J.T.; Tan, L.; Hardy, J. Apolipoprotein E in Alzheimer’s disease: An update. Annu. Rev. Neurosci. 2014, 37, 79–100. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.T.; Seo, J.; Gao, F.; Feldman, H.M.; Wen, H.L.; Penney, J.; Cam, H.P.; Gjoneska, E.; Raja, W.K.; Cheng, J.; et al. APOE4 causes widespread molecular and cellular alterations associated with Alzheimer’s disease phenotypes in human iPSC-derived brain cell types. Neuron 2018, 98, 1141–1154.e7. [Google Scholar] [CrossRef] [PubMed]
- György, B.; Lööv, C.; Zaborowski, M.P.; Takeda, S.; Kleinstiver, B.P.; Commins, C.; Kastanenka, K.; Mu, D.; Volak, A.; Giedraitis, V.; et al. CRISPR/Cas9 mediated disruption of the Swedish APP allele as a therapeutic approach for early-onset Alzheimer’s disease. Mol. Ther.-Nucleic Acids 2018, 11, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.L.; Hinrich, A.J.; Roman, B.; Norrbom, M.; Rigo, F.; Marr, R.A.; Norstrom, E.M.; Hastings, M.L. Targeting amyloid-β precursor protein, APP, splicing with antisense oligonucleotides reduces toxic amyloid-β production. Mol. Ther. 2018, 26, 1539–1551. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Carlson-Stevermer, J.; Das, U.; Shen, M.; Delenclos, M.; Snead, A.M.; Koo, S.Y.; Wang, L.; Qiao, D.; Loi, J.; et al. CRISPR/Cas9 editing of APP C-terminus attenuates β-cleavage and promotes α-cleavage. Nat. Commun. 2019, 10, 53. [Google Scholar] [CrossRef]
- Mittermeyer, G.; Christine, C.W.; Rosenbluth, K.H.; Baker, S.L.; Starr, P.; Larson, P.; Kaplan, P.L.; Forsayeth, J.; Aminoff, M.J.; Bankiewicz, K.S. Long-term evaluation of a phase 1 study of AADC gene therapy for Parkinson’s disease. Hum. Gene Ther. 2012, 23, 377–381. [Google Scholar] [CrossRef]
- Sebastian, W.S.; Richardson, R.M.; Kells, A.P.; Lamarre, C.; Bringas, J.; Pivirotto, P.; Salegio, E.A.; DeArmond, S.J.; Forsayeth, J.; Bankiewicz, K.S. Safety and tolerability of magnetic resonance imaging-guided convection-enhanced delivery of AAV2-hAADC with a novel delivery platform in nonhuman primate striatum. Hum. Gene Ther. 2012, 23, 210–217. [Google Scholar] [CrossRef]
- Palfi, S.; Gurruchaga, J.M.; Ralph, G.S.; Lepetit, H.; Lavisse, S.; Buttery, P.C.; Watts, C.; Miskin, J.; Kelleher, M.; Deeley, S.; et al. Long-term safety and tolerability of ProSavin, a lentiviral vector-based gene therapy for Parkinson’s disease: A dose escalation, open-label, phase 1/2 trial. Lancet 2014, 383, 1138–1146. [Google Scholar] [CrossRef]
- Emborg, M.E.; Carbon, M.; E Holden, J.; During, M.J.; Ma, Y.; Tang, C.; Moirano, J.; Fitzsimons, H.; Roitberg, B.Z.; Tuccar, E.; et al. Subthalamic glutamic acid decarboxylase gene therapy: Changes in motor function and cortical metabolism. J. Cereb. Blood Flow Metab. 2007, 27, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Niethammer, M.; Tang, C.C.; LeWitt, P.A.; Rezai, A.R.; Leehey, M.A.; Ojemann, S.G.; Flaherty, A.W.; Eskandar, E.N.; Kostyk, S.K.; Sarkar, A.; et al. Long-term follow-up of a randomized AAV2-GAD gene therapy trial for Parkinson’s disease. J. Clin. Investig. 2017, 2, e90133. [Google Scholar] [CrossRef] [PubMed]
- Alarcón-Arís, D.; Pavia-Collado, R.; Miquel-Rio, L.; Coppola-Segovia, V.L.; Ferrés-Coy, A.; Ruiz-Bronchal, E.; Galofré, M.; Paz, V.; Campa, L.; Revilla, R.; et al. Anti-α-synuclein ASO delivered to monoamine neurons prevents α-synuclein accumulation in a Parkinson’s disease-like mouse model and in monkeys. EBioMedicine 2020, 59, 102944. [Google Scholar] [CrossRef]
- Izco, M.; Blesa, J.; Schleef, M.; Schmeer, M.; Porcari, R.; Al-Shawi, R.; Ellmerich, S.; de Toro, M.; Gardiner, C.; Seow, Y.; et al. Systemic exosomal delivery of shRNA minicircles prevents parkinsonian pathology. Mol. Ther. 2019, 27, 2111–2122. [Google Scholar] [CrossRef]
- Inoue, S.; Nishimura, K.; Gima, S.; Nakano, M.; Takata, K. CRISPR-Cas9-Edited SNCA Knockout Human Induced Pluripotent Stem Cell-Derived Dopaminergic Neurons and Their Vulnerability to Neurotoxicity. Biol. Pharm. Bull. 2023, 46, 517–522. [Google Scholar] [CrossRef]
- Benskey, M.J.; Sellnow, R.C.; Sandoval, I.M.; Sortwell, C.E.; Lipton, J.W.; Manfredsson, F.P. Silencing alpha synuclein in mature nigral neurons results in rapid neuroinflammation and subsequent toxicity. Front. Mol. Neurosci. 2018, 11, 36. [Google Scholar] [CrossRef]
- Rocha, E.M.; Smith, G.A.; Park, E.; Cao, H.; Brown, E.; Hayes, M.A.; Beagan, J.; McLean, J.R.; Izen, S.C.; Perez-Torres, E.; et al. Glucocerebrosidase gene therapy prevents α-synucleinopathy of midbrain dopamine neurons. Neurobiol. Dis. 2015, 82, 495–503. [Google Scholar] [CrossRef]
- Morabito, G.; Giannelli, S.G.; Ordazzo, G.; Bido, S.; Castoldi, V.; Indrigo, M.; Cabassi, T.; Cattaneo, S.; Luoni, M.; Cancellieri, C.; et al. AAV-PHP.B-mediated global-scale expression in the mouse nervous system enables GBA1 gene therapy for wide protection from synucleinopathy. Mol. Ther. 2017, 25, 2727–2742. [Google Scholar] [CrossRef]
- Fuji, R.N.; Flagella, M.; Baca, M.; Baptista, M.A.S.; Brodbeck, J.; Chan, B.K.; Fiske, B.K.; Honigberg, L.; Jubb, A.M.; Katavolos, P.; et al. Effect of selective LRRK2 kinase inhibition on nonhuman primate lung. Sci. Transl. Med. 2015, 7, ra15–ra273. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.T.; John, N.; Delic, V.; Ikeda-Lee, K.; Kim, A.; Weihofen, A.; Swayze, E.E.; Kordasiewicz, H.B.; West, A.B.; Volpicelli-Daley, L.A. LRRK2 antisense oligonucleotides ameliorate α-synuclein inclusion formation in a Parkinson’s disease mouse model. Mol. Ther.-Nucleic Acids 2017, 8, 508–519. [Google Scholar] [CrossRef] [PubMed]
- McCampbell, A.; Cole, T.; Wegener, A.J.; Tomassy, G.S.; Setnicka, A.; Farley, B.J.; Schoch, K.M.; Hoye, M.L.; Shabsovich, M.; Sun, L.; et al. Antisense oligonucleotides extend survival and reverse decrement in muscle response in ALS models. J. Clin. Investig. 2018, 128, 3558–3567. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.M.; Pestronk, A.; David, W.; Rothstein, J.; Simpson, E.; Appel, S.H.; Andres, P.L.; Mahoney, K.; Allred, P.; Alexander, K.; et al. A phase I, randomised, first-in-human study of an antisense oligonucleotide directed against SOD1 delivered intrathecally in SOD1-familial ALS patients. Lancet Neurol. 2013, 12, 435. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.; Cudkowicz, M.; Shaw, P.J.; Andersen, P.M.; Atassi, N.; Bucelli, R.C.; Genge, A.; Glass, J.; Ladha, S.; Ludolph, A.L.; et al. Phase 1–2 trial of antisense oligonucleotide tofersen for SOD1 ALS. N. Engl. J. Med. 2020, 383, 109–119. [Google Scholar] [CrossRef]
- Chen, Y.A.; Kankel, M.W.; Hana, S.; Lau, S.K.; Zavodszky, M.I.; McKissick, O.; Mastrangelo, N.; Dion, J.; Wang, B.; Ferretti, D.; et al. In vivo genome editing using novel AAV-PHP variants rescues motor function deficits and extends survival in a SOD1-ALS mouse model. Gene Ther. 2023, 30, 443–454. [Google Scholar] [CrossRef]
- Mueller, C.; Berry, J.D.; McKenna-Yasek, D.M.; Gernoux, G.; Owegi, M.A.; Pothier, L.M.; Douthwright, C.L.; Gelevski, D.; Luppino, S.D.; Blackwood, M.; et al. SOD1 suppression with adeno-associated virus and microRNA in familial ALS. N. Engl. J. Med. 2020, 383, 151–158. [Google Scholar] [CrossRef]
- Jiang, J.; Zhu, Q.; Gendron, T.F.; Saberi, S.; McAlonis-Downes, M.; Seelman, A.; Stauffer, J.E.; Jafar-Nejad, P.; Drenner, K.; Schulte, D.; et al. Gain of toxicity from ALS/FTD-linked repeat expansions in C9ORF72 is alleviated by antisense oligonucleotides targeting GGGGCC-containing RNAs. Neuron 2016, 90, 535–550. [Google Scholar] [CrossRef]

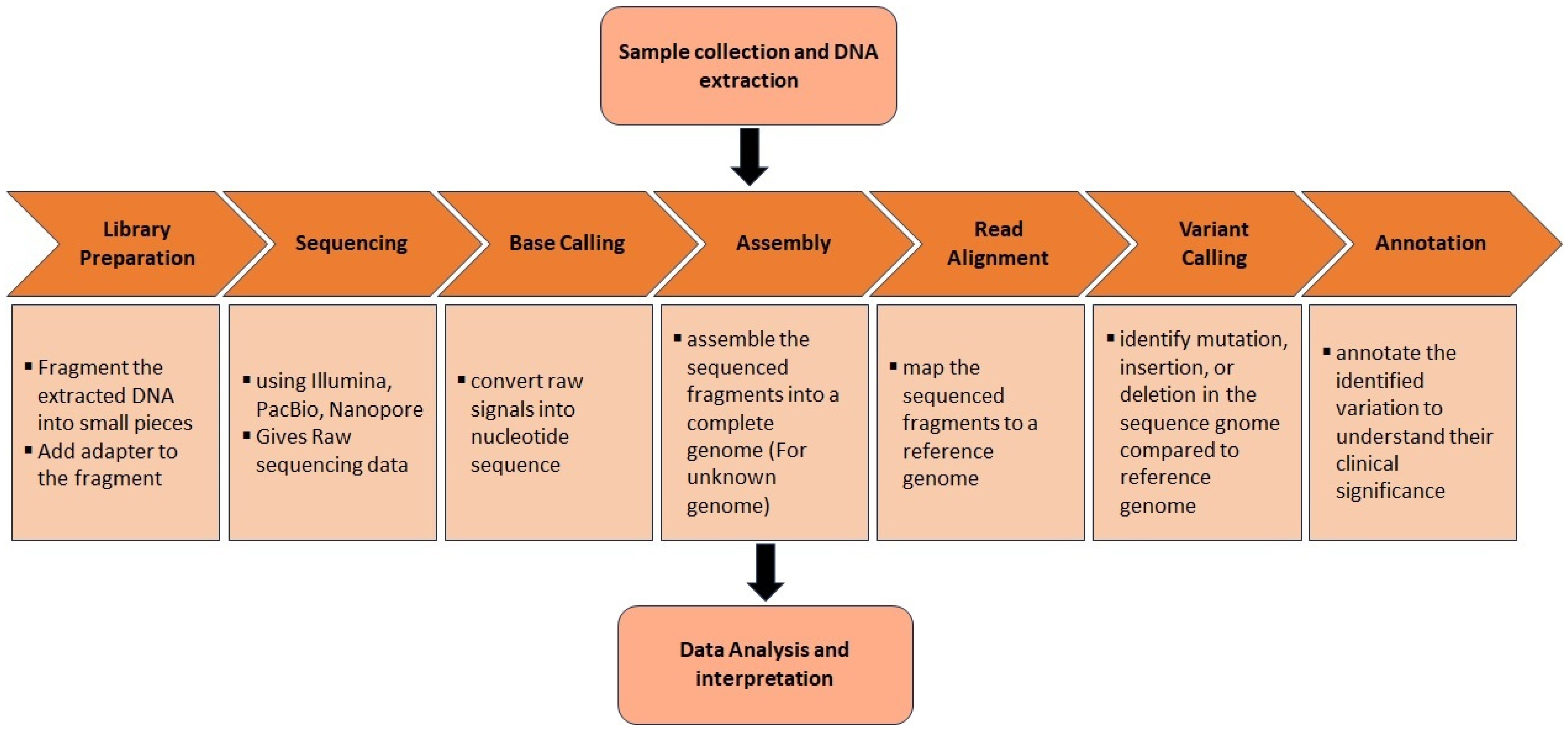

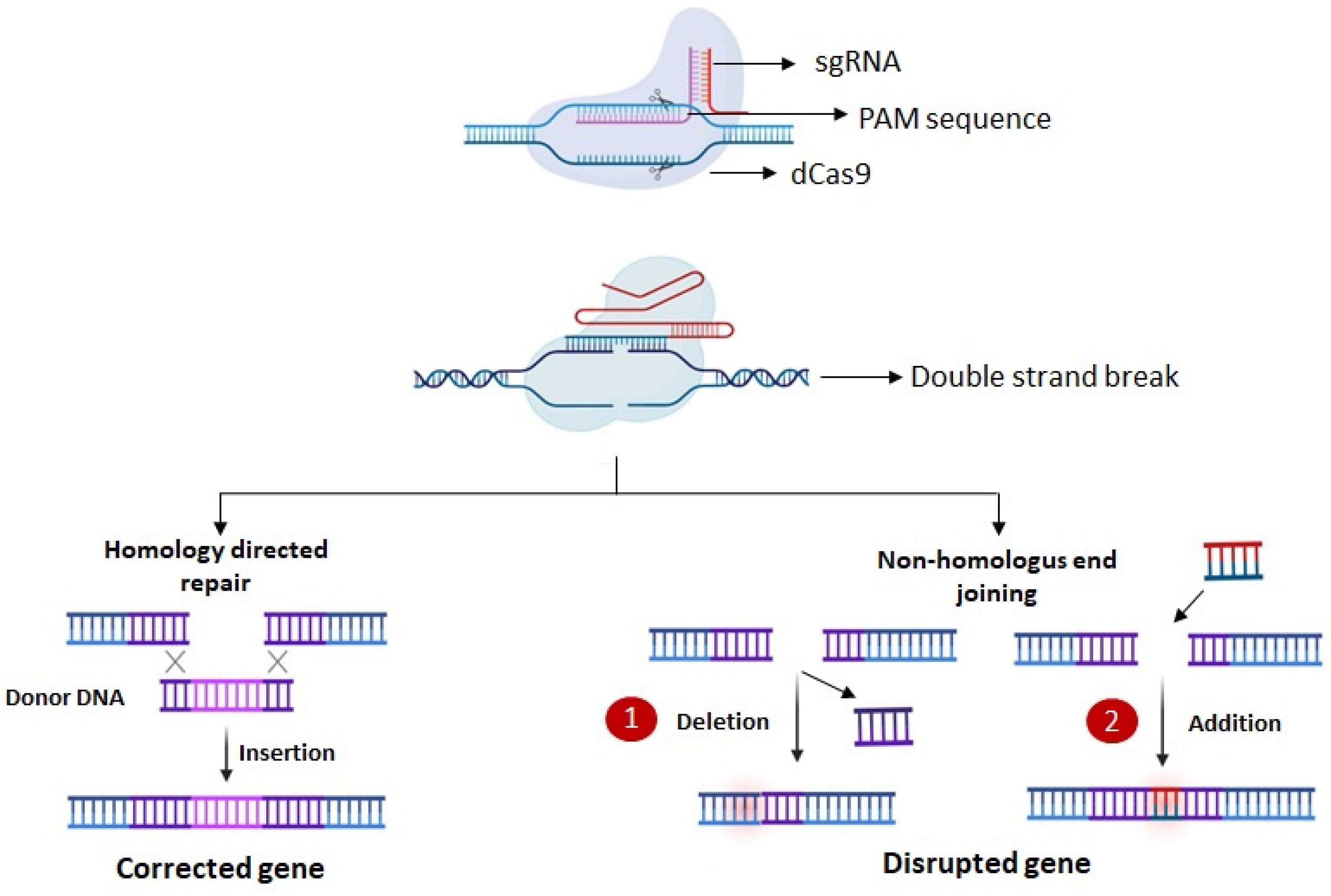
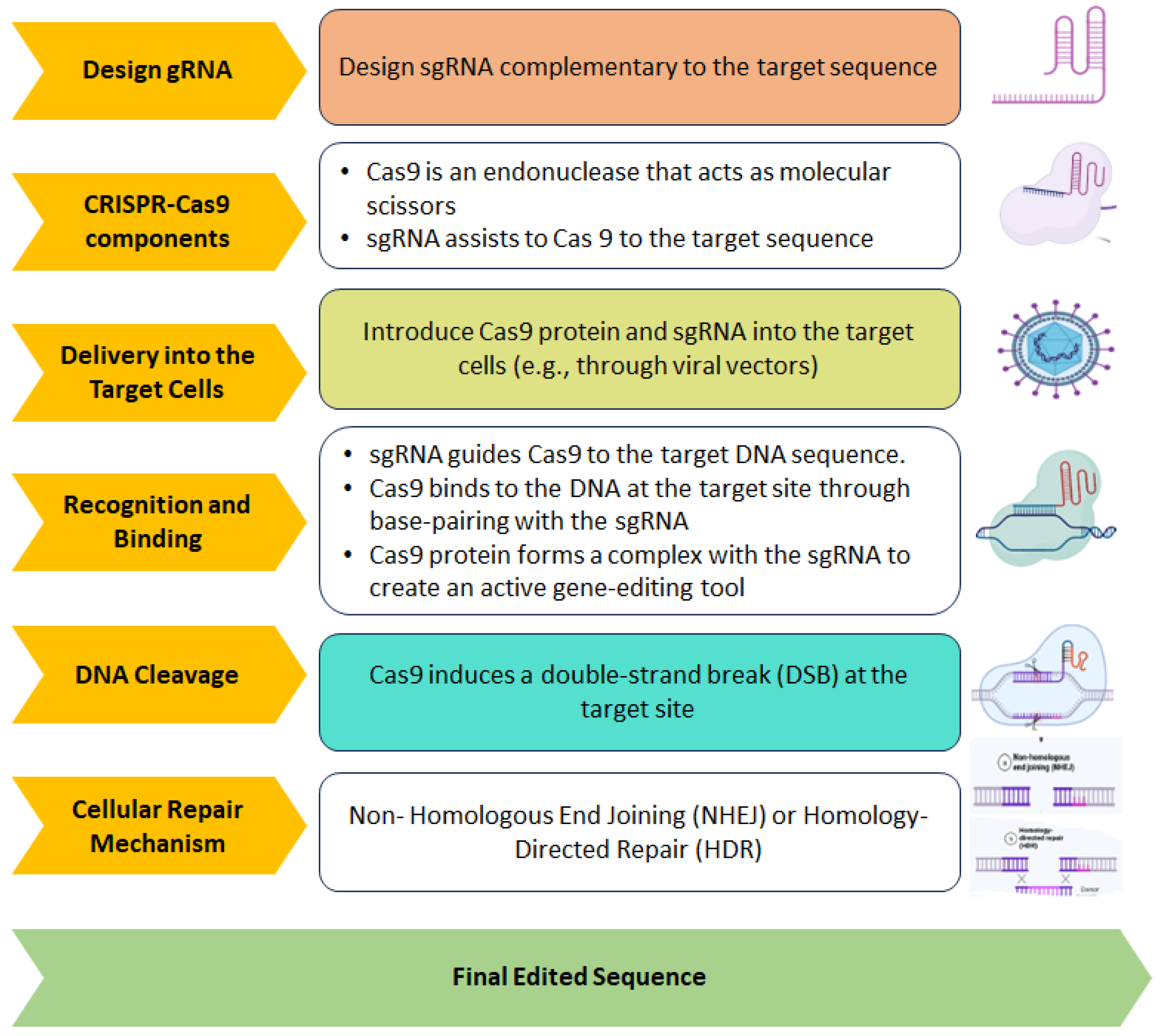

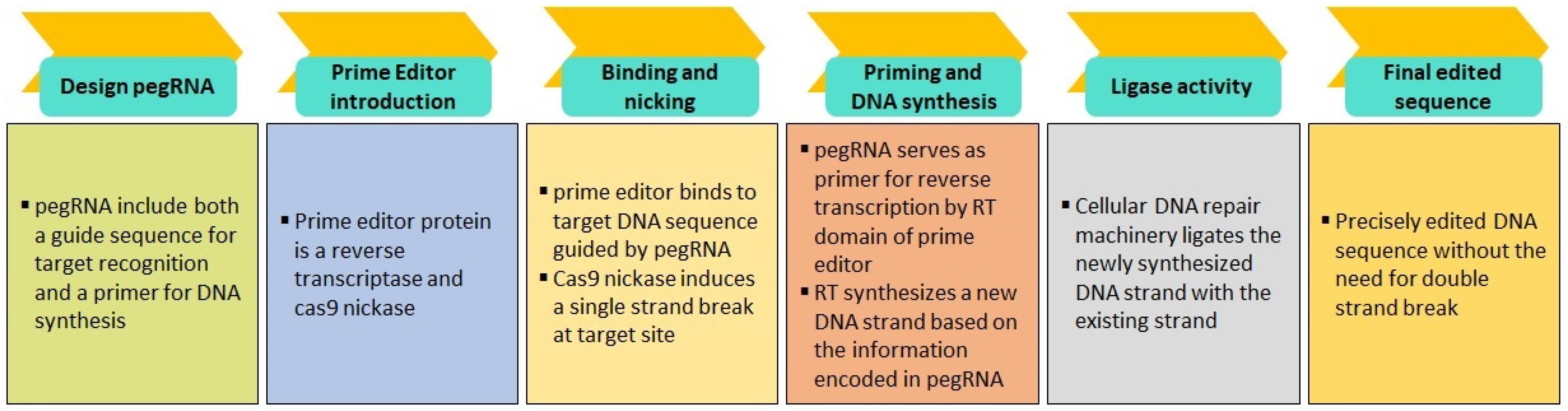

| S.No. | Techniques | Methods | Benefits | Disadvantages |
|---|---|---|---|---|
| 1 | Sanger Sequencing | Traditional method involving the sequencing of DNA fragments using chain-termination dideoxy nucleotides. |
|
|
| 2 | GWAS (Genome-wide association studies) | Analyze the genetic variation across the entire genome to identify the link between specific genetic variants and a particular trait or disease. |
|
|
| 3 | WGS (Whole Genome Sequencing) | Involves sequencing the entire genome to identify both coding and non-coding variants associated with disease. |
|
|
| 4 | WES (Whole Exome Sequencing) | Focus on sequencing the protein coding region of genome to identify disease-associated variants. |
|
|
| 5 | NGS (Next-Generation Sequencing) | Utilizes high throughput sequencing technologies to sequence DNA or RNA molecule in parallel. |
|
|
| 6 | LRS (Long Read Sequencing) | Employs sequencing platforms that generate reads spanning hundreds to thousands of base pairs, providing more contiguous sequence information. |
|
|
| S.No. | Gene (ALS) | Function | Disease Mechanism |
|---|---|---|---|
| 1 | C9orf72 | Regulates vesicular transport and autophagy | C9ORF72 haploinsufficiency (loss of function) Sense and antisense RNA (GGGGCC)n the function of RNA binding protein (gain of function) |
| 2 | UNC13A | Facilitates Neurotransmission | Impaired synaptic transmission [95] |
| 3 | SOD1 | Antioxidant role | Oxidative stress, mitochondrial dysfunction, and excitotoxicity |
| 4 | SCFD1 | Regulates ER to Golgi anterograde vesicular transport | Protein misfolding and aggregation [96] |
| 5 | MOBP-RPSA | Neurons myelination | Demyelination of neurons [97] |
| 6 | HLA | Antigen presentation and immune response | Inflammation due to suppressed immune response [98] |
| 7 | KIF5A | Engaged in anterograde transport of cargos along the microtubule rails in neurons | Impaired axonal transport, synaptic transmission, and motor neuronal toxicity [99] |
| 8 | CFAP410 | Cytoskeletal organization and ciliary function | Decreased stability and increased degradation of mutant protein causes dysfunction of primary cilium [100] |
| 9 | GPX3-TNIP | Antioxidant | Oxidative stress, mitochondrial dysfunction, and excitotoxicity [101] |
| 10 | SLC9A8 | Na/H exchanger | Excitotoxicity and axonal degeneration [102] |
| 11 | TBK1 | Requires in cargo recruitment during autophagy | Neuroinflammation and autophagy [103] |
| 12 | ERGIC1 | Maintains ER-Golgi structure | Disintegration of ER and mitophagy [104] |
| 13 | NEK1 | A protein kinase that regulates cell cycle, DNA damage repair, apoptosis, and ciliary function | Induces DNA damage [105] |
| 14 | COG3 | Regulating Golgi processes, protein trafficking, and glycosylation in neurons | Protein trafficking by Golgi fragmentation [106] |
| 15 | PTPRN2 | Involved in vesicle-mediated secretory process in hippocampus [107] | Not clear. Probably motor neuron dysfunction [108] |
| S.No. | Gene (AD) | Function | Disease Mechanism |
|---|---|---|---|
| 1 | SORT1 | Directs trafficking of APP into recycling pathways | Low level of SORT1 in AD causes increased Aβ deposition [157] |
| 2 | CR1 | Immune complement cascade | Regulates Aβ metabolism [158] |
| 3 | ADAM17 | Alpha-secretase imparts a role in APP processing | Causes increased APP production [159] |
| 4 | PRKD3 | Cell proliferation | Causes neuroinflammation [160] |
| 5 | NCK2 | Axon growth and synapse formation and Epinephrin-mediated axon guidance | Disturbs motor axon trajectory selection [161] |
| 6 | WDR12 | Ribosome biogenesis and cell proliferation | Possibly causing neuroinflammation [162] |
| 7 | BIN1 | Endocytosis and intracellular trafficking | Endosome defect [163] |
| 8 | INPP5D | Immune signaling | Inflammasome activation in microglia [164] |
| 9 | MME | Cleaves and degrades beta-amyloid | Increased Aβ deposition and axonal neuropathy [165] |
| 10 | IDUA | Lysosomal protein acts in degradation of misfolded protein | Lysosomal dysfunction and increased proteinopathy [166] |
| 11 | RHOH | Regulation of actin cytoskeleton, and dendrites formation | Synaptic loss and spinal dysfunction [167] |
| 12 | CLNK | Immunomodulatory function | Disturbed immune signaling and neuroinflammation [168] |
| 13 | ANKH | Regulating inflammation | NF-κB-mediated neuroinflammation [162] |
| 14 | COX7C | Mitochondrial bioenergetics | Mitochondrial respiratory defects [169] |
| 15 | TNIP1 | Inhibition of the TNF-α signaling pathway and NF-κB activation/translocation | Microglial activation and inflammation [170] |
| 16 | RASGEF1C | Associated with immune function | Neuroinflammation [171] |
| 17 | HS3ST5 | Cellular uptake and distribution of molecules like growth factors and morphogens | Promotes tau fibrillation into NFTs [172] |
| 18 | HLA-DQA1 | Dendritic cells, macrophages and B cells and involved in adaptive immune responses | Stimulates adaptive immune signaling in AD and also activates PKC and TLR signaling [173] |
| 19 | UNC5CL | Involved in mediating axon growth, neuronal migration in neuronal development, regulation of cell apoptosis. | Contributes to AD pathogenesis by activating DAPK1 which in turn causes aberrant tau, Aβ and neuronal apoptosis/autophagy |
| 20 | TREM2 | Regulates microglia proliferation, survival, migration, and phagocytosis. | Downregulation induces neuroinflammation [174] |
| 21 | TREML2 | Regulates microglial proliferation | Immune-related neuroinflammatory and increased tau deposition [175] |
| 22 | CD2AP | Early endosome formation and protein trafficking | Regulates Aβ generation by a neuron-specific polarization of Aβ in dendritic early endosomes [176] |
| 23 | UMAD1 | Involved in endosome-ubiquitin homeostasis [177] | Possibly defects in protein degradation cascade and increased deposit of Aβ and tau |
| 24 | ICA1 | ICA1 regulates AMPA receptor trafficking [178] | Possibly disturb synaptic signaling |
| 25 | TMEM106B | Brain lipid metabolism, | Disturbed lipid homeostasis [179] |
| 26 | JAZF1 | Lipid/cholesterol metabolism and microglial efferocytosis [180] | Neuroinflammation by defective efferocytosis and defective lipid metabolism (not clear) |
| 27 | SEC61G | Protein trafficking, ER calcium leak channel [181] | - |
| 28 | EPDR1 | Neurogenesis and synaptic signaling [162] | Not clear |
| 29 | SPDYE3 | Cell cycle regulator [182] | - |
| 30 | EPHA1 | Immune response, cholesterol metabolism, and synaptic function | Spine morphology abnormalities and synaptic dysfunction [183] |
| 31 | CTSB | Regulates apoptosis, neuroinflammation, and autophagy | lysosomal leakage of cathepsin B to the cytosol leads to neurodegeneration and behavioral deficits [184] |
| 32 | SHARPIN | Inflammation and immune system activation Synaptic signaling | Attenuated inflammatory/immune response [185] |
| 33 | PTK2B | Ca2+-activated non-receptor tyrosine kinase, involved in synaptic plasticity | Neuronal hyperexcitability by neuronal differentiation and electrical maturation [186] |
| 34 | CLU | Secreted by glia binds to Aβ and plays a protective role by preventing Aβ aggregation | Aβ clearance [187] |
| 35 | ABCA1 | Cholesterol mobilization | Defective lipid metabolism, and neuroinflammation [188] |
| 36 | ANK3 | Scaffolding proteins recruit diverse membrane proteins, (ion channels and cell adhesion molecules) into subcellular membrane domains | Altered neuronal excitability and altered neuronal connectivity [138] |
| 37 | TSPAN14 | Regulates maturation and trafficking of the transmembrane metalloprotease ADAM10 [189] | Not clear |
| 38 | BLNK | Participates in the regulation of PLC-γ activity and the activation of Ras pathway [190] | Not clear. Possibly involved in immune regulation |
| 39 | PLEKHA1 | Adaptive immunity | Inflammatory responses [191] |
| 40 | USP6NL | GTPase-activating protein involved in control of endocytosis | Dysfunction of the myeloid endolysosomal system [192] |
| 41 | SPI1 | Controls microglial development and function | Regulating neuroinflammation [193] |
| 42 | EED | Catalyzes the methylation of histone and mediates the repressive chromatin | Synaptic dysfunction due to upregulation of synapse related gene [194] |
| 43 | SORL1 | Regulates the recycling of the APP out of the endosome | Endosomal swelling and APP misprocessing [195] |
| 44 | TPCN1 | Encodes a voltage-dependent calcium channel and involved in long-term potentiation in hippocampal neurons, | Altered calcium signaling and cognitive dysfunction [196] |
| 45 | IGH gene cluster | Immune response [197] | Not clear |
| 46 | FERMT2 | APP metabolism and axonal growth | Impaired synaptic connectivity, and long-term potentiation in an APP-dependent manner [198] |
| 47 | SLC24A4 | Neural development and cholesterol metabolism | Increased deposition of Ab and tau [199] |
| 48 | SPPL2A | Engaged in the function of B-cells and dendritic cells. | Activates TNF-α signaling [156] |
| 49 | MINDY2 | Deubiquitination | Not clear |
| 50 | APH1B | γ-secretase | Brain atrophy and amyloid-β deposition [200] |
| 51 | SNX1 | Endosome trafficking | Prevents BACE1 trafficking to the lysosomal degradation system, resulting in increased production of Aβ [201] |
| 52 | CTSH | Immune regulation | Role in neuroinflammation and amyloid β production [202] |
| 53 | BCKDK | Regulation of neurotransmitter synthesis, and mTOR activity. | Causes hyperexcitability, neuroinflammation, and dysregulation of neurotrophic factors [203] |
| 54 | IL34 | Stimulates proliferation of monocytes and macrophages | Triggers neuroinflammation via colony-stimulating factor-1 receptor (CSF-1r) [204]. |
| 55 | PLCG2 | Present on microglia and function as immune regulator | Upregulated and activates inflammation related pathway [205] |
| 56 | DOC2A | A calcium sensor, facilitates neurotranbsmitter release in Ca2+-dependent manner | Abnormality in synaptic transmission [206] |
| 57 | MAF | Regulates T-cell susceptibility to apoptosis | Probably immune cell dysfunction and neuroinflammation [207] |
| 58 | FOXF1 | Cell proliferation, cell cycle, and regulatory protein | Activated by PI3K/AKT and stress response and may cause inflammation [208] |
| 59 | PRDM7 | Methyltransferases induce trimethylation | Possibly suppresses the synaptic gene [209] |
| 60 | WDR81 | Facilitates the recruitment of autophagic protein aggregates and promotes autophagic clearance | Impaired autophagy [210] |
| 61 | MYO15A | A myosin involved in actin organization | Retromer dysfunction [211] |
| 62 | GRN | Regulates lysosomal biogenesis, inflammation, repair, stress response, and aging. | Neuroinflammation [211] |
| 63 | SCIMP | Immune regulation via major histocompatibility complex class II signaling. | Neuroinflammation [212] |
| 64 | WNT3 | Synaptic function and immune regulation | Causes synaptic dysfunction and inflammation via Wnt3/β-catenin/GSK3β signaling pathway [213] |
| 65 | ABI3 | Regulator of microglia | Neuroinflammation [214] |
| 66 | TSPOAP1 | TSPO-associated protein 1, interacts with translocator protein (TSPO) and act indirectly to activate microglia | Neuroinflammation [215] |
| 67 | ACE | An endopeptidase | ACE has been shown to cleave amyloid-β (Aβ) [216] |
| 68 | KLF16 | Regulates dopamine receptors | Modulates dopaminergic transmission in the brain [217] |
| 69 | SIGLEC11 | Immune regulation | Proinflammation and phagocytosis [218] |
| 70 | LILRB2 | Aβ receptor | Perturbance in synaptic signaling and cognitive impairment [219] |
| 71 | ABCA7 | Lipid homeostasis and phagocytosis. | Disturbed lipid metabolism, ER stress. Impaired microglial response to inflammation |
| 72 | RBCK1 | Involved in ubiquitination | TNF-α-mediated activation of NF-κB pathway. |
| 73 | SLC2A4RG | Encodes solute carrier protein. Involved in cell cycle via CDK1 pathway [220] | Not clear. Possibly induces proliferation |
| 74 | CASS4 | Role in inflammation, calcium signaling, and microtubule stabilization. | Disturbed synaptic signaling and neuroinflammation [221] |
| 75 | APP | Proliferation, differentiation, and maturation of neural stem cells. | Abnormal cleavage causes plaque deposition [222] |
| 76 | ADAMTS1 | APP hydrolysis | Increased Aβ generation through β-secretase-mediated cleavage [223] |
| S.No. | Gene (PD) | Function | Mechanism |
|---|---|---|---|
| 1 | KRTCAP2 | Dementia-related gene | Inflammation and neurodegeneration [260] |
| 2 | PMVK | Involved in mevalonate pathway | Not clear. Possibly same as GBA [261] |
| 3 | GBAP1 | Encodes for the enzyme glucocerebrosidase (GCase), a lysosomal enzyme | Lysosomal dysfunction [262] |
| 4 | FCGR2A | Phagocytosis and modulates inflammatory responses | Binds with IgG-specific immune complexes and activates signaling [263] |
| 5 | VAMP4 | Endosomal trafficking of synaptic proteins | Impaired synaptic signaling and lysosomal degradation [264] |
| 6 | NUCKS1 | Cell growth and proliferation [265] | Not clear |
| 7 | RAB29 | Lysosome-related organelle biogenesis | Lysosomal dysfunction Axon termination [266] |
| 8 | ITPKB | Involved in inositol metabolism and calcium release from ER | Causes α-synuclein aggregation by dysregulated calcium release from ER-to-mitochondria [267] |
| 9 | SIPA1L2 | Controls protein trafficking and BDNF/TrkB signaling [268] | Not clear; possibly abrupt synaptic signaling |
| 10 | KCNS3 | Potassium channel | Neuroinflammation [269] |
| 11 | KCNIP3 | Associated with inositol biosynthetic pathway [270] | Not clear |
| 12 | MAP4K4 | Cell proliferation, inflammation, and stress response | Cytokine activation and neuroinflammation [271] |
| 13 | TMEM163 | Influx or efflux transporter particularly Zn transport [272] | Not clear |
| 14 | STK39 | Immune regulation | Inflammatory pathway [273] |
| 15 | SATB1 | Transcriptional response in dopaminergic neurons | Senescence-mediated neuroinflammation [274] |
| 16 | LINC00693 | Involved in miRNA processing complex [275] | Might affect protein expression and accumulation |
| 17 | IP6K2 | Mitochondrial respiration | Mitophagy via PINK1 signaling [276] |
| 18 | KPNA1 | Encodes importin α5 and is involved in lysosomal biogenesis and autophagy | Disturbed protein degradation [277] |
| 19 | MED12L | Transcriptional coactivation of nearly all RNA polymerase II-dependent genes, Wnt/beta-catenin pathway, and immune response [278] | Possibly transcriptional defects |
| 20 | SPTSSB | Regulates de novo synthesis of ceramides | Neuronal signaling, synaptic transmission, cell metabolism [279] |
| 21 | MCCC1 | Mitochondrial enzyme and involved in leucine catabolism [280] | Possibly associated with mitochondrial dysfunction |
| 22 | GAK | Associated with lysosomal and chaperons | Defected lysosomal-mediated protein degradation [281] |
| 23 | TMEM175 | Proton channel to maintain optimum pH in lysosomes | Downregulation of TMEM175 causes lysosomal over-acidification, impaired proteolytic activity, and facilitated a-synuclein aggregation [282] |
| 24 | BST1 | Serves as a receptor that regulates leukocyte adhesion and migration | Immune regulation and inflammation [283] |
| 25 | LCORL | - | - |
| 26 | SCARB2 | Encodes a receptor responsible for the transport of glucocerebrosidase (GCase) to the lysosome | Associated with lysosomal defects [284] |
| 27 | FAM47E | Present in close proximity to SCARB2 | Lysosome/autophagy dysfunction |
| 28 | FAM47E-STBD1 | - | - |
| 29 | SNCA | Dopamine release and transport, fibrillization of MAPT, and suppression of both p53 expression and transactivation of proapoptotic genes leading to decreased caspase-3 activation | Synuclein aggregation and induction of apoptosis [285] |
| 30 | CAMK2D | Calcium/calmodulin-dependent protein kinase ii delta. Involved in synaptic plasticity | Disturbed calcium signaling and synaptic function [286] |
| 31 | CLCN3 | Ion channel transporter and neurotransmitter signaling [239]. | Disturbed synaptic signaling |
| 32 | ELOVL7 | Catalyzing the elongation of very long-chain fatty acids. | Possibly disturbed lipid metabolism and oxidative stress [287] |
| 33 | PAM | Glutamate receptor at parasynapses, associated with anxiety and hyperexcitation. | Disturbed glutamatergic and GABA signaling [288] |
| 34 | C5orf24 | - | Function and mechanism are not clear. However, upregulated expression and DNA methylation in disease condition [289] |
| 35 | LOC100131289 | - | - |
| 36 | TRIM40 | E3 ubiquitin-protein ligase and inhibits NF-κB activity | Protein degradation and inflammation [290] |
| 37 | HLA-DRB5 | Immune regulation | Inflammation [291] |
| 38 | RIMS1 | Encodes a synaptic protein and involved in neurotransmitter release and synaptic transmission [277] | Possibly perturbance in synaptic signaling |
| 39 | FYN | Ion channel function, growth factor receptor signaling, immune system regulation. | Activates BDNF/TrkB, PKCδ, NF-κB, MAPK, Nrf2, and NMDAR signaling pathway and induces synuclein phosphorylation, inflammation and excitotoxicity [292] |
| 40 | RPS12 | A special function in cell competition that defines the competitiveness of cells. | Not clear in PD; however, reported to cause inflammation [293] |
| 41 | GPNMB | Cell differentiation, migration, proliferation | Interacts with a-synuclein and induces its phosphorylation, cellular internalization, and fibrillization [294] |
| 42 | GS1-124K5.11 | - | - |
| 43 | CTSB | Lysosomal hydrolase cathepsin B involved in waste degradation in cells | Autophagy and lysosomal dysfunction causing a-synuclein aggregation [295] |
| 44 | FGF20 | Maintenance of dopaminergic neurons | Affects dopaminergic neurons in paracrine manner [296] |
| 45 | BIN3 | Cytokinesis and RNA methyltransferase | Probably target transcription and translation step [297] |
| 46 | FAM49B | Regulates mitochondrial function | Mitochondrial fission, oxidative stress, and inflammation [298] |
| 47 | SH3GL2 | Encodes Endophilin A which regulates autophagy in calcium dependent manner | Autophagy dysfunction at synapses [299] |
| 48 | UBAP2 | Synapse formation, maintenance, and signaling | - |
| 49 | ITGA8 | Alpha8 integrin, cell adhesion, cell signaling, and cytoskeletal organization | Increases cell-to-cell transfer of a-Syn [300] |
| 50 | GBF1 | Maintenance and function of the Golgi apparatus, and mitochondria migration and positioning | Increase in Golgi fragmentation [301] |
| 51 | BAG3 | A chaperone and regulates autophagy | Its downregulation promotes autophagy dysfunction and disease progression [302] |
| 52 | INPP5F | PI4P-phosphatase Involved in endocytic pathway | Disturbed endocytosis [303] |
| 53 | RNF141 | - | - |
| 54 | DLG2 | DLG2-encoded protein involved in glutamate receptor phosphorylation | Phosphorylation of NR2 subunit and hyperexcitability [280] |
| 55 | IGSF9B | Cell adhesion molecule at inhibitory synapses and plays role in neuroplasticity and synaptic transmission | Any disturbance in inhibitory synapses causes dysregulation of information flow and cognitive defects [304] |
| 56 | LRRK2 | Associated with intracellular membranes and vesicular structures | Causes accumulation of a-synuclein, which activates MAPK signaling and microglial activation leading to inflammation [305] |
| 57 | SCAF11 | Encodes a caspase | - |
| 58 | HIP1R | Clathrin-mediated endocytosis, actin dynamics, intrinsic cell death pathway [306] | Not clear. Probably affects the endocytosis of a-synuclein and activate caspase response |
| 59 | FBRSL1 | - | - |
| 60 | CAB39L | Encodes calcium binding 39-like protein | - |
| 61 | MBNL2 | Encodes for the muscleblind-like protein 2, which belongs to a conserved family of RNA-binding proteins | Reduced MBNL2 expression accompanied by the reduction in a developmental RNA processing [307] |
| 62 | MIPOL1 | - | - |
| 63 | GCH1 | Essential for dopamine production | Affects dopaminergic signaling [308] |
| 64 | RPS6KL1 | - | - |
| 65 | GALC | Encodes galactocerebrosidase | Impaired autophagy and disturbed protein trafficking causes a-synuclein deposition [309] |
| 66 | VPS13C | Localized to the outer membrane of mitochondria | PINK1/Parkin-dependent mitophagy [310] |
| 67 | SYT17 | Encodes synaptotagmin-17, associated with vesicle trafficking and transport at synapses | Disturbed synaptic trafficking [311] |
| 68 | CD19 | Immune regulatory molecule presents on B lymphocyte | Neuroinflammation by suppressing local immune response [312] |
| 69 | SETD1A | Histone methyltransferase | Might affect synaptic signaling, excitation and glutamatergic signaling [313] |
| 70 | NOD2 | Immune homeostasis | Nox-2-mediated oxidative stress and neuroinflammation followed by loss of dopaminergic neurons [314] |
| 71 | CASC16 | - | - |
| 72 | CHD9 | Activates transcription factor CREBPP, Involve in Notch signaling | Aberrant survival signaling pathway [315] |
| 73 | CHRNB1 | Encodes subunit of the n-acetylcholine receptor, Ion channels, transporters, and neurotransmitter signaling [239] | Disturbed cholinergic signaling |
| 74 | RETREG3 | Involved in ER autophagy | Activation of ER autophagy by mTOR signaling [316] |
| 75 | UBTF | Transcription factor associated with ds-DNA break and apoptosis | Altered protein expression [317] |
| 76 | BRIP1 | Facilitates repair of SSBs and DSBs | Excitotoxicity, mitochondrial damage, and cell death [318] |
| 77 | DNAH17 | Encodes dynein axonemal heavy chain 17 involved in cytokinesis, microtubule-based movement, mitotic spindle organization, meiotic nuclear division [319] | - |
| 78 | ASXL3 | - | - |
| 79 | RIT2 | Involved in lysosomal activity | Activate LRRK2 gene and lysosomal dysfunction and leads to a-synuclein deposition [320] |
| 80 | MEX3C | Encodes RNF 194, an RNA binding protein impart immunoregulatory role | Neuroinflammation [321] |
| 81 | SPPL2B | - | - |
| 82 | CRLS1 | Encodes cardiolipin synthase 1, involved in mitochondrial membrane formation | Mitophagy [322] |
| 83 | DYRK1A | Synaptic and nuclear proteins, including transcription factors | Causes phosphorylation of a-synuclein and downregulates PI3K/AKT pathway to induce apoptosis [323] |
| 84 | FAM171A2 | Downstream of GRN, is a novel genetic regulator of progranulin production expressed on microglial surface | Downregulates progranulin level in brain [324] |
| 85 | CRHR1 | Encodes corticotropin-releasing hormone receptor, involved in regulation of stress and immune responses | Downregulates CREB signaling [325] |
| 86 | WNT3 | Immune regulation | PD-related gene expression in immune cells [291] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Firdaus, Z.; Li, X. Unraveling the Genetic Landscape of Neurological Disorders: Insights into Pathogenesis, Techniques for Variant Identification, and Therapeutic Approaches. Int. J. Mol. Sci. 2024, 25, 2320. https://doi.org/10.3390/ijms25042320
Firdaus Z, Li X. Unraveling the Genetic Landscape of Neurological Disorders: Insights into Pathogenesis, Techniques for Variant Identification, and Therapeutic Approaches. International Journal of Molecular Sciences. 2024; 25(4):2320. https://doi.org/10.3390/ijms25042320
Chicago/Turabian StyleFirdaus, Zeba, and Xiaogang Li. 2024. "Unraveling the Genetic Landscape of Neurological Disorders: Insights into Pathogenesis, Techniques for Variant Identification, and Therapeutic Approaches" International Journal of Molecular Sciences 25, no. 4: 2320. https://doi.org/10.3390/ijms25042320
APA StyleFirdaus, Z., & Li, X. (2024). Unraveling the Genetic Landscape of Neurological Disorders: Insights into Pathogenesis, Techniques for Variant Identification, and Therapeutic Approaches. International Journal of Molecular Sciences, 25(4), 2320. https://doi.org/10.3390/ijms25042320





