Impact of Histone Lysine Methyltransferase SUV4-20H2 on Cancer Onset and Progression with Therapeutic Potential
Abstract
:1. Introduction
1.1. DNA Methylation: A Key Epigenetic Mechanism in Cancer
1.2. The Role of Histone Modifications in Cancer
2. Biochemical Aspects of SUV4-20H2
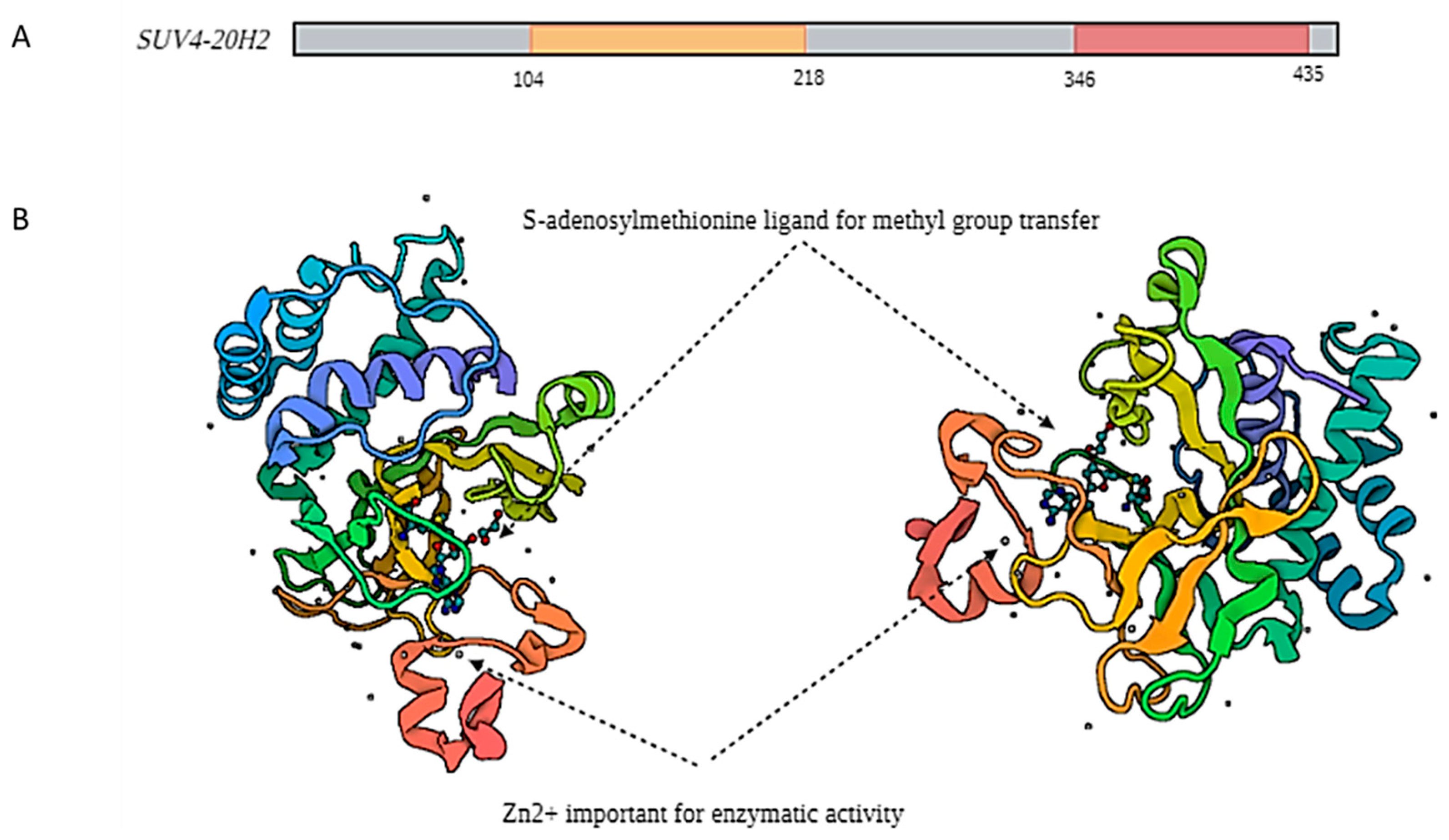
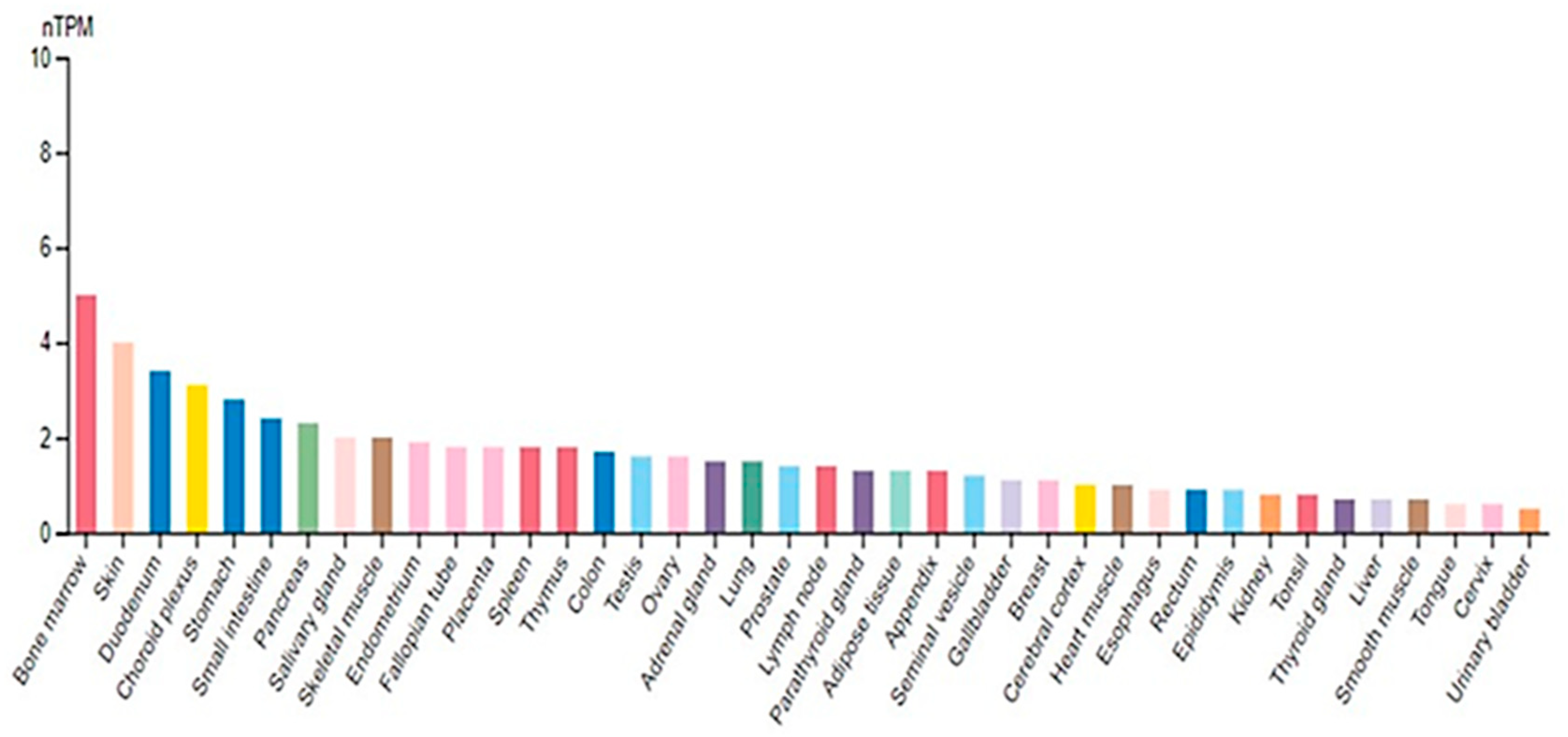
2.1. SUV4-20H2 Structural Characteristics
2.2. SUV4-20H2 Enzymatic Properties
3. SUV4-20H2 Functions in Normal Physiology
3.1. SUV4-20H2′s Role in DNA Replication
3.2. SUV4-20H2 Implication in B Cell and Erythrocyte Maturation
3.3. SUV4-20H2 Implication in Heterochromatin Formation and Chromosomal Integrity
3.4. SUV4-20H2 Participation in Neuronal Differentiation
4. Involvement of SUV4-20H2 in Cancer
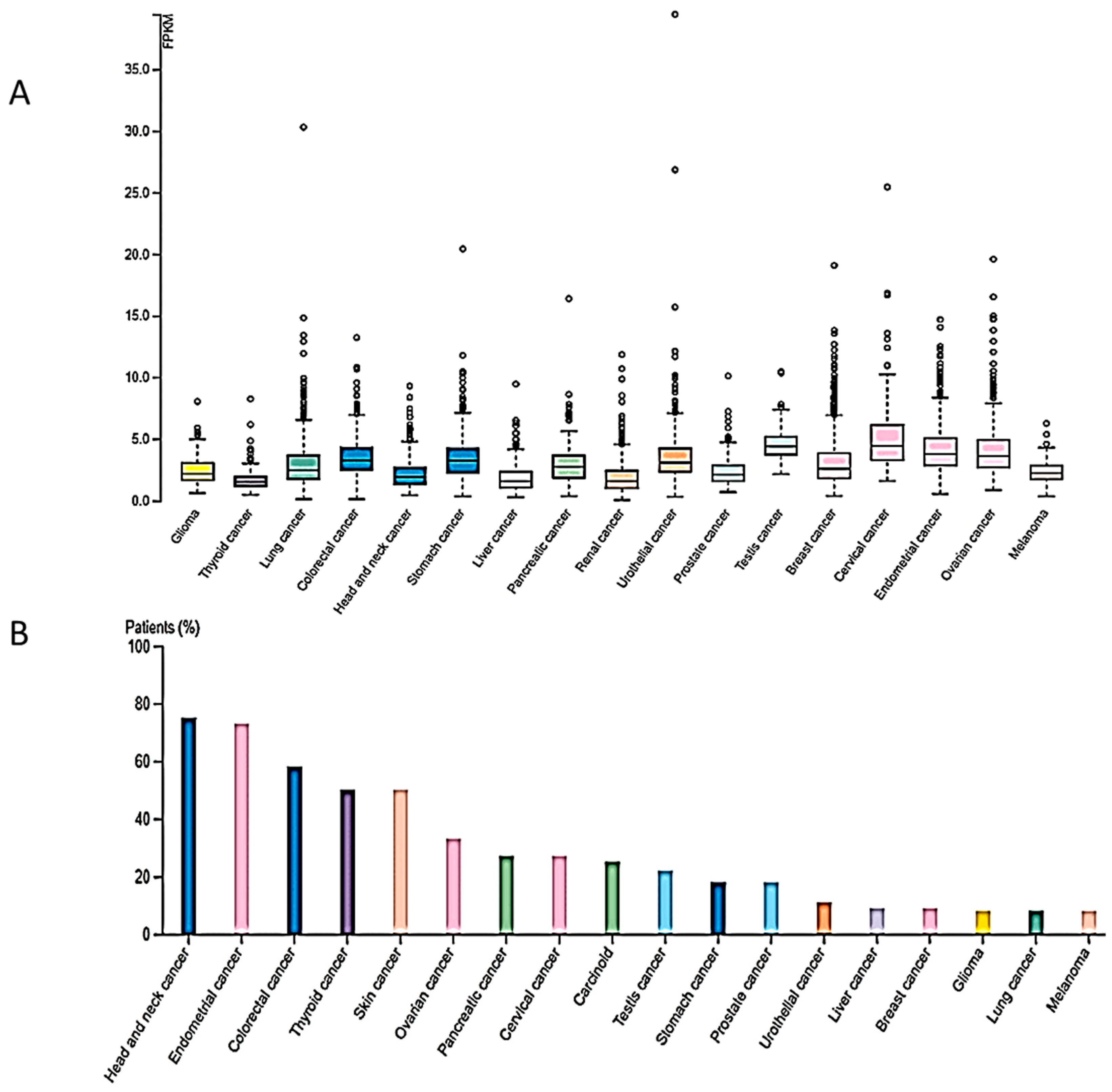
4.1. Cancer Types Exhibiting Reduced SUV4-20H2 Expression
4.1.1. Breast Cancer
4.1.2. Colon Cancer
4.1.3. Lung Cancer
4.2. Cancer Types Exhibiting Increased SUV4-20H2 Expression
4.2.1. Pancreatic Cancer
4.2.2. Renal Cell Carcinoma
4.2.3. Hepatocellular Carcinoma
5. Targeting Options of SUV4-20H2
6. Clinical Trials Based on Histone Methyltransferase Activity
6.1. Targeting of KMT2A and DOT1L to Combat Different Forms of Myeloid Leukemia
6.2. Targeting of EZH2 in Various Cancers
7. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Shamsi, M.B.; Firoz, A.S.; Imam, S.N.; Alzaman, N.; Samman, M.A. Epigenetics of Human Diseases and Scope in Future Therapeutics. J. Taibah Univ. Med. Sci. 2017, 12, 205–211. [Google Scholar] [CrossRef]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in Cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar] [CrossRef]
- Leick, M.B.; Shoff, C.J.; Wang, E.C.; Congress, J.L.; Gallicano, G.I. Loss of Imprinting of IGF2 and the Epigenetic Progenitor Model of Cancer. Am. J. Stem Cells 2012, 1, 59–74. [Google Scholar]
- Chen, J.F.; Yan, Q. The Roles of Epigenetics in Cancer Progression and Metastasis. Biochem. J. 2021, 478, 3373–3393. [Google Scholar] [CrossRef]
- Cheng, Y.; He, C.; Wang, M.; Ma, X.; Mo, F.; Yang, S.; Han, J.; Wei, X. Targeting Epigenetic Regulators for Cancer Therapy: Mechanisms and Advances in Clinical Trials. Signal Transduct. Target. Ther. 2019, 4, 62. [Google Scholar] [CrossRef]
- Ahuja, N.; Sharma, A.R.; Baylin, S.B. Epigenetic Therapeutics: A New Weapon in the War Against Cancer. Annu. Rev. Med. 2016, 67, 73–89. [Google Scholar] [CrossRef]
- Martisova, A.; Holcakova, J.; Izadi, N.; Sebuyoya, R.; Hrstka, R.; Bartosik, M. DNA Methylation in Solid Tumors: Functions and Methods of Detection. Int. J. Mol. Sci. 2021, 22, 4247. [Google Scholar] [CrossRef]
- Nishiyama, A.; Nakanishi, M. Navigating the DNA Methylation Landscape of Cancer. Trends Genet. 2021, 37, 1012–1027. [Google Scholar] [CrossRef]
- Xie, W.; Schultz, M.D.; Lister, R.; Hou, Z.; Rajagopal, N.; Ray, P.; Whitaker, J.W.; Tian, S.; Hawkins, R.D.; Leung, D.; et al. Epigenomic Analysis of Multilineage Differentiation of Human Embryonic Stem Cells. Cell 2013, 153, 1134–1148. [Google Scholar] [CrossRef]
- Pappalardo, X.G.; Barra, V. Losing DNA Methylation at Repetitive Elements and Breaking Bad. Epigenetics Chromatin 2021, 14, 25. [Google Scholar] [CrossRef]
- Messerschmidt, D.M.; Knowles, B.B.; Solter, D. DNA Methylation Dynamics during Epigenetic Reprogramming in the Germline and Preimplantation Embryos. Genes Dev. 2014, 28, 812–828. [Google Scholar] [CrossRef]
- Cusack, M.; King, H.W.; Spingardi, P.; Kessler, B.M.; Klose, R.J.; Kriaucionis, S. Distinct Contributions of DNA Methylation and Histone Acetylation to the Genomic Occupancy of Transcription Factors. Genome Res. 2020, 30, 1393–1406. [Google Scholar] [CrossRef]
- Nan, X.; Ng, H.H.; Johnson, C.A.; Laherty, C.D.; Turner, B.M.; Eisenman, R.N.; Bird, A. Transcriptional Repression by the Methyl-CpG-Binding Protein MeCP2 Involves a Histone Deacetylase Complex. Nature 1998, 393, 386–389. [Google Scholar] [CrossRef]
- Wade, P.A.; Gegonne, A.; Jones, P.L.; Ballestar, E.; Aubry, F.; Wolffe, A.P. Mi-2 Complex Couples DNA Methylation to Chromatin Remodelling and Histone Deacetylation. Nat. Genet. 1999, 23, 62–66. [Google Scholar] [CrossRef]
- Lee, H.-T.; Oh, S.; Ro, D.H.; Yoo, H.; Kwon, Y.-W. The Key Role of DNA Methylation and Histone Acetylation in Epigenetics of Atherosclerosis. J. Lipid Atheroscler. 2020, 9, 419–434. [Google Scholar] [CrossRef]
- Kim, S. New and Emerging Factors in Tumorigenesis: An Overview. Cancer Manag. Res. 2015, 7, 225–239. [Google Scholar] [CrossRef]
- Lakshminarasimhan, R.; Liang, G. The Role of DNA Methylation in Cancer. In DNA Methyltransferases—Role and Function; Jeltsch, A., Jurkowska, R.Z., Eds.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2016; Volume 945, pp. 151–172. [Google Scholar] [CrossRef]
- Vitorakis, N.; Piperi, C. Insights into the Role of Histone Methylation in Brain Aging and Potential Therapeutic Interventions. Int. J. Mol. Sci. 2023, 24, 17339. [Google Scholar] [CrossRef]
- Markouli, M.; Strepkos, D.; Piperi, C. Structure, Activity and Function of the SETDB1 Protein Methyltransferase. Life 2021, 11, 817. [Google Scholar] [CrossRef]
- Markouli, M.; Strepkos, D.; Basdra, E.K.; Papavassiliou, A.G.; Piperi, C. Prominent Role of Histone Modifications in the Regulation of Tumor Metastasis. Int. J. Mol. Sci. 2021, 22, 2778. [Google Scholar] [CrossRef]
- Angelopoulou, E.; Pyrgelis, E.-S.; Ahire, C.; Suman, P.; Mishra, A.; Piperi, C. Functional Implications of Protein Arginine Methyltransferases (PRMTs) in Neurodegenerative Diseases. Biology 2023, 12, 1257. [Google Scholar] [CrossRef]
- Jarrold, J.; Davies, C.C. PRMTs and Arginine Methylation: Cancer’s Best-Kept Secret? Trends Mol. Med. 2019, 25, 993–1009. [Google Scholar] [CrossRef]
- Liu, L.; Kimball, S.; Liu, H.; Holowatyj, A.; Yang, Z.-Q. Genetic Alterations of Histone Lysine Methyltransferases and Their Significance in Breast Cancer. Oncotarget 2015, 6, 2466–2482. [Google Scholar] [CrossRef]
- Zhu, Z.; Jiang, L.; Ding, X. Advancing Breast Cancer Heterogeneity Analysis: Insights from Genomics, Transcriptomics and Proteomics at Bulk and Single-Cell Levels. Cancers 2023, 15, 4164. [Google Scholar] [CrossRef]
- Yu, W.; Liu, N.; Song, X.; Chen, L.; Wang, M.; Xiao, G.; Li, T.; Wang, Z.; Zhang, Y. EZH2: An Accomplice of Gastric Cancer. Cancers 2023, 15, 425. [Google Scholar] [CrossRef]
- Chen, Y.; Ren, B.; Yang, J.; Wang, H.; Yang, G.; Xu, R.; You, L.; Zhao, Y. The Role of Histone Methylation in the Development of Digestive Cancers: A Potential Direction for Cancer Management. Signal Transduct. Target. Ther. 2020, 5, 143. [Google Scholar] [CrossRef]
- Q86Y97-KMT5C_HUMAN. The UniProt Consortium, UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. Available online: https://www.uniprot.org/uniprotkb/Q86Y97/entry#sequences (accessed on 17 November 2023). [CrossRef]
- Del Rizzo, P.A.; Trievel, R.C. Molecular Basis for Substrate Recognition by Lysine Methyltransferases and Demethylases. Biochim. Biophys. Acta BBA-Gene Regul. Mech. 2014, 1839, 1404–1415. [Google Scholar] [CrossRef]
- Southall, S.M.; Cronin, N.B.; Wilson, J.R. A novel route to product specificity in the Suv4-20 family of histone H4K20 methyltransferases. Nucleic Acids Res. 2014, 42, 661–671. [Google Scholar] [CrossRef]
- Gabellini, D.; Pedrotti, S. The SUV4-20H Histone Methyltransferases in Health and Disease. Int. J. Mol. Sci. 2022, 23, 4736. [Google Scholar] [CrossRef]
- Cheng, X.; Collins, R.E.; Zhang, X. Structural and Sequence Motifs of Protein (Histone) Methylation Enzymes. Annu. Rev. Biophys. Biomol. Struct. 2005, 34, 267–294. [Google Scholar] [CrossRef]
- Wu, H.; Siarheyeva, A.; Zeng, H.; Lam, R.; Dong, A.; Wu, X.-H.; Li, Y.; Schapira, M.; Vedadi, M.; Min, J. Crystal Structures of the Human Histone H4K20 Methyltransferases SUV420H1 and SUV420H2. FEBS Lett. 2013, 587, 3859–3868. [Google Scholar] [CrossRef]
- Weirich, S.; Kudithipudi, S.; Jeltsch, A. Specificity of the SUV4–20H1 and SUV4–20H2 Protein Lysine Methyltransferases and Methylation of Novel Substrates. J. Mol. Biol. 2016, 428, 2344–2358. [Google Scholar] [CrossRef]
- Beck, D.B.; Burton, A.; Oda, H.; Ziegler-Birling, C.; Torres-Padilla, M.-E.; Reinberg, D. The Role of PR-Set7 in Replication Licensing Depends on Suv4-20h. Genes Dev. 2012, 26, 2580–2589. [Google Scholar] [CrossRef]
- Schotta, G.; Sengupta, R.; Kubicek, S.; Malin, S.; Kauer, M.; Callén, E.; Celeste, A.; Pagani, M.; Opravil, S.; De La Rosa-Velazquez, I.A.; et al. A Chromatin-Wide Transition to H4K20 Monomethylation Impairs Genome Integrity and Programmed DNA Rearrangements in the Mouse. Genes Dev. 2008, 22, 2048–2061. [Google Scholar] [CrossRef]
- Brustel, J.; Kirstein, N.; Izard, F.; Grimaud, C.; Prorok, P.; Cayrou, C.; Schotta, G.; Abdelsamie, A.F.; Déjardin, J.; Méchali, M.; et al. Histone H4K20 Tri-Methylation at Late-Firing Origins Ensures Timely Heterochromatin Replication. EMBO J. 2017, 36, 2726–2741. [Google Scholar] [CrossRef]
- Eid, A.; Rodriguez-Terrones, D.; Burton, A.; Torres-Padilla, M.-E. SUV4-20 Activity in the Preimplantation Mouse Embryo Controls Timely Replication. Genes Dev. 2016, 30, 2513–2526. [Google Scholar] [CrossRef]
- Kurup, J.T.; Han, Z.; Jin, W.; Kidder, B.L. H4K20me3 Methyltransferase SUV420H2 Shapes the Chromatin Landscape of Pluripotent Embryonic Stem Cells. Dev. Camb. Engl. 2020, 147, dev188516. [Google Scholar] [CrossRef]
- Rodríguez-Cortez, V.C.; Martínez-Redondo, P.; Català-Moll, F.; Rodríguez-Ubreva, J.; Garcia-Gomez, A.; Poorani-Subramani, G.; Ciudad, L.; Hernando, H.; Pérez-García, A.; Company, C.; et al. Activation-Induced Cytidine Deaminase Targets SUV4-20-Mediated Histone H4K20 Trimethylation to Class-Switch Recombination Sites. Sci. Rep. 2017, 7, 7594. [Google Scholar] [CrossRef]
- Kapoor-Vazirani, P.; Kagey, J.D.; Vertino, P.M. SUV420H2-Mediated H4K20 Trimethylation Enforces RNA Polymerase II Promoter-Proximal Pausing by Blocking hMOF-Dependent H4K16 Acetylation. Mol. Cell. Biol. 2011, 31, 1594–1609. [Google Scholar] [CrossRef]
- Gillinder, K.R.; Tuckey, H.; Bell, C.C.; Magor, G.W.; Huang, S.; Ilsley, M.D.; Perkins, A.C. Direct Targets of pSTAT5 Signalling in Erythropoiesis. PLoS ONE 2017, 12, e0180922. [Google Scholar] [CrossRef]
- Benetti, R.; Gonzalo, S.; Jaco, I.; Schotta, G.; Klatt, P.; Jenuwein, T.; Blasco, M.A. Suv4-20h Deficiency Results in Telomere Elongation and Derepression of Telomere Recombination. J. Cell Biol. 2007, 178, 925–936. [Google Scholar] [CrossRef]
- Ballmer, D.; Tardat, M.; Ortiz, R.; Graff-Meyer, A.; Ozonov, E.A.; Genoud, C.; Peters, A.H.; Fanourgakis, G. HP1 Proteins Regulate Nucleolar Structure and Function by Secluding Pericentromeric Constitutive Heterochromatin. Nucleic Acids Res. 2023, 51, 117–143. [Google Scholar] [CrossRef]
- Makrantoni, V.; Marston, A.L. Cohesin and Chromosome Segregation. Curr. Biol. CB 2018, 28, R688–R693. [Google Scholar] [CrossRef]
- Hahn, M.; Dambacher, S.; Dulev, S.; Kuznetsova, A.Y.; Eck, S.; Wörz, S.; Sadic, D.; Schulte, M.; Mallm, J.-P.; Maiser, A.; et al. Suv4-20h2 Mediates Chromatin Compaction and Is Important for Cohesin Recruitment to Heterochromatin. Genes Dev. 2013, 27, 859–872. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Ma, L.; Nie, M.; Ju, J.; Liu, M.; Deng, Y.; Yao, B.; Gui, T.; Li, X.; et al. Heterochromatin Protein 1γ Is a Novel Epigenetic Repressor of Human Embryonic Ɛ-Globin Gene Expression. J. Biol. Chem. 2017, 292, 4811–4817. [Google Scholar] [CrossRef]
- Bosch-Presegué, L.; Raurell-Vila, H.; Thackray, J.K.; González, J.; Casal, C.; Kane-Goldsmith, N.; Vizoso, M.; Brown, J.P.; Gómez, A.; Ausió, J.; et al. Mammalian HP1 Isoforms Have Specific Roles in Heterochromatin Structure and Organization. Cell Rep. 2017, 21, 2048–2057. [Google Scholar] [CrossRef]
- Nicetto, D.; Hahn, M.; Jung, J.; Schneider, T.D.; Straub, T.; David, R.; Schotta, G.; Rupp, R.A.W. Suv4-20h Histone Methyltransferases Promote Neuroectodermal Differentiation by Silencing the Pluripotency-Associated Oct-25 Gene. PLoS Genet. 2013, 9, e1003188. [Google Scholar] [CrossRef]
- Rhodes, C.T.; Zunino, G.; Huang, S.-W.A.; Cardona, S.M.; Cardona, A.E.; Berger, M.S.; Lemmon, V.P.; Lin, C.-H.A. Region Specific Knock-out Reveals Distinct Roles of Chromatin Modifiers in Adult Neurogenic Niches. Cell Cycle 2018, 17, 377–389. [Google Scholar] [CrossRef]
- Wickramasekara, R.; Stessman, H. Histone 4 Lysine 20 Methylation: A Case for Neurodevelopmental Disease. Biology 2019, 8, 11. [Google Scholar] [CrossRef]
- Tryndyak, V.P.; Kovalchuk, O.; Pogribny, I.P. Loss of DNA Methylation and Histone H4 Lysine 20 Trimethylation in Human Breast Cancer Cells Is Associated with Aberrant Expression of DNA Methyltransferase 1, Suv4-20h2 Histone Methyltransferase and Methyl-Binding Proteins. Cancer Biol. Ther. 2006, 5, 65–70. [Google Scholar] [CrossRef]
- Isin, H.; Özgür, E.; Kelten Talu, C.; Can Trabulus, D.; Karaçetin, D.; Gezer, U. Impact of Histone Methyltransferase SUV420H2 in Breast Cancer. Biomed. Rep. 2020. [Google Scholar] [CrossRef]
- Shinchi, Y.; Hieda, M.; Nishioka, Y.; Matsumoto, A.; Yokoyama, Y.; Kimura, H.; Matsuura, S.; Matsuura, N. SUV420H2 Suppresses Breast Cancer Cell Invasion through down Regulation of the SH2 Domain-Containing Focal Adhesion Protein Tensin-3. Exp. Cell Res. 2015, 334, 90–99. [Google Scholar] [CrossRef]
- Wu, Y.; Shi, W.; Tang, T.; Wang, Y.; Yin, X.; Chen, Y.; Zhang, Y.; Xing, Y.; Shen, Y.; Xia, T.; et al. miR-29a Contributes to Breast Cancer Cells Epithelial–Mesenchymal Transition, Migration, and Invasion via down-Regulating Histone H4K20 Trimethylation through Directly Targeting SUV420H2. Cell Death Dis. 2019, 10, 176. [Google Scholar] [CrossRef]
- Boonsanay, V.; Mosa, M.H.; Looso, M.; Weichenhan, D.; Ceteci, F.; Pudelko, L.; Lechel, A.; Michel, C.S.; Künne, C.; Farin, H.F.; et al. Loss of SUV420H2-Dependent Chromatin Compaction Drives Right-Sided Colon Cancer Progression. Gastroenterology 2023, 164, 214–227. [Google Scholar] [CrossRef]
- Van Den Broeck, A.; Brambilla, E.; Moro-Sibilot, D.; Lantuejoul, S.; Brambilla, C.; Eymin, B.; Gazzeri, S. Loss of Histone H4K20 Trimethylation Occurs in Preneoplasia and Influences Prognosis of Non-Small Cell Lung Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 7237–7245. [Google Scholar] [CrossRef]
- Viotti, M.; Wilson, C.; McCleland, M.; Koeppen, H.; Haley, B.; Jhunjhunwala, S.; Klijn, C.; Modrusan, Z.; Arnott, D.; Classon, M.; et al. SUV420H2 Is an Epigenetic Regulator of Epithelial/Mesenchymal States in Pancreatic Cancer. J. Cell Biol. 2018, 217, 763–777. [Google Scholar] [CrossRef]
- Ryu, T.Y.; Lee, J.; Kang, Y.; Son, M.-Y.; Kim, D.-S.; Lee, Y.S.; Kim, M.-Y.; Cho, H.-S. Epigenetic Regulation of DHRS2 by SUV420H2 Inhibits Cell Apoptosis in Renal Cell Carcinoma. Biochem. Biophys. Res. Commun. 2023, 663, 41–46. [Google Scholar] [CrossRef]
- Pogribny, I.P.; Ross, S.A.; Tryndyak, V.P.; Pogribna, M.; Poirier, L.A.; Karpinets, T.V. Histone H3 Lysine 9 and H4 Lysine 20 Trimethylation and the Expression of Suv4-20h2 and Suv-39h1 Histone Methyltransferases in Hepatocarcinogenesis Induced by Methyl Deficiency in Rats. Carcinogenesis 2006, 27, 1180–1186. [Google Scholar] [CrossRef]
- Phoyen, S.; Sanpavat, A.; Ma-on, C.; Stein, U.; Hirankarn, N.; Tangkijvanich, P.; Jindatip, D.; Whongsiri, P.; Boonla, C. H4K20me3 Upregulated by Reactive Oxygen Species Is Associated with Tumor Progression and Poor Prognosis in Patients with Hepatocellular Carcinoma. Heliyon 2023, 9, e22589. [Google Scholar] [CrossRef]
- Markouli, M.; Strepkos, D.; Papavassiliou, K.A.; Papavassiliou, A.G.; Piperi, C. Crosstalk of Epigenetic and Metabolic Signaling Underpinning Glioblastoma Pathogenesis. Cancers 2022, 14, 2655. [Google Scholar] [CrossRef]
- Skouras, P.; Markouli, M.; Strepkos, D.; Piperi, C. Advances on Epigenetic Drugs for Pediatric Brain Tumors. Curr. Neuropharmacol. 2023, 21, 1519–1535. [Google Scholar] [CrossRef]
- Kunadis, E.; Lakiotaki, E.; Korkolopoulou, P.; Piperi, C. Targeting Post-Translational Histone Modifying Enzymes in Glioblastoma. Pharmacol. Ther. 2021, 220, 107721. [Google Scholar] [CrossRef]
- Kaniskan, H.Ü.; Martini, M.L.; Jin, J. Inhibitors of Protein Methyltransferases and Demethylases. Chem. Rev. 2018, 118, 989–1068. [Google Scholar] [CrossRef]
- Wang, M.Y.; Liow, P.; Guzman, M.I.T.; Qi, J. Exploring Methods of Targeting Histone Methyltransferases and Their Applications in Cancer Therapeutics. ACS Chem. Biol. 2022, 17, 744–755. [Google Scholar] [CrossRef]
- Feoli, A.; Viviano, M.; Cipriano, A.; Milite, C.; Castellano, S.; Sbardella, G. Lysine Methyltransferase Inhibitors: Where We Are Now. RSC Chem. Biol. 2022, 3, 359–406. [Google Scholar] [CrossRef]
- Bromberg, K.D.; Mitchell, T.R.H.; Upadhyay, A.K.; Jakob, C.G.; Jhala, M.A.; Comess, K.M.; Lasko, L.M.; Li, C.; Tuzon, C.T.; Dai, Y.; et al. The SUV4-20 Inhibitor A-196 Verifies a Role for Epigenetics in Genomic Integrity. Nat. Chem. Biol. 2017, 13, 317–324. [Google Scholar] [CrossRef]
- Pavlidis, A.; Piperi, C.; Papadavid, E. Novel Therapeutic Approaches for Cutaneous T Cell Lymphomas. Expert Rev. Clin. Immunol. 2021, 17, 629–641. [Google Scholar] [CrossRef]
- Maksimova, V.; Makus, J.; Popova, V.; Prus, A.; Usalka, O.; Trapeznikova, E.; Zhidkova, E.; Belitsky, G.; Yakubovskaya, M.; Kirsanov, K. Histone Methyltransferases as a New Target for Epigenetic Action of Vorinostat. Biochem. Mosc. 2023, 88, 968–978. [Google Scholar] [CrossRef]
- Morera, L.; Lübbert, M.; Jung, M. Targeting Histone Methyltransferases and Demethylases in Clinical Trials for Cancer Therapy. Clin. Epigenetics 2016, 8, 57. [Google Scholar] [CrossRef]
- Górecki, M.; Kozioł, I.; Kopystecka, A.; Budzyńska, J.; Zawitkowska, J.; Lejman, M. Updates in KMT2A Gene Rearrangement in Pediatric Acute Lymphoblastic Leukemia. Biomedicines 2023, 11, 821. [Google Scholar] [CrossRef]
- Patel, A.; Dharmarajan, V.; Vought, V.E.; Cosgrove, M.S. On the Mechanism of Multiple Lysine Methylation by the Human Mixed Lineage Leukemia Protein-1 (MLL1) Core Complex. J. Biol. Chem. 2009, 284, 24242–24256. [Google Scholar] [CrossRef]
- Dou, Y.; Milne, T.A.; Tackett, A.J.; Smith, E.R.; Fukuda, A.; Wysocka, J.; Allis, C.D.; Chait, B.T.; Hess, J.L.; Roeder, R.G. Physical Association and Coordinate Function of the H3 K4 Methyltransferase MLL1 and the H4 K16 Acetyltransferase MOF. Cell 2005, 121, 873–885. [Google Scholar] [CrossRef]
- Mims, A.S.; Mishra, A.; Orwick, S.; Blachly, J.; Klisovic, R.B.; Garzon, R.; Walker, A.R.; Devine, S.M.; Walsh, K.J.; Vasu, S.; et al. A Novel Regimen for Relapsed/Refractory Adult Acute Myeloid Leukemia Using a KMT2A Partial Tandem Duplication Targeted Therapy: Results of Phase 1 Study NCI 8485. Haematologica 2018, 103, 982–987. [Google Scholar] [CrossRef]
- Dzhoshibaev, S.D.; Lisiukov, N.V.; Rysbekov, A.N.; Tumasov, V.M.; Pegov, N.A. Plastika khord stvorki mitral’nogo klapana pri ego nedostatochnosti [Plastic surgery of the mitral valve chordae tendineae in mitral valve insufficiency]. Grudn Khir. 1987, 90–91. [Google Scholar]
- Feng, Q.; Wang, H.; Ng, H.H.; Erdjument-Bromage, H.; Tempst, P.; Struhl, K.; Zhang, Y. Methylation of H3-Lysine 79 Is Mediated by a New Family of HMTases without a SET Domain. Curr. Biol. CB 2002, 12, 1052–1058. [Google Scholar] [CrossRef]
- Stein, E.M.; Garcia-Manero, G.; Rizzieri, D.A.; Tibes, R.; Berdeja, J.G.; Savona, M.R.; Jongen-Lavrenic, M.; Altman, J.K.; Thomson, B.; Blakemore, S.J.; et al. The DOT1L Inhibitor Pinometostat Reduces H3K79 Methylation and Has Modest Clinical Activity in Adult Acute Leukemia. Blood 2018, 131, 2661–2669. [Google Scholar] [CrossRef]
- McCabe, M.T.; Graves, A.P.; Ganji, G.; Diaz, E.; Halsey, W.S.; Jiang, Y.; Smitheman, K.N.; Ott, H.M.; Pappalardi, M.B.; Allen, K.E.; et al. Mutation of A677 in Histone Methyltransferase EZH2 in Human B-Cell Lymphoma Promotes Hypertrimethylation of Histone H3 on Lysine 27 (H3K27). Proc. Natl. Acad. Sci. USA 2012, 109, 2989–2994. [Google Scholar] [CrossRef]
- Izutsu, K.; Ando, K.; Nishikori, M.; Shibayama, H.; Teshima, T.; Kuroda, J.; Kato, K.; Imaizumi, Y.; Nosaka, K.; Sakai, R.; et al. Phase II Study of Tazemetostat for Relapsed or Refractory B-Cell Non-Hodgkin Lymphoma with EZH2 Mutation in Japan. Cancer Sci. 2021, 112, 3627–3635. [Google Scholar] [CrossRef]
- Zauderer, M.G.; Szlosarek, P.W.; Le Moulec, S.; Popat, S.; Taylor, P.; Planchard, D.; Scherpereel, A.; Koczywas, M.; Forster, M.; Cameron, R.B.; et al. EZH2 Inhibitor Tazemetostat in Patients with Relapsed or Refractory, BAP1-Inactivated Malignant Pleural Mesothelioma: A Multicentre, Open-Label, Phase 2 Study. Lancet Oncol. 2022, 23, 758–767. [Google Scholar] [CrossRef]
- Song, Y.; Liu, Y.; Li, Z.-M.; Li, L.; Su, H.; Jin, Z.; Zuo, X.; Wu, J.; Zhou, H.; Li, K.; et al. SHR2554, an EZH2 Inhibitor, in Relapsed or Refractory Mature Lymphoid Neoplasms: A First-in-Human, Dose-Escalation, Dose-Expansion, and Clinical Expansion Phase 1 Trial. Lancet Haematol. 2022, 9, e493–e503. [Google Scholar] [CrossRef]
| Molecular Process | Role of SUV4-20H2 | Cell Type | Reference |
|---|---|---|---|
| Recombination | Maintenance of telomeric length homeostasis | Μouse embryonic fibroblasts, murine embryonic stem cells | [43] |
| Immunoglobulin class switch recombination | Μature B cells | [40,41] | |
| Transcriptional regulation | RNA pol II promoter proximal pausing | Μature erythrocytes | [40,41,42] |
| Direct regulation of Oct-25 gene during neuroectodermal differentiation | Μelanocytes and eye cells from Xenopus laevis, neural progenitor cells of murine brains from the subgranular zone of dante gyrus and the subventricular zone adjacent to the lateral ventricle | [49,50,51] | |
| Repression of lineage-specific genes in ES cells, embryonic stem cell differentiation | Εmbryonic stem cells | [38,39] | |
| Control of chromatin architecture, namely the establishment of heterochromatin | Ιn diverse cell types, very important for red cell maturation | [44,46,47,48] | |
| DNA replication | Recruitment of origin of recognition complex (ORC) to replication origin sites | Mouse embryonic fibroblasts | [35] |
| Assistance in the correct replication time in heterochromatic regions | - | [37] |
| Cancer Type | SUV420H2 Expression | H4K20me3 Expression | Main Effects | Reference |
|---|---|---|---|---|
| Breast | High levels in low-grade tumors Low levels in high-grade tumors | Low | Repression of genes responsible for cell adhesion, tensin-3 blocked by miR-29a | [50,51] |
| Colon | Low levels | Low | Enrichment of genes implicated in Wnt pathway and dissociation of LAD areas in mouse embryonic fibroblasts, attenuating gene repression | [52] |
| Lung | Low levels | Low | Loss of H4K20me3 correlates with worse survival in stage I adenocarcinoma patients | [53] |
| Pancreatic | High levels | High | Regulation of MET-associated transcriptional factors, FOXA1 and OVOL 1/2, regulation of epithelial or mesenchymal state | [54] |
| Renal cell carcinoma | High levels | High | Maintenance of H4K20me3 levels on DHRS2 promoter to maintain growth and avoid apoptosis | [55] |
| Hepatocellular | High levels in early stages, which drop with disease progression | Low | Reduction in epithelial marker E-cadherin and increase in mesenchymal markers SMA and MMP-9 | [57] |
| Clinical Trial Identifier | Targeted HMT | Drug | Diagnosis/Cancer Type | Patients (Number, Genetic Characteristics, Previous Treatments) | Results | Reference |
|---|---|---|---|---|---|---|
| NCT1130506 | KMT2A (lysine methyltransferase 2A) | Dacitabine (intravenously at 20 mg/m2/day on days 1–10), vorinostat (oral administration at 400 mg/day on days 5–10), cytarabine (intravenous infusion twice a day on days 12, 14 and 16 with gradual increase in dosage from 1.5 g to 3 g/m2. | Acute myeloid leukemia (AML) relapsed or refractory | 17 adults with rearrangements in KMT2A gene and a median of 2 prior treatments (range 1–3) | 35% overall response with 6/17 presenting complete response. 5/6 relapsed | [75] |
| NCT04065399 | KMT2A | Revumenib (oral administration in two parallel dose escalation cohorts, one receiving a CYP3A4 inhibitor and the other not) | Acute myeloid leukemia (AML) relapsed or refractory | 60 patients with mutations in KMT2A and NPM1 gene all heavily medicated with a median of 4 previous treatments, 44% received allogeneic stem cell transplant before participation | 53% overall response with 32/60 presenting complete response and complete remission or remission with hematologic response reaching 30% | [76] |
| NCT01684150 | DOT1L (disruptor of telomeric silencing 1) | Pinometostat (EPZ-5676) (intravenous administration at two different doses of 54 mg/m2 and 94 mg/m2 per day) | Advanced acute leukemia, mainly mixed lineage | 51 patients enrolled in two cohorts (n = 26 for the 6 dose escalation phase and n = 25 for the expansion cohort), translocations for the MLL gene (KMT2A) were present in the patients but not all of them had the same or similar translocation | Only 2 patients achieved complete remission bearing translocations (11:19) but the duration of remission was different and both relapsed eventually | [78] |
| NCT03456726 | EZH2 (enhancer of zeste homologue 2) | Tamezostat (administration twice a day at 800 mg) | B cell non-Hodgkin lymphoma | 20 patients bearing mutations in EZH2, divided in two cohorts (n = 17 with follicular lymphoma and n = 3 with diffuse large B cell lymphoma) | Cohort 1: objective response rate 76.5% with 35.5% amounting to complete recovery and 41.2% to partial response, with progression without events at 12 months being 94.1% and at 15 months, 73.2%; Cohort 2: only partial response | [80] |
| NCT02860286 | EZH2 | Tamezostat | Malignant pleural mesothelioma | 74 patients (13 for the first part of the trial and 61 for the second), at least one form of treatment prior to participation, 99% had BAP1 inactivated tumors | No patient presented complete response to treatment but two presented partial response that lasted different time intervals (18 and 42 weeks) | [81] |
| NCT03603951 | EZH2 | SHR2554 | Mature lymphoid neoplasms, B and T cell lymphomas, classical Hodgkin lymphoma | 113 patients with 107 used for analysis | 46/107, 43%, developed an overall response to treatment | [82] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papadaki, S.; Piperi, C. Impact of Histone Lysine Methyltransferase SUV4-20H2 on Cancer Onset and Progression with Therapeutic Potential. Int. J. Mol. Sci. 2024, 25, 2498. https://doi.org/10.3390/ijms25052498
Papadaki S, Piperi C. Impact of Histone Lysine Methyltransferase SUV4-20H2 on Cancer Onset and Progression with Therapeutic Potential. International Journal of Molecular Sciences. 2024; 25(5):2498. https://doi.org/10.3390/ijms25052498
Chicago/Turabian StylePapadaki, Stela, and Christina Piperi. 2024. "Impact of Histone Lysine Methyltransferase SUV4-20H2 on Cancer Onset and Progression with Therapeutic Potential" International Journal of Molecular Sciences 25, no. 5: 2498. https://doi.org/10.3390/ijms25052498







