Maternal Age at Menarche Genes Determines Fetal Growth Restriction Risk
Abstract
1. Introduction
2. Results
2.1. FGR and FGR Free Group Characteristics
2.2. SNP/Haplotype Association Analysis
| Parameters | FGR Patients ± SD/%(n) | Controls ± SD/%(n) | p-Value |
|---|---|---|---|
| N | 273 | 631 | |
| Age, years | 27.18 ± 4.84 | 26.57 ± 4.94 | 0.017 |
| Height, m | 1.65 ± 0.06 | 1.65 ± 0.06 | 0.52 |
| Weight, kg | 63.53 ± 11.81 | 63.50 ± 11.22 | 0.78 |
| Pre-pregnancy BMI, kg/m2 | 23.40 ± 4.31 | 23.40 ± 3.47 | 0.23 |
| Underweight (<18.50) Normal weight (18.50–24.99) Overweight (25.00–29.99) Obesity (>30) | 8.69 (24) 66.30 (181) 14.29 (39) 10.62 (29) | 5.55 (35) 66.72 (421) 22.19 (140) 5.55 (35) | 0.0001 |
| Age at menarche and menstrual cycle | |||
| Age at menarche, years | 12.80 ± 1.14 | 12.83 ± 1.17 | 0.51 |
| Early (<12) Average (12–14) Late (>14) | 3.66 (10) 90.11 (246) 6.23 (17) | 6.5 (41) 86.69 (547) 6.81 (43) | 0.23 |
| Menstrual cycle length, days | 28.11 ± 1.64 | 28.05 ± 1.93 | 0.19 |
| Duration of menstrual bleeding, days | 5.02 ± 0.85 | 5.04 ± 1.03 | 0.93 |
| Reproductive characteristic | |||
| First pregnancy | 34.93 (95) | 40.41 (255) | 0.19 |
| No. of gravidity | 1.47 ± 1.55 | 1.07 ± 2.04 | 0.004 |
| No. of births | 0.59 ± 0.69 | 0.56 ± 0.81 | 0.43 |
| Stillbirth | 0.01 ± 0.10 | 0.007 ± 0.08 | 0.62 |
| No. of spontaneous abortions | 0.23 ± 0.53 | 0.13 ± 0.35 | 0.07 |
| No. of induced abortions | 0.64 ± 0.99 | 0.37 ± 0.78 | 0.0001 |
| Risk factors | |||
| Smoking * | 61.54 (168) | 63.71 (402) | 0.87 |
| Alcohol ** | 81.68 (223) | 79.72 (503) | 0.79 |
| History of arterial hypertension | 8.79 (24) | 1.74 (11) | 0.0001 |
| History of sexually transmitted diseases | 35.56 (16) | 26.62 (168) | 0.28 |
| History of preeclampsia | 10.99 (30) | 3.80 (24) | 0.001 |
| History of FGR | 19.78 (54) | 0.79 (5) | 0.00001 |
2.3. Association Analysis of SNP Interactions
| Minore Allele (SNP) | Gene | Chr | N | Genetic Models | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allelic | Additive | Dominant | Recessive | ||||||||||||||||
| OR | 95%CI | p | OR | 95%CI | p | OR | 95%CI | p | OR | 95%CI | p | ||||||||
| L95 | U95 | L95 | U95 | L95 | U95 | L95 | U95 | ||||||||||||
| T (rs1514175) | TNNI3K | 1 | 896 | 1.08 | 0.88 | 1.33 | 0.454 | 1.04 | 0.83 | 1.30 | 0.759 | 1.04 | 0.75 | 1.44 | 0.827 | 1.07 | 0.69 | 1.65 | 0.764 |
| T (rs466639) | RXRG | 1 | 899 | 0.91 | 0.66 | 1.24 | 0.534 | 0.89 | 0.63 | 1.25 | 0.505 | 0.90 | 0.61 | 1.33 | 0.613 | 0.65 | 0.20 | 2.08 | 0.464 |
| G (rs7538038) | KISS1 | 1 | 898 | 0.63 | 0.49 | 0.81 | 0.0002 | 0.61 | 0.46 | 0.82 | 0.001 | 0.56 | 0.40 | 0.79 | 0.001 | 0.57 | 0.24 | 1.37 | 0.208 |
| C (rs713586) | RBJ | 2 | 899 | 0.97 | 0.79 | 1.19 | 0.775 | 0.97 | 0.78 | 1.21 | 0.777 | 0.97 | 0.69 | 1.36 | 0.838 | 0.95 | 0.64 | 1.40 | 0.790 |
| A (rs2164808) | POMC | 2 | 898 | 1.09 | 0.89 | 1.34 | 0.382 | 1.06 | 0.84 | 1.32 | 0.641 | 1.14 | 0.80 | 1.62 | 0.475 | 1.00 | 0.68 | 1.48 | 0.995 |
| A (rs7589318) | POMC | 2 | 900 | 0.98 | 0.79 | 1.22 | 0.867 | 0.96 | 0.75 | 1.22 | 0.739 | 1.16 | 0.84 | 1.59 | 0.364 | 0.49 | 0.27 | 0.91 | 0.025 |
| C (rs4374421) | LHCGR | 2 | 874 | 0.93 | 0.74 | 1.16 | 0.504 | 0.94 | 0.73 | 1.21 | 0.637 | 0.99 | 0.71 | 1.37 | 0.941 | 0.76 | 0.42 | 1.36 | 0.356 |
| T (rs7579411) | LHCGR | 2 | 891 | 0.91 | 0.74 | 1.11 | 0.351 | 0.98 | 0.78 | 1.25 | 0.890 | 0.85 | 0.60 | 1.20 | 0.347 | 1.19 | 0.80 | 1.79 | 0.392 |
| C (rs4953616) | LHCGR | 2 | 892 | 0.96 | 0.76 | 1.20 | 0.697 | 0.93 | 0.72 | 1.20 | 0.569 | 0.89 | 0.65 | 1.23 | 0.478 | 1.00 | 0.54 | 1.84 | 0.993 |
| G (rs6732220) | FSHR | 2 | 898 | 0.93 | 0.74 | 1.17 | 0.538 | 0.87 | 0.67 | 1.14 | 0.321 | 0.93 | 0.68 | 1.28 | 0.664 | 0.52 | 0.23 | 1.14 | 0.104 |
| G (rs4953655) | FSHR | 2 | 898 | 0.87 | 0.68 | 1.10 | 0.248 | 0.81 | 0.61 | 1.06 | 0.129 | 0.84 | 0.60 | 1.16 | 0.277 | 0.50 | 0.22 | 1.15 | 0.104 |
| A (rs12617311) | PLCL1 | 2 | 895 | 0.99 | 0.80 | 1.23 | 0.954 | 1.05 | 0.82 | 1.33 | 0.714 | 1.03 | 0.75 | 1.43 | 0.838 | 1.12 | 0.68 | 1.84 | 0.649 |
| C (rs6438424) | IGSF11 | 3 | 888 | 0.95 | 0.78 | 1.17 | 0.647 | 0.93 | 0.74 | 1.16 | 0.518 | 0.96 | 0.67 | 1.36 | 0.800 | 0.85 | 0.58 | 1.25 | 0.408 |
| A (rs2013573) | UGT2B4 | 4 | 899 | 1.23 | 0.95 | 1.60 | 0.120 | 1.20 | 0.88 | 1.62 | 0.249 | 1.22 | 0.87 | 1.70 | 0.252 | 1.27 | 0.42 | 3.83 | 0.676 |
| A (rs13111134) | UGT2B4 | 4 | 896 | 1.27 | 1.00 | 1.62 | 0.052 | 1.28 | 0.96 | 1.71 | 0.095 | 1.35 | 0.98 | 1.87 | 0.068 | 1.06 | 0.40 | 2.81 | 0.905 |
| C (rs222003) | GC | 4 | 900 | 1.21 | 0.80 | 1.83 | 0.360 | 1.07 | 0.67 | 1.71 | 0.765 | 1.05 | 0.65 | 1.72 | 0.833 | 2.01 | 0.18 | 22.37 | 0.571 |
| C (rs222020) | GC | 4 | 899 | 0.96 | 0.70 | 1.32 | 0.806 | 1.08 | 0.75 | 1.55 | 0.666 | 1.09 | 0.75 | 1.60 | 0.649 | 0.98 | 0.14 | 7.01 | 0.985 |
| G (rs3756261) | EGF | 4 | 898 | 1.00 | 0.69 | 1.47 | 0.983 | 1.01 | 0.66 | 1.56 | 0.954 | 1.01 | 0.64 | 1.58 | 0.963 | 1.11 | 0.09 | 14.24 | 0.935 |
| T (rs757647) | KDM3B | 5 | 889 | 0.94 | 0.74 | 1.19 | 0.621 | 0.99 | 0.75 | 1.31 | 0.955 | 1.08 | 0.78 | 1.49 | 0.658 | 0.59 | 0.25 | 1.41 | 0.237 |
| G (rs7766109) | F13A1 | 6 | 898 | 1.16 | 0.95 | 1.42 | 0.141 | 1.06 | 0.85 | 1.33 | 0.619 | 1.10 | 0.77 | 1.58 | 0.600 | 1.06 | 0.73 | 1.54 | 0.778 |
| A (rs4946651) | LIN28B | 6 | 900 | 1.00 | 0.81 | 1.22 | 0.972 | 1.14 | 0.90 | 1.44 | 0.274 | 1.20 | 0.86 | 1.69 | 0.288 | 1.16 | 0.75 | 1.77 | 0.504 |
| C (rs7759938) | LIN28B | 6 | 899 | 1.04 | 0.83 | 1.30 | 0.718 | 1.18 | 0.92 | 1.52 | 0.180 | 1.31 | 0.95 | 1.81 | 0.104 | 1.05 | 0.59 | 1.88 | 0.861 |
| T (rs314280) | LIN28B | 6 | 888 | 0.99 | 0.80 | 1.22 | 0.918 | 1.14 | 0.89 | 1.44 | 0.301 | 1.18 | 0.84 | 1.67 | 0.334 | 1.17 | 0.74 | 1.83 | 0.505 |
| A (rs314276) | LIN28B | 6 | 874 | 0.97 | 0.78 | 1.21 | 0.791 | 1.10 | 0.86 | 1.41 | 0.445 | 1.15 | 0.83 | 1.59 | 0.398 | 1.08 | 0.63 | 1.85 | 0.789 |
| G (rs3020394) | ESR1 | 6 | 899 | 1.24 | 1.00 | 1.54 | 0.049 | 1.17 | 0.92 | 1.49 | 0.202 | 1.07 | 0.78 | 1.47 | 0.687 | 1.74 | 1.05 | 2.88 | 0.033 |
| G (rs1884051) | ESR1 | 6 | 900 | 1.22 | 0.99 | 1.52 | 0.068 | 1.14 | 0.89 | 1.45 | 0.291 | 1.04 | 0.76 | 1.43 | 0.812 | 1.67 | 1.00 | 2.80 | 0.051 |
| C (rs7753051) | IGF2R | 6 | 898 | 1.01 | 0.81 | 1.26 | 0.910 | 0.93 | 0.72 | 1.20 | 0.579 | 1.05 | 0.76 | 1.44 | 0.772 | 0.53 | 0.27 | 1.05 | 0.068 |
| C (rs1079866) | INHBA | 7 | 899 | 0.89 | 0.68 | 1.16 | 0.386 | 0.88 | 0.65 | 1.19 | 0.403 | 0.91 | 0.64 | 1.28 | 0.579 | 0.55 | 0.19 | 1.60 | 0.275 |
| T (rs2288696) | FGFR1 | 8 | 899 | 1.12 | 0.87 | 1.44 | 0.367 | 1.08 | 0.81 | 1.44 | 0.588 | 1.19 | 0.85 | 1.65 | 0.307 | 0.59 | 0.22 | 1.60 | 0.298 |
| A (rs10980926) | ZNF483 | 9 | 899 | 0.86 | 0.69 | 1.07 | 0.185 | 0.85 | 0.66 | 1.08 | 0.186 | 0.85 | 0.62 | 1.18 | 0.334 | 0.68 | 0.38 | 1.21 | 0.193 |
| C (rs10441737) | ZNF483 | 9 | 880 | 0.81 | 0.65 | 1.02 | 0.068 | 0.79 | 0.61 | 1.01 | 0.058 | 0.79 | 0.57 | 1.09 | 0.144 | 0.60 | 0.33 | 1.07 | 0.083 |
| C (rs10769908) | STK33 | 11 | 885 | 1.14 | 0.93 | 1.40 | 0.207 | 1.11 | 0.89 | 1.39 | 0.354 | 1.35 | 0.94 | 1.95 | 0.106 | 0.97 | 0.66 | 1.42 | 0.860 |
| G (rs555621) | FSHB | 11 | 896 | 1.13 | 0.92 | 1.39 | 0.232 | 1.08 | 0.85 | 1.36 | 0.531 | 1.16 | 0.82 | 1.64 | 0.395 | 1.02 | 0.66 | 1.57 | 0.937 |
| A (rs11031010) | FSHB | 11 | 892 | 1.11 | 0.82 | 1.51 | 0.484 | 1.12 | 0.80 | 1.57 | 0.498 | 1.16 | 0.80 | 1.69 | 0.445 | 0.99 | 0.29 | 3.37 | 0.987 |
| C (rs1782507) | FSHB | 11 | 898 | 1.08 | 0.88 | 1.33 | 0.476 | 1.19 | 0.94 | 1.51 | 0.151 | 1.11 | 0.80 | 1.54 | 0.531 | 1.59 | 1.01 | 2.51 | 0.047 |
| A (rs6589964) | BSX | 11 | 900 | 0.87 | 0.71 | 1.06 | 0.165 | 0.80 | 0.64 | 1.01 | 0.058 | 0.78 | 0.55 | 1.11 | 0.170 | 0.70 | 0.47 | 1.04 | 0.080 |
| A (rs1544410) | VDR | 12 | 896 | 1.18 | 0.96 | 1.45 | 0.121 | 1.20 | 0.95 | 1.53 | 0.125 | 1.28 | 0.92 | 1.78 | 0.150 | 1.26 | 0.79 | 1.99 | 0.332 |
| A (rs999460) | NKX2-1 | 14 | 898 | 1.37 | 1.11 | 1.69 | 0.003 | 1.45 | 1.14 | 1.83 | 0.002 | 1.33 | 0.96 | 1.83 | 0.088 | 2.41 | 1.53 | 3.82 | 0.0002 |
| A (rs4986938) | ESR2 | 14 | 898 | 0.95 | 0.77 | 1.18 | 0.657 | 0.93 | 0.73 | 1.19 | 0.576 | 0.90 | 0.65 | 1.24 | 0.510 | 0.96 | 0.58 | 1.59 | 0.883 |
| A (rs2241423) | MAP2K5 | 15 | 895 | 1.21 | 0.93 | 1.57 | 0.160 | 1.13 | 0.83 | 1.52 | 0.434 | 1.13 | 0.80 | 1.59 | 0.480 | 1.30 | 0.50 | 3.38 | 0.597 |
| T (rs12444979) | GPRC5B | 16 | 895 | 1.67 | 1.27 | 2.20 | 0.0002 | 1.56 | 1.14 | 2.13 | 0.005 | 1.59 | 1.11 | 2.27 | 0.011 | 2.56 | 0.94 | 6.92 | 0.065 |
| A (rs9939609) | FTO | 16 | 898 | 1.10 | 0.90 | 1.35 | 0.348 | 1.07 | 0.85 | 1.34 | 0.584 | 1.06 | 0.75 | 1.49 | 0.748 | 1.13 | 0.75 | 1.70 | 0.547 |
| A (rs12324955) | FTO | 16 | 898 | 0.97 | 0.78 | 1.22 | 0.813 | 0.94 | 0.74 | 1.21 | 0.636 | 1.05 | 0.76 | 1.44 | 0.786 | 0.61 | 0.33 | 1.14 | 0.120 |
| G (rs1398217) | SKOR2 | 18 | 891 | 0.97 | 0.79 | 1.19 | 0.778 | 1.06 | 0.84 | 1.34 | 0.618 | 0.99 | 0.70 | 1.38 | 0.936 | 1.26 | 0.82 | 1.94 | 0.296 |
| G (rs2252673) | INSR | 19 | 897 | 1.04 | 0.81 | 1.33 | 0.787 | 0.98 | 0.74 | 1.30 | 0.891 | 1.06 | 0.76 | 1.47 | 0.751 | 0.56 | 0.21 | 1.46 | 0.233 |
| A (rs1073768) | GHRH | 20 | 899 | 1.08 | 0.88 | 1.32 | 0.473 | 1.01 | 0.81 | 1.26 | 0.932 | 1.17 | 0.82 | 1.68 | 0.397 | 0.86 | 0.58 | 1.27 | 0.446 |
| C (rs4633) | COMT | 22 | 898 | 1.08 | 0.88 | 1.32 | 0.442 | 1.12 | 0.90 | 1.40 | 0.318 | 1.19 | 0.83 | 1.70 | 0.346 | 1.14 | 0.79 | 1.66 | 0.488 |
| A (rs5930973) | CD40LG | 23 | 893 | 1.10 | 0.73 | 1.65 | 0.658 | 1.10 | 0.69 | 1.74 | 0.690 | ||||||||
| T (rs3092921) | CD40LG | 23 | 901 | 1.13 | 0.78 | 1.62 | 0.524 | 1.28 | 0.85 | 1.92 | 0.230 | ||||||||
| Haploblocks (Genes) and Included SNPs | Haplotypes | Frequency | OR | p | |
|---|---|---|---|---|---|
| FGR (n = 273) | Controls (n = 631) | ||||
| H1 (FSHB) rs555621-rs11031010-rs1782507 | ACC | 0.368 | 0.349 | 1.20 | 0.131 |
| GAA | 0.131 | 0.120 | 1.11 | 0.529 | |
| GCA | 0.308 | 0.290 | 1.02 | 0.881 | |
| ACA | 0.194 | 0.242 | 0.71 | 0.017 | |
| H2 (ZNF483) rs1098092-rs10441737 | AC | 0.284 | 0.323 | 0.80 | 0.083 |
| GT | 0.716 | 0.677 | 1.23 | 0.104 | |
| H3 (ESR1) rs3020394-rs1884051 | GG | 0.333 | 0.289 | 1.15 | 0.268 |
| AA | 0.667 | 0.711 | 0.87 | 0.235 | |
| H4 (LIN28B) rs4946651-rs7759938 | AC | 0.288 | 0.280 | 1.18 | 0.185 |
| AT | 0.120 | 0.130 | 0.96 | 0.803 | |
| GT | 0.592 | 0.590 | 0.88 | 0.283 | |
| H5 (UGT2B4) rs2013573-rs13111134 | AA | 0.191 | 0.159 | 1.22 | 0.202 |
| GA | 0.042 | 0.033 | 1.39 | 0.255 | |
| GG | 0.767 | 0.808 | 0.77 | 0.078 | |
| H6 (FSHR) rs6732220-rs4953655 | GG | 0.224 | 0.246 | 0.84 | 0.200 |
| GA | 0.018 | 0.011 | 1.60 | 0.301 | |
| CA | 0.757 | 0.743 | 1.17 | 0.243 | |
| H7 (LHCGR) rs7579411-rs4953616 | TC | 0.276 | 0.286 | 0.93 | 0.580 |
| TT | 0.156 | 0.169 | 1.10 | 0.534 | |
| CT | 0.568 | 0.546 | 1.02 | 0.896 | |
| H8 (POMC) rs7579411-rs4953616 | AA | 0.297 | 0.299 | 0.97 | 0.785 |
| AG | 0.178 | 0.153 | 1.17 | 0.317 | |
| GG | 0.526 | 0.548 | 0.95 | 0.658 | |
| N | Models | NH | beta H | WH | NL | beta L | WL | pperm |
|---|---|---|---|---|---|---|---|---|
| Two-order interaction (p < 5 × 10−6) | ||||||||
| 1 | rs3020394 ESR1 × rs7538038 KISS1 | 2 | 0.494 | 10.21 | 3 | −0.762 | 20.93 | <0.001 |
| 2 | rs3020394 ESR1 × rs12444979 GPRC5B | 3 | 0.836 | 22.05 | 1 | −0.571 | 13.00 | <0.001 |
| 3 | rs7538038 KISS1 × rs999460 NKX2-1 | 3 | 0.732 | 23.74 | 1 | −0.773 | 11.42 | <0.001 |
| Three-order interaction (p < 1 × 10−8) | ||||||||
| 1 | rs222003 GC × rs7538038 KISS1 × rs999460 NKX2-1 | 5 | 0.871 | 32.84 | 1 | −1.072 | 16.80 | <0.001 |
| 2 | rs713586 RBJ × rs3020394 ESR1 × rs12444979 GPRC5B | 5 | 1.194 | 33.03 | 3 | −0.794 | 18.91 | <0.001 |
| 3 | rs3020394 ESR1 × rs999460 NKX2-1 × rs12444979 GPRC5B | 3 | 0.736 | 14.48 | 3 | −0.872 | 32.13 | <0.001 |
| 4 | rs11031010 FSHB × rs999460 NKX2-1 × rs12444979 GPRC5B | 4 | 0.801 | 17.78 | 3 | −0.816 | 30.32 | <0.001 |
| 5 | rs7538038 KISS1 × rs999460 NKX2-1 × rs12444979 GPRC5B | 6 | 0.809 | 29.90 | 3 | −0.809 | 27.65 | <0.001 |
| Four-order interaction (p < 3 × 10−12) | ||||||||
| 1 | rs1544410 VDR × rs1398217 SKOR2 × rs7579411 LHCGR × rs314280 LIN28B | 8 | 1.547 | 49.59 | 0 | - | - | <0.001 |
| 2 | rs314280 LIN28B × rs10769908 STK33 × rs13111134 UGT2B4 × rs7579411 LHCGR | 8 | 1.280 | 47.32 | 1 | −0.665 | 2.77 | <0.001 |
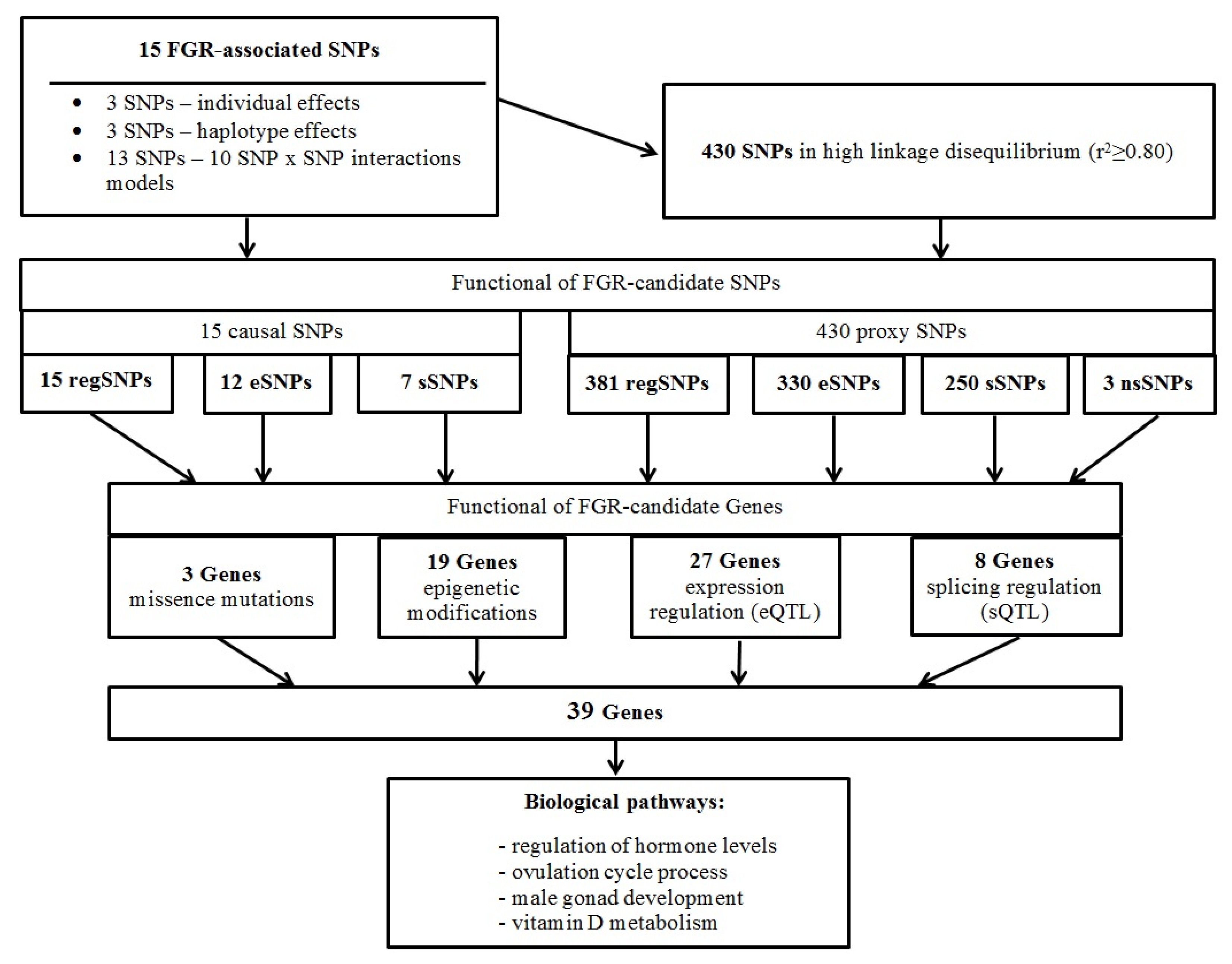
2.4. FGR-Significant Locus/Gene Probable Functions
2.4.1. Missense Mutations and FGR-Linked SNPs
2.4.2. Link FGR-Involved Loci with Deoxyribonucleic Acid (DNA) Epigenetic Changes
2.4.3. Possible Gene Expression Regulatory Effects of FGR-Linked Polymorphisms
2.4.4. Potential Opportunity of Splicing Regulation of FGR-Correlated Loci
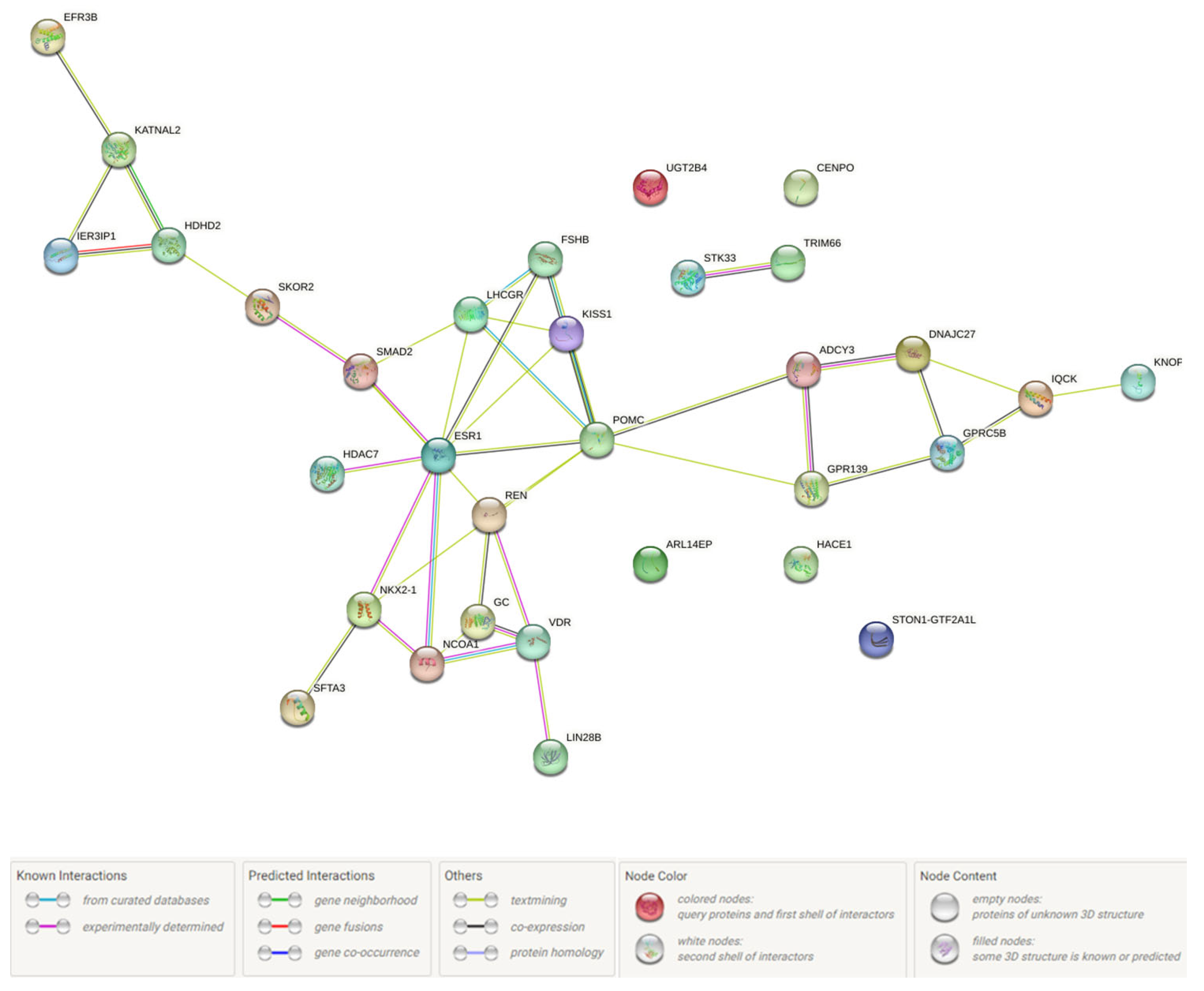
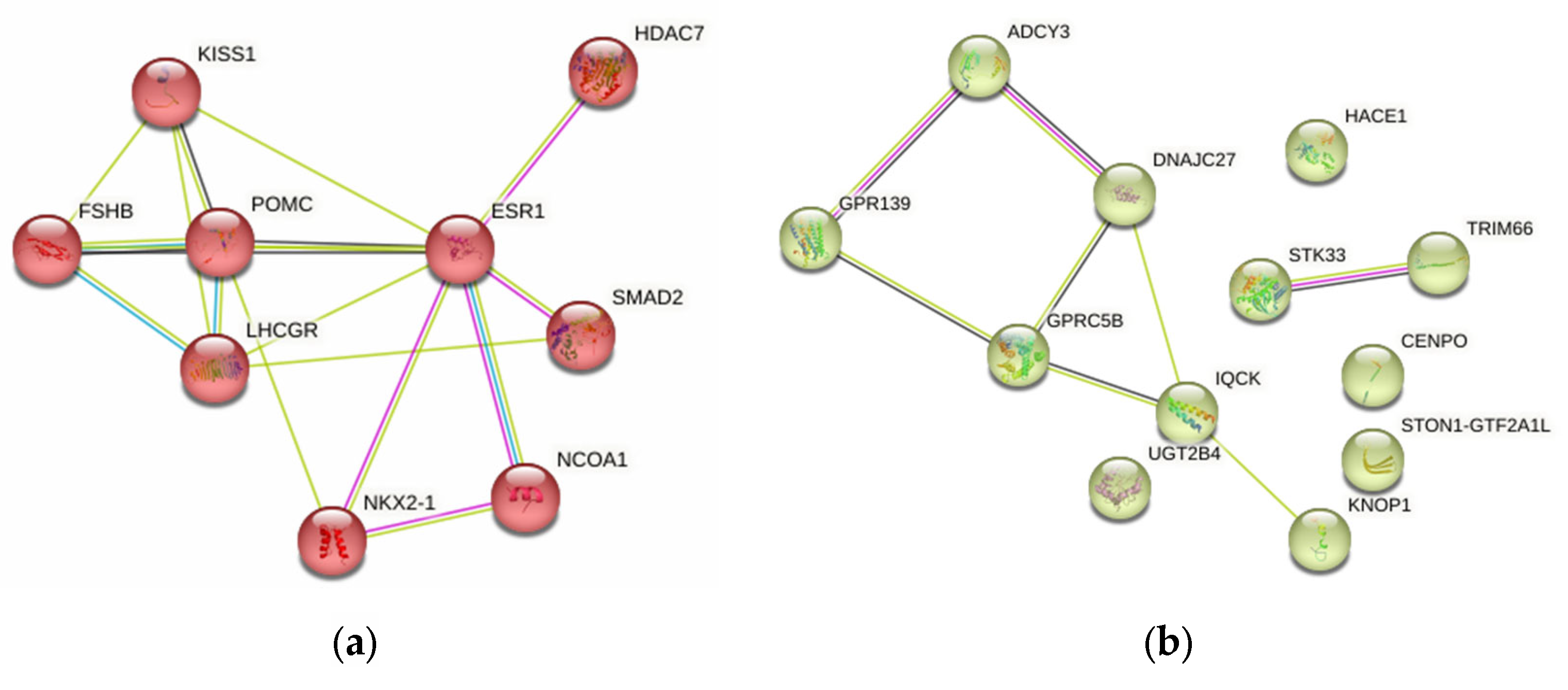
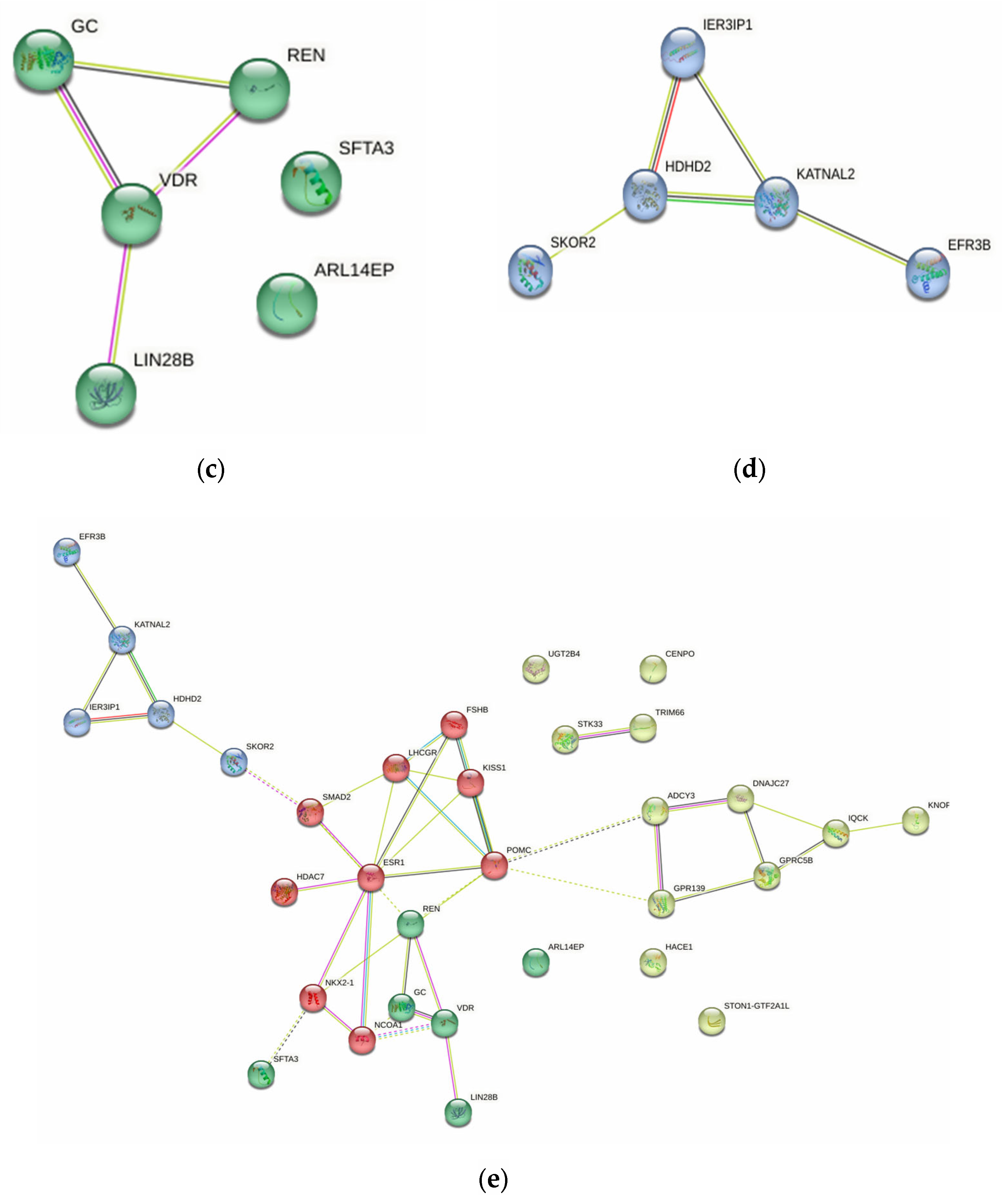
2.4.5. Probable FGR-Related Pathways
2.5. Syntropic Effects of mAAM-Involved Genes in FGR, mAAM, Pre-Pregnancy mBMI and Offspring BW
3. Discussion
4. Materials and Methods
4.1. Study Design/Subjects
4.2. Genetic Laboratory Analysis

4.3. Statistical Genetic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Society for Maternal-Fetal Medicine (SMFM). Electronic address: Pubs@smfm.org; Martins, J.G.; Biggio, J.R.; Abuhamad, A. Society for Maternal-Fetal Medicine Consult Series #52: Diagnosis and management of fetal growth restriction: (Replaces Clinical Guideline Number 3, April 2012). Am. J. Obstet. Gynecol. 2020, 223, B2–B17. [Google Scholar] [CrossRef]
- Melamed, N.; Baschat, A.; Yinon, Y.; Athanasiadis, A.; Mecacci, F.; Figueras, F.; Berghella, V.; Nazareth, A.; Tahlak, M.; McIntyre, H.D.; et al. FIGO (international Federation of Gynecology and obstetrics) initiative on fetal growth: Best practice advice for screening, diagnosis, and management of fetal growth restriction. Int. J. Gynaecol. Obstet. 2021, 152 (Suppl. S1), 3–57. [Google Scholar] [CrossRef] [PubMed]
- Bamfo, J.E.; Odibo, A.O. Diagnosis and management of fetal growth restriction. J. Pregnancy 2011, 2011, 640715. [Google Scholar] [CrossRef] [PubMed]
- Crispi, F.; Miranda, J.; Gratacós, E. Long-term cardiovascular consequences of fetal growth restriction: Biology, clinical implications, and opportunities for prevention of adult disease. Am. J. Obstet. Gynecol. 2018, 218, S869–S879. [Google Scholar] [CrossRef] [PubMed]
- Pels, A.; Beune, I.M.; van Wassenaer-Leemhuis, A.G.; Limpens, J.; Ganzevoort, W. Early-onset fetal growth restriction: A systematic review on mortality and morbidity. Acta Obstet. Gynecol. Scand. 2020, 99, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Mierzynski, R.; Dluski, D.; Darmochwal-Kolarz, D.; Poniedziałek-Czajkowska, E.; Leszczynska-Gorzelak, B.; Kimber-Trojnar, Z.; Agnieszka-Wankowicz; Oleszczuk, J. Intra-uterine Growth Retardation as a Risk Factor of Postnatal Metabolic Disorders. Curr. Pharm. Biotechnol. 2016, 17, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, A.; Allison, B.J.; Castillo-Melendez, M.; Jenkin, G.; Polglase, G.R.; Miller, S.L. Neonatal Morbidities of Fetal Growth Restriction: Pathophysiology and Impact. Front. Endocrinol. 2019, 10, 55. [Google Scholar] [CrossRef] [PubMed]
- Barker, D.J.; Osmond, C.; Forsén, T.J.; Kajantie, E.; Eriksson, J.G. Trajectories of growth among children who have coronary events as adults. N. Engl. J. Med. 2005, 353, 1802–1809. [Google Scholar] [CrossRef]
- Simeoni, U.; Armengaud, J.B.; Siddeek, B.; Tolsa, J.F. Perinatal Origins of Adult Disease. Neonatology 2018, 113, 393–399. [Google Scholar] [CrossRef]
- D’Agostin, M.; Di Sipio Morgia, C.; Vento, G.; Nobile, S. Long-term implications of fetal growth restriction. World J. Clin. Cases 2023, 11, 2855–2863. [Google Scholar] [CrossRef]
- Sawant, L.D.; Venkat, S. Comparative Analysis of Normal versus Fetal Growth Restriction in Pregnancy: The Significance of Maternal Body Mass Index, Nutritional Status, Anemia, and Ultrasonography Screening. Int. J. Reprod. Med. 2013, 2013, 671954. [Google Scholar] [CrossRef]
- Voskamp, B.J.; Kazemier, B.M.; Ravelli, A.C.; Schaaf, J.; Mol, B.W.; Pajkrt, E. Recurrence of small-for-gestational-age pregnancy: Analysis of first and subsequent singleton pregnancies in The Netherlands. Am. J. Obstet. Gynecol. 2013, 208, 374.e1–374.e6. [Google Scholar] [CrossRef]
- Albu, A.R.; Anca, A.F.; Horhoianu, V.V.; Horhoianu, I.A. Predictive factors for intrauterine growth restriction. J. Med. Life 2014, 7, 165–171. [Google Scholar] [PubMed]
- Nardozza, L.M.; Caetano, A.C.; Zamarian, A.C.; Mazzola, J.B.; Silva, C.P.; Marçal, V.M.; Lobo, T.F.; Peixoto, A.B.; Araujo Júnior, E. Fetal growth restriction: Current knowledge. Arch. Gynecol. Obstet. 2017, 295, 1061–1077. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.Y.; Wang, Y.F.; Huang, K.; Yan, S.Q.; Ge, X.; Chen, M.L.; Hao, J.H.; Tong, S.L.; Tao, F.B. [The effect of pre-pregnancy weight and the increase of gestational weight on fetal growth restriction: A cohort study]. Zhonghua Yu Fang Yi Xue Za Zhi 2017, 51, 1074–1078. (In Chinese) [Google Scholar] [CrossRef] [PubMed]
- Beune, I.M.; Bloomfield, F.H.; Ganzevoort, W.; Embleton, N.D.; Rozance, P.J.; van Wassenaer-Leemhuis, A.G.; Wynia, K.; Gordijn, S.J. Consensus Based Definition of Growth Restriction in the Newborn. J. Pediatr. 2018, 196, 71–76.e1. [Google Scholar] [CrossRef] [PubMed]
- Tamura, N.; Hanaoka, T.; Ito, K.; Araki, A.; Miyashita, C.; Ito, S.; Minakami, H.; Cho, K.; Endo, T.; Sengoku, K.; et al. Different Risk Factors for Very Low Birth Weight, Term-Small-for-Gestational-Age, or Preterm Birth in Japan. Int. J. Environ. Res. Public Health 2018, 15, 369. [Google Scholar] [CrossRef]
- Ortega, M.A.; Fraile-Martínez, O.; García-Montero, C.; Sáez, M.A.; Álvarez-Mon, M.A.; Torres-Carranza, D.; Álvarez-Mon, M.; Bujan, J.; García-Honduvilla, N.; Bravo, C.; et al. The Pivotal Role of the Placenta in Normal and Pathological Pregnancies: A Focus on Preeclampsia, Fetal Growth Restriction, and Maternal Chronic Venous Disease. Cells 2022, 11, 568. [Google Scholar] [CrossRef] [PubMed]
- Gaccioli, F.; Lager, S. Placental Nutrient Transport and Intrauterine Growth Restriction. Front. Physiol. 2016, 7, 40. [Google Scholar] [CrossRef]
- Yermachenko, A.; Dvornyk, V. Nongenetic determinants of age at menarche: A systematic review. Biomed. Res. Int. 2014, 2014, 371583. [Google Scholar] [CrossRef]
- Plant, T.M. The hypothalamo–pituitary–gonadal axis. J. Endocrinol. 2015, 226, 41–54. [Google Scholar] [CrossRef]
- Rudra, C.L.; Williams, M.A. BMI as a modifying factor in the relations between age at menarche, menstrual cycle characteristics, and risk of preeclampsia. Gynecol. Endocrinol. 2005, 21, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Abetew, D.F.; Enquobahrie, D.A.; Dishi, M.; Rudra, C.B.; Miller, R.S.; Williams, M.A. Age at menarche, menstrual characteristics, and risk of preeclampsia. ISRN Obstet. Gynecol. 2011, 2011, 472083. [Google Scholar] [CrossRef]
- Li, H.; Song, L.; Shen, L.; Liu, B.; Zheng, X.; Zhang, L.; Li, Y.; Xia, W.; Lu, B.; Zhang, B.; et al. Age at menarche and prevalence of preterm birth: Results from the Healthy Baby Cohort study. Sci. Rep. 2017, 7, 12594. [Google Scholar] [CrossRef]
- Petry, C.J.; Ong, K.K.; Hughes, I.A.; Acerini, C.L.; Dunger, D.B. Age at Menarche and Blood Pressure in Pregnancy. Pregnancy Hypertens. 2019, 15, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Deng, P.; Yu, Q.; Tang, H.; Lu, Y.; He, Y. Age at Menarche Mediating Visceral Adipose Tissue’s Influence on Pre-eclampsia: A Mendelian Randomization Study. J. Clin. Endocrinol. Metab. 2023, 108, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Wan, B.; Sun, M. Mendelian randomization identifies age at menarche as an independent causal effect factor for gestational diabetes mellitus. Diabetes Obes. Metab. 2023, 25, 248–260. [Google Scholar] [CrossRef]
- D’Aloisio, A.A.; DeRoo, L.A.; Baird, D.D.; Weinberg, C.R.; Sandler, D.P. Prenatal and infant exposures and age at menarche. Epidemiology 2013, 24, 277–284. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Y.; Tian, Y.; Li, F.; Yan, C.; Wang, H.; Luo, Z.; Jiang, F.; Zhang, J. Maternal age at menarche and offspring body mass index in childhood. BMC Pediatr. 2019, 19, 312. [Google Scholar] [CrossRef]
- Coall, D.A.; Chisholm, J.S. Evolutionary perspectives on pregnancy: Maternal age at menarche and infant birth weight. Soc. Sci. Med. 2003, 57, 1771–1781. [Google Scholar] [CrossRef]
- Ponomarenko, I.; Reshetnikov, E.; Polonikov, A.; Sorokina, I.; Yermachenko, A.; Dvornyk, V.; Churnosov, M. Candidate genes for age at menarche are associated with endometrial hyperplasia. Gene 2020, 757, 4933. [Google Scholar] [CrossRef]
- Ponomarenko, I.; Reshetnikov, E.; Polonikov, A.; Verzilina, I.; Sorokina, I.; Elgaeva, E.E.; Tsepilov, Y.A.; Yermachenko, A.; Dvornyk, V.; Churnosov, M. Candidate genes for age at menarche are associated with endometriosis. Reprod. Biomed. Online 2020, 41, 943–956. [Google Scholar] [CrossRef]
- Ponomarenko, I.; Reshetnikov, E.; Polonikov, A.; Verzilina, I.; Sorokina, I.; Yermachenko, A.; Dvornyk, V.; Churnosov, M. Candidate genes for age at menarche are associated with uterine leiomyoma. Front. Genet. 2021, 11, 512940. [Google Scholar] [CrossRef] [PubMed]
- Elks, C.E.; Perry, J.R.B.; Sulem, P.; Chasman, D.I.; Franceschini, N.; He, C.; Lunetta, K.L.; Visser, J.A.; Byrne, E.M.; Cousminer, D.L.; et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat. Genet. 2010, 42, 1077–1085. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.R.; Day, F.; Elks, C.E.; Sulem, P.; Thompson, D.J.; Ferreira, T.; He, C.; Chasman, D.I.; Esko, T.; Thorleifsson, G.; et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature 2014, 514, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Ponomarenko, I.; Reshetnikov, E.; Altuchova, O.; Polonikov, A.; Sorokina, I.; Yermachenko, A.; Dvornyk, V.; Golovchenko, O.; Churnosov, M. Association of genetic polymorphisms with age at menarche in Russian women. Gene 2019, 686, 228–236. [Google Scholar] [CrossRef]
- Gill, D.; Brewer, C.F.; Del Greco, M.F.; Sivakumaran, P.; Bowden, J.; Sheehan, N.A.; Minelli, C. Age at menarche and adult body mass index: A Mendelian randomization study. Int. J. Obes. 2018, 42, 1574–1581. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.A.; Carslake, D.; Wade, K.H.; Richmond, R.C.; Langdon, R.J.; Vincent, E.E.; Holmes, M.V.; Timpson, N.J.; Davey Smith, G. Influence of puberty timing on adiposity and cardiometabolic traits: A Mendelian randomisation study. PLoS Med. 2018, 15, e1002641. [Google Scholar] [CrossRef] [PubMed]
- Jo, E.J.; Han, S.; Wang, K. Estimation of Causal Effect of Age at Menarche on Pubertal Height Growth Using Mendelian Randomization. Genes 2022, 13, 710. [Google Scholar] [CrossRef] [PubMed]
- Reshetnikova, Y.; Churnosova, M.; Stepanov, V.; Bocharova, A.; Serebrova, V.; Trifonova, E.; Ponomarenko, I.; Sorokina, I.; Efremova, O.; Orlova, V.; et al. Maternal Age at Menarche Gene Polymorphisms Are Associated with Offspring Birth Weight. Life 2023, 13, 1525. [Google Scholar] [CrossRef]
- Fernandez-Rhodes, L.; Demerath, E.W.; Cousminer, D.L.; Tao, R.; Dreyfus, J.G.; Esko, T.; Smith, A.V.; Gudnason, V.; Harris, T.B.; Launer, L.; et al. Association of adiposity genetic variants with menarche timing in 92,105 women of European descent. Am. J. Epidemiol. 2013, 178, 451–460. [Google Scholar] [CrossRef]
- Yermachenko, A.; Dvornyk, V. UGT2B4 previously implicated in the risk of breast cancer is associated with menarche timing in Ukrainian females. Gene 2016, 1, 85–89. [Google Scholar] [CrossRef]
- Huang, J.; Huffman, J.E.; Huang, Y.; Do Valle, Í.; Assimes, T.L.; Raghavan, S.; Voight, B.F.; Liu, C.; Barabási, A.L.; Huang, R.D.L.; et al. Genomics and phenomics of body mass index reveals a complex disease network. Nat. Commun. 2022, 13, 7973. [Google Scholar] [CrossRef] [PubMed]
- Speliotes, E.K.; Willer, C.J.; Berndt, S.I.; Monda, K.L.; Thorleifsson, G.; Jackson, A.U.; Allen, H.L.; Lindgren, C.M.; Luan, J.; Mägi, R.; et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010, 42, 937–948. [Google Scholar] [CrossRef]
- Yang, T.L.; Guo, Y.; Li, S.M.; Li, S.K.; Tian, Q.; Liu, Y.J.; Deng, H.W. Ethnic differentiation of copy number variation on chromosome 16p12.3 for association with obesity phenotypes in European and Chinese populations. Int. J. Obes. 2013, 37, 188–1890. [Google Scholar] [CrossRef] [PubMed]
- Abadi, A.; Peralta-Romero, J.; Suarez, F.; Gomez-Zamudio, J.; Burguete-Garcia, A.I.; Cruz, M.; Meyre, D. Assessing the effects of 35 European-derived BMI-associated SNPs in Mexican children. Obesity 2016, 24, 1989–1995. [Google Scholar] [CrossRef]
- GeneCards: The Human Gene Database. Available online: https://www.genecards.org/ (accessed on 22 February 2023).
- Jiang, X.; Xu, X.; Wu, M.; Guan, Z.; Su, X.; Chen, S.; Wang, H.; Teng, L. GPRC5A: An Emerging Biomarker in Human Cancer. Biomed Res. Int. 2018, 2018, 1823726. [Google Scholar] [CrossRef]
- Kim, Y.J.; Greimel, P.; Hirabayashi, Y. GPRC5B-Mediated Sphingomyelin Synthase 2 Phosphorylation Plays a Critical Role in Insulin Resistance. iScience 2018, 8, 250–266. [Google Scholar] [CrossRef] [PubMed]
- Atanes, P.; Ruz-Maldonado, I.; Hawkes, R.; Liu, B.; Persaud, S.J.; Amisten, S. Identifying Signalling Pathways Regulated by GPRC5B in β-Cells by CRISPR-Cas9-Mediated Genome Editing. Cell Physiol. Biochem. 2018, 45, 656–666. [Google Scholar] [CrossRef]
- Kim, Y.J.; Sano, T.; Nabetani, T.; Asano, Y.; Hirabayashi, Y. GPRC5B activates obesity-associated inflammatory signaling in adipocytes. Sci. Signal 2012, 5, ra85. [Google Scholar] [CrossRef]
- Grasberger, H.; Bell, G.I. Subcellular recruitment by TSG118 and TSPYL implicates a role for zinc finger protein 106 in a novel developmental pathway. Int. J. Biochem. Cell Biol. 2005, 37, 1421–1437. [Google Scholar] [CrossRef]
- He, C.; Kraft, P.; Chasman, D.I.; Buring, J.E.; Chen, C.; Hankinson, S.E.; Paré, G.; Chanock, S.; Ridker, P.M.; Hunter, D.J. A large-scale candidate gene association study of age at menarche and age at natural menopause. Hum. Genet. 2010, 128, 515–527. [Google Scholar] [CrossRef]
- Li, Y.; Tao, N.; Chen, M.; Chu, J.; Huang, X.; Kong, X. Gene Polymorphisms Associated with Central Precocious Puberty and Hormone Levels in Chinese Girls. Int. J. Endocrinol. 2022, 2022, 9450663. [Google Scholar] [CrossRef]
- de Roux, N.; Genin, E.; Carel, J.C.; Matsuda, F.; Chaussain, J.L.; Milgrom, E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc. Natl. Acad. Sci. USA 2003, 100, 10972–10976. [Google Scholar] [CrossRef] [PubMed]
- Hameed, S.; Jayasena, C.N.; Dhillo, W.S. Kisspeptin and fertility. J. Endocrinol. 2011, 208, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Clarke, H.; Dhillo, W.S.; Jayasena, C.N. Comprehensive Review on Kisspeptin and Its Role in Reproductive Disorders. Endocrinol. Metab. 2015, 30, 124–141. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.L.; Chang, H.M.; Zhao, H.C.; Yu, Y.; Li, R.; Qiao, J. Potential roles for the kisspeptin/kisspeptin receptor system in implantation and placentation. Hum. Reprod. Update 2019, 25, 326–343. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, R.A.; Reynolds, R.M.; Leask, R.; Shearing, C.H.; Calder, A.A.; Riley, S.C. Decreased serum levels of kisspeptin in early pregnancy are associated with intra-uterine growth restriction and pre-eclampsia. Prenat. Diagn. 2009, 29, 982–985. [Google Scholar] [CrossRef] [PubMed]
- Cetković, A.; Miljic, D.; Ljubić, A.; Patterson, M.; Ghatei, M.; Stamenković, J.; Nikolic-Djurovic, M.; Pekic, S.; Doknic, M.; Glišić, A.; et al. Plasma kisspeptin levels in pregnancies with diabetes and hypertensive disease as a potential marker of placental dysfunction and adverse perinatal outcome. Endocr. Res. 2012, 37, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Abbara, A.; Al-Memar, M.; Phylactou, M.; Daniels, E.; Patel, B.; Eng, P.C.; Nadir, R.; Izzi-Engbeaya, C.; Clarke, S.A.; Mills, E.G.; et al. Changes in Circulating Kisspeptin Levels During Each Trimester in Women with Antenatal Complications. J. Clin. Endocrinol. Metab. 2022, 107, e71–e83. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, X.; Zhou, S.; Liao, L.; Jiang, R.; Xu, J. The histone H3K9 demethylase Kdm3b is required for somatic growth and female reproductive function. Int. J. Biol. Sci. 2015, 11, 494–507. [Google Scholar] [CrossRef]
- Guan, L.; Zhao, X.; Tang, L.; Chen, J.; Zhao, J.; Guo, M.; Chen, C.; Zhou, Y.; Xu, L. Thyroid Transcription Factor-1: Structure, Expression, Function and Its Relationship with Disease. Biomed. Res. Int. 2021, 2021, 9957209. [Google Scholar] [CrossRef]
- Kusakabe, T.; Kawaguchi, A.; Hoshi, N.; Kawaguchi, R.; Hoshi, S.; Kimura, S. Thyroid-specific enhancer-binding protein/NKX2.1 is required for the maintenance of ordered architecture and function of the differentiated thyroid. Mol. Endocrinol. 2006, 20, 1796–1809. [Google Scholar] [CrossRef]
- Tsialikas, J.; Romer-Seibert, J. LIN28: Roles and regulation in development and beyond. Development 2015, 14, 2397–2404. [Google Scholar] [CrossRef]
- Ong, K.K.; Elks, C.E.; Li, S.; Zhao, J.H.; Luan, J.A.; Andersen, L.B.; Bingham, S.A.; Brage, S.; Smith, G.D.; Ekelund, U.; et al. Genetic variation in LIN28B is associated with the timing of puberty. Nat. Genet. 2009, 41, 729–733. [Google Scholar] [CrossRef]
- Zhu, H.; Shyh-Chang, N.; Segrè, A.V.; Shinoda, G.; Shah, S.P.; Einhorn, W.S.; Takeuchi, A.; Engreitz, J.M.; Hagan, J.P.; Kharas, M.G.; et al. The Lin28/let-7 axis regulates glucose metabolism. Cell 2011, 147, 81–94. [Google Scholar] [CrossRef]
- Zhu, H.; Shah, S.; Shyh-Chang, N.; Shinoda, G.; Einhorn, W.S.; Viswanathan, S.R.; Takeuchi, A.; Grasemann, C.; Rinn, J.L.; Lopez, M.F.; et al. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat. Genet. 2010, 42, 626–630. [Google Scholar] [CrossRef]
- Ong, K.K.; Elks, C.E.; Wills, A.K.; Wong, A.; Wareham, N.J.; Loos, R.J.; Kuh, D.; Hardy, R. Associations between the pubertal timing-related variant in LIN28B and BMI vary across the life course. J. Clin. Endocrinol. Metab. 2011, 96, E125–E129. [Google Scholar] [CrossRef] [PubMed]
- Cousminer, D.L.; Stergiakouli, E.; Berry, D.J.; Ang, W.; Groen-Blokhuis, M.M.; Körner, A.; Siitonen, N.; Ntalla, I.; Marinelli, M.; Perry, J.R.; et al. Genome-wide association study of sexual maturation in males and females highlights a role for body mass and menarche loci in male puberty. Hum. Mol. Genet. 2014, 23, 4452–4464. [Google Scholar] [CrossRef] [PubMed]
- Berndt, S.I.; Gustafsson, S.; Mägi, R.; Ganna, A.; Wheeler, E.; Feitosa, M.F.; Justice, A.E.; Monda, K.L.; Croteau-Chonka, D.C.; Day, F.R.; et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat. Genet. 2013, 45, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.R.; Esko, T.; Yang, J.; Vedantam, S.; Pers, T.H.; Gustafsson, S.; Chu, A.Y.; Estrada, K.; Luan, J.; Kutalik, Z.; et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 2014, 46, 1173–1186. [Google Scholar] [CrossRef] [PubMed]
- Rashina, O.V. Associations of polymorphic variants of candidate genes with the development of H. pylori-negative duodenal ulcer in residents of the Central Chernozem region of Russia. Res. Results Biomed. 2023, 9, 333–346. [Google Scholar] [CrossRef]
- Pasenov, K.N. Features of associations of SHBG-related genes with breast cancer in women, depending on the presence of hereditary burden and mutations in the BRCA1/CHEK2 genes. Res. Results Biomed. 2024, 10, 69–88. [Google Scholar] [CrossRef]
- Reshetnikov, E.; Zarudskaya, O.; Polonikov, A.; Bushueva, O.; Orlova, V.; Krikun, E.; Dvornyk, V.; Churnosov, M. Genetic markers for inherited thrombophilia are associated with fetal growth retardation in the population of Central Russia. J. Obstet. Gynaecol. Res. 2017, 43, 1139–1144. [Google Scholar] [CrossRef] [PubMed]
- Golovchenko, O.; Abramova, M.; Ponomarenko, I.; Reshetnikov, E.; Aristova, I.; Polonikov, A.; Dvornyk, V.; Churnosov, M. Functionally significant polymorphisms of ESR1and PGR and risk of intrauterine growth restriction in population of Central Russia. Eur. J. Obstet. Gynecol. Reprod. Biol. 2020, 253, 52–57. [Google Scholar] [CrossRef]
- Hadlock, F.P.; Deter, R.L.; Harrist, R.B. Sonographic detection of abnormal fetal growth patterns. Clin. Obstet. Gynecol. 1984, 27, 342–351. [Google Scholar] [CrossRef]
- Golovchenko, O.V. Molecular genetic determinants of pre-eclampsia. Res. Results Biomed. 2019, 5, 139–149. [Google Scholar] [CrossRef]
- Abramova, M.; Churnosova, M.; Efremova, O.; Aristova, I.; Reshetnikov, E.; Polonikov, A.; Churnosov, M.; Ponomarenko, I. Effects of pre-pregnancy over-weight/obesity on the pattern of association of hypertension susceptibility genes with preeclampsia. Life 2022, 12, 2018. [Google Scholar] [CrossRef]
- Reshetnikov, E.; Ponomarenko, I.; Golovchenko, O.; Sorokina, I.; Batlutskaya, I.; Yakunchenko, T.; Dvornyk, V.; Polonikov, A.; Churnosov, M. The VNTR polymorphism of the endothelial nitric oxide synthase gene and blood pressure in women at the end of pregnancy. Taiwan J. Obstet. Gynecol. 2019, 58, 390–395. [Google Scholar] [CrossRef]
- Abramova, M.Y. Genetic markers of severe preeclampsia. Res. Results Biomed. 2022, 8, 305–316. (In Russian) [Google Scholar] [CrossRef]
- Churnosov, M.; Abramova, M.; Reshetnikov, E.; Lyashenko, I.V.; Efremova, O.; Churnosova, M.; Ponomarenko, I. Polymorphisms of hypertension susceptibility genes as a risk factors of preeclampsia in the Caucasian population of central Russia. Placenta 2022, 129, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Graff, M.; Ngwa, J.S.; Workalemahu, T.; Homuth, G.; Schipf, S.; Teumer, A.; Völzke, H.; Wallaschofski, H.; Abecasis, G.R.; Edward, L.; et al. Genome–wide analysis of BMI in adolescents and young adults reveals additional insight into the effects of genetic loci over the life course. Hum. Mol. Genet. 2013, 22, 3597–3607. [Google Scholar] [CrossRef]
- Hong, K.W.; Oh, B. Recapitulation of genome-wide association studies on body mass index in the Korean population. Int. J. Obes. 2012, 36, 1127–1130. [Google Scholar] [CrossRef] [PubMed]
- Delahanty, R.J.; Beeghly-Fadiel, A.; Long, J.R.; Gao, Y.T.; Lu, W.; Xiang, Y.B.; Zheng, Y.; Ji, B.T.; Wen, W.Q.; Cai, Q.Y.; et al. Evaluation of GWAS-identified genetic variants for age at menarche among Chinese women. Hum. Reprod. 2013, 28, 1135–1143. [Google Scholar] [CrossRef] [PubMed]
- Pyun, J.A.; Kim, S.; Cho, N.H.; Koh, I.; Lee, J.Y.; Shin, C.; Kwack, K. Genome-wide association studies and epistasis analyses of candidate genes related to age at menarche and age at natural menopause in a Korean population. Menopause 2014, 21, 522–529. [Google Scholar] [CrossRef]
- Engelman, C.D.; Fingerlin, T.E.; Langefeld, C.D.; Hicks, P.J.; Rich, S.S.; Wagenknecht, L.E.; Bowden, D.W.; Norris, J.M. Genetic and environmental determinants of 25-hydroxyvitamin D and 1,25-dihydroxyvitamin D levels in Hispanic and African Americans. J. Clin. Endocrinol. Metab. 2008, 93, 3381–3388. [Google Scholar] [CrossRef]
- Xu, X.H.; Xiong, D.H.; Liu, X.G.; Guo, Y.; Chen, Y.; Zhao, J.; Recker, R.R.; Deng, H.W. Association analyses of vitamin D-binding protein gene with compression strength index variation in Caucasian nuclear families. Osteoporos. Int. 2010, 21, 99–107. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Liu, Y.; Qu, H.; Qu, S.; Wang, W.; Ren, L. The GC, CYP2R1 and DHCR7 genes are associated with vitamin D levels in northeastern Han Chinese children. Swiss Med. Wkly. 2012, 142, w13636. [Google Scholar] [CrossRef]
- Wang, H.J.; Hinney, A.; Song, J.Y.; Scherag, A.; Meng, X.R.; Grallert, H.; Illig, T.; Hebebrand, J.; Wang, Y.; Ma, J. Association of common variants identified by recent genome-wide association studies with obesity in Chinese children: A case-control study. BMC Med. Genet. 2016, 17, 7. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chenthuran, T.; Galhenagey, G.H.; Jayasekara, R.W.; Dissanayake, V.H. Polymorphism in the epidermal growth factor gene is associated with pre-eclampsia and low birthweight. J. Obstet. Gynaecol. Res. 2014, 5, 1235–1242. [Google Scholar] [CrossRef] [PubMed]
- Schweighofer, N.; Lerchbaum, E.; Trummer, O.; Schwetz, V.; Pilz, S.; Pieber, T.R.; Obermayer-Pietsch, B. Androgen levels and metabolic parameters are associated with a genetic variant of F13A1 in women with polycystic ovary syndrome. Gene 2012, 504, 133–139. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Kraft, P.; Chen, C.; Buring, J.E.; Paré, G.; Hankinson, S.E.; Chanock, S.J.; Ridker, P.M.; David, J.; Chasman, D.I. Genome-wide association studies identify novel loci associated with age at menarche and age at natural menopause. Nat. Genet. 2009, 41, 724–728. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.R.; Stolk, L.; Franceschini, N.; Lunetta, K.L.; Zhai, G.; McArdle, P.F.; Smith, A.V.; Aspelund, T.; Bandinelli, S.; Boerwinkle, E.; et al. Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nat. Genet. 2009, 41, 648–650. [Google Scholar] [CrossRef] [PubMed]
- Lango Allen, H.; Estrada, K.; Lettre, G.; Berndt, S.I.; Weedon, M.N.; Rivadeneira, F.; Willer, C.J.; Jackson, A.U.; Vedantam, S.; Raychaudhuri, S.; et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 2010, 7317, 832–838. [Google Scholar] [CrossRef]
- Cousminer, D.L.; Berry, D.J.; Timpson, N.J.; Ang, W.; Thiering, E.; Byrne, E.M.; Taal, H.R.; Huikari, V.; Bradfield, J.P.; Kerkhof, M.; et al. Genome-wide association and longitudinal analyses reveal genetic loci linking pubertal height growth, pubertal timing and childhood adiposity. Hum. Mol. Genet. 2013, 13, 2735–2747. [Google Scholar] [CrossRef]
- Leinonen, J.T.; Surakka, I.; Havulinna, A.S.; Kettunen, J.; Luoto, R.; Salomaa, V.; Widen, E. Association of LIN28B with Adult Adiposity-Related Traits in Females. PLoS ONE 2012, 11, e48785. [Google Scholar] [CrossRef]
- Widén, E.; Ripatti, S.; Cousminer, D.L.; Surakka, I.; Lappalainen, T.; Järvelin, M.R.; Eriksson, J.G.; Raitakari, O.; Salomaa, V.; Sovio, U.; et al. Distinct variants at LIN28B influence growth in height from birth to adulthood. Am. J. Hum. Genet. 2010, 86, 773–782. [Google Scholar] [CrossRef]
- Carroll, J.; Saxena, R.; Welt, C.K. Environmental and genetic factors influence age at menarche in women with polycystic ovary syndrome. J. Pediatr. Endocrinol. Metab. 2012, 25, 459–466. [Google Scholar] [CrossRef]
- Sulem, P.; Gudbjartsson, D.F.; Rafnar, T.; Holm, H.; Olafsdottir, E.J.; Olafsdottir, G.H.; Jonsson, T.; Alexandersen, P.; Feenstra, B.; Boyd, H.A.; et al. Genome-wide association study identifies sequence variants on 6q21 associated with age at menarche. Nat. Genet. 2009, 41, 734–738. [Google Scholar] [CrossRef]
- Carty, C.L.; Spencer, K.L.; Setiawan, V.W.; Fernandez-Rhodes, L.; Malinowski, J.; Buyske, S.; Young, A.; Jorgensen, N.W.; Cheng, I.; Carlson, C.S.; et al. Replication of genetic loci for ages at menarche and menopause in the multi-ethnic Population Architecture using Genomics and Epidemiology (PAGE) study. Hum. Reprod. 2013, 28, 1695–1706. [Google Scholar] [CrossRef]
- Wang, J.T.; Guo, Y.; Yang, T.L.; Xu, X.H.; Dong, S.S.; Li, M.; Li, T.Q.; Chen, Y.; Deng, H.W. Polymorphisms in the estrogen receptor genes are associated with hip fractures in Chinese. Bone 2008, 43, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Doo, M.; Kim, Y. Association between ESR1 rs1884051 polymorphism and dietary total energy and plant protein intake on obesity in Korean men. Nutr. Res. Pract. 2011, 5, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Dahlman, I.; Vaxillaire, M.; Nilsson, M.; Lecoeur, C.; Gu, H.F.; Cavalcanti-Proença, C.; Efendic, S.; Ostenson, C.G.; Brismar, K.; Charpentier, G.; et al. Estrogen receptor alpha gene variants associate with type 2 diabetes and fasting plasma glucose. Pharmacogenetics Genom. 2008, 18, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.T.; Fernández-Rhodes, L.; Brzyski, R.G.; Carlson, C.S.; Chen, Z.; Heiss, G.; North, K.E.; Woods, N.F.; Rajkovic, A.; Kooperberg, C.; et al. Replication of loci influencing ages at menarche and menopause in Hispanic women: The Women’s Health Initiative SHARe Study. Hum. Mol. Genet. 2012, 21, 1419–1432. [Google Scholar] [CrossRef] [PubMed]
- Demerath, E.W.; Liu, C.T.; Franceschini, N.; Chen, G.; Palmer, J.R.; Smith, E.N.; Chen, C.T.; Ambrosone, C.B.; Arnold, A.M.; Bandera, E.V.; et al. Genome-wide association study of age at menarche in African-American women. Hum. Mol. Genet. 2013, 22, 3329–3346. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zhao, H.; Chen, H.; Peng, Y.; Cui, L.; Du, Y.; Wang, Z.; Xu, J.; Chen, Z.-J. Variants in FSHB are associated with polycystic ovary syndrome and luteinizing hormone level in han chinese women. J. Clin. Endocrinol. Metab. 2016, 101, 2178–2184. [Google Scholar] [CrossRef] [PubMed]
- Kitanaka, S.; Isojima, T.; Takaki, M.; Numakura, C.; Hayasaka, K.; Igarashi, T. Association of vitamin D-related gene polymorphisms with manifestation of vitamin D deficiency in children. Endocr. J. 2012, 59, 1007–1014. [Google Scholar] [CrossRef]
- Laczmanski, L.; Lwow, F.; Mossakowska, M.; Puzianowska-Kuznicka, M.; Szwed, M.; Kolackov, K.; Krzyzanowska-Swiniarska, B.; Bar-Andziak, E.; Chudek, J.; Sloka, N.; et al. Association between vitamin D concentration and levels of sex hormones in an elderly Polish population with different genotypes of VDR polymorphisms (rs10735810, rs1544410, rs7975232, rs731236). Gene 2015, 559, 73–76. [Google Scholar] [CrossRef]
- Cobayashi, F.; Lourenço, B.H.; Cardoso, M.A. 25-Hydroxyvitamin D3 levels, BsmI polymorphism and insulin resistance in Brazilian Amazonian children. Int. J. Mol. Sci. 2015, 16, 12531–12546. [Google Scholar] [CrossRef]
- Jia, F.; Sun, R.F.; Li, Q.H.; Wang, D.X.; Zhao, F.; Li, J.M.; Pu, Q.; Zhang, Z.Z.; Jin, Y.; Liu, B.L.; et al. Vitamin D receptor BsmI polymorphism and osteoporosis risk: A meta-analysis from 26 studies. Genet. Test. Mol. Biomark. 2013, 1, 30–34. [Google Scholar] [CrossRef]
- Suh, K.T.; Eun, I.S.; Lee, J.S. Polymorphism in vitamin D receptor is associated with bone mineral density in patients with adolescent idiopathic scoliosis. Eur. Spine J. 2010, 19, 1545–1550. [Google Scholar] [CrossRef] [PubMed]
- Stavrou, I.; Zois, C.; Chatzikyriakidou, A.; Georgiou, I.; Tsatsoulis, A. Combined estrogen receptor α and estrogen receptor β genotypes influence the age of menarche. Hum. Reprod. 2006, 21, 554–557. [Google Scholar] [CrossRef] [PubMed]
- Rask-Andersen, M.; Jacobsson, J.A.; Moschonis, G.; Ek, A.E.; Chrousos, G.P.; Marcus, C.; Manios, Y.; Fredriksson, R.; Schiöth, H.B. The MAP2K5-linked SNP rs2241423 is associated with BMI and obesity in two cohorts of Swedish and Greek children. BMC Med. Genet. 2012, 13, 36. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.; Zhang, D.D.; Wang, H.; Zhang, Y.; Liang, L.; Fu, J.F.; Xiong, F.; Liu, G.L.; Gong, C.X.; Luo, F.H.; et al. Genetic variations in SEC16B, MC4R, MAP2K5 and KCTD15 were associated with childhood obesity and interacted with dietary behaviors in Chinese school-age population. Gene 2015, 560, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Mei, H.; Chen, W.; Jiang, F.; He, J.; Srinivasan, S.; Smith, E.N.; Schork, N.; Murray, S.; Berenson, G.S. Longitudinal replication studies of GWAS risk SNPs influencing body mass index over the course of childhood and adulthood. PLoS ONE 2012, 7, e31470. [Google Scholar] [CrossRef] [PubMed]
- Willer, C.J.; Schmidt, E.M.; Sengupta, S.; Peloso, G.M.; Gustafsson, S.; Kanoni, S.; Ganna, A.; Chen, J.; Buchkovich, M.L.; Mora, S.; et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013, 45, 1274–1283. [Google Scholar]
- Frayling, T.M.; Timpson, N.J.; Weedon, M.N.; Zeggini, E.; Freathy, R.M.; Lindgren, C.M.; Perry, J.R.; Elliott, K.S.; Lango, H.; Rayner, N.W.; et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 2007, 316, 889–894. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.R.; Voight, B.F.; Yengo, L.; Amin, N.; Dupuis, J.; Ganser, M.; Grallert, H.; Navarro, P.; Li, M.; Qi, L.; et al. Stratifying type 2 diabetes cases by BMI identifies genetic risk variants in LAMA1 and enrichment for risk variants in lean compared to obese cases. PLoS Genet. 2012, 8, e1002741. [Google Scholar] [CrossRef]
- Hsiao, T.J.; Lin, E. Association of a common rs9939609 variant in the fat mass and obesity-associated (FTO) gene with obesity and metabolic phenotypes in a Taiwanese population: A replication study. J. Genet. 2016, 95, 595–601. [Google Scholar] [CrossRef]
- Duicu, C.; Mărginean, C.O.; Voidăzan, S.; Tripon, F.; Bănescu, C. FTO rs9939609 SNP is associated with adiponectin and leptin levels and the risk of obesity in a cohort of Romanian children population. Medicine 2016, 20, e3709. [Google Scholar] [CrossRef]
- Petkeviciene, J.; Smalinskiene, A.; Klumbiene, J.; Petkevicius, V.; Kriaucioniene, V.; Lesauskaite, V. Physical activity, but not dietary intake, attenuates the effect of the FTO rs9939609 polymorphism on obesity and metabolic syndrome in Lithuanian adult population. Public Health 2016, 135, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Quan, L.L.; Wang, H.; Tian, Y.; Mu, X.; Zhang, Y.; Tao, K. Association of fat-mass and obesity-associated gene FTO rs9939609 polymorphism with the risk of obesity among children and adolescents: A meta-analysis. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 614–623. [Google Scholar] [PubMed]
- Tu, W.; Wagner, E.K.; Eckert, G.J.; Yu, Z.; Hannon, T.; Pratt, J.H.; He, C. Associations between menarche-related genetic variants and pubertal growth in male and female adolescents. J. Adolesc. Health 2015, 56, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Cotsapas, C.; Speliotes, E.K.; Hatoum, I.J.; Greenawalt, D.M.; Dobrin, R.; Lum, P.Y.; Suver, C.; Chudin, E.; Kemp, D.; Reitman, M.; et al. Common body mass index-associated variants confer risk of extreme obesity. Hum. Mol. Genet. 2009, 18, 3502–3507. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, M.O.; Louwers, Y.V.; Taylor, K.D.; Jones, M.R.; Cui, J.; Kwon, S.; Chen, Y.D.; Guo, X.; Stolk, L.; Uitterlinden, A.G.; et al. Replication of association of a novel insulin receptor gene polymorphism with polycystic ovary syndrome. Fertil. Steril. 2011, 95, 1736–1741.e11. [Google Scholar] [CrossRef]
- Du, J.; Wang, Z.; Zhang, J.; Jia, L.; Zhang, F.; Shi, Y.; Chen, Z. Association between single nucleotide polymorphism of rs2252673 of INSR gene and polycystic ovarian syndrome. Zhonghua Fu Chan Ke Za Zhi 2014, 12, 919–924. [Google Scholar]
- Ward, L.D.; Kellis, M. HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016, 44, D877–D881. [Google Scholar] [CrossRef]
- Mathew, C.G. The isolation of high molecular weight eukaryotic DNA. Methods Mol. Biol. 1985, 2, 31–34. [Google Scholar] [CrossRef]
- Pavlova, N.; Demin, S.; Churnosov, M.; Reshetnikov, E.; Aristova, I.; Churnosova, M.; Ponomarenko, I. The Modifying Effect of Obesity on the Association of Matrix Metalloproteinase Gene Polymorphisms with Breast Cancer Risk. Biomedicines 2022, 10, 2617. [Google Scholar] [CrossRef]
- Ivanova, T.A. Sex-specific features of interlocus interactions determining susceptibility to hypertension. Res. Results Biomed. 2024, 10, 53–68. [Google Scholar] [CrossRef]
- Nielsen, D.M.; Ehm, M.G.; Weir, B.S. Detecting marker-disease association by testing for Hardy-Weinberg disequilibrium at a marker locus. Am. J. Hum. Genet. 1998, 63, 1531–1540. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Novakov, V.; Novakova, O.; Churnosova, M.; Sorokina, I.; Aristova, I.; Polonikov, A.; Reshetnikov, E.; Churnosov, M. Intergenic Interactions of SBNO1, NFAT5 and GLT8D1 Determine the Susceptibility to Knee Osteoarthritis among Europeans of Russia. Life 2023, 13, 405. [Google Scholar] [CrossRef] [PubMed]
- Calle, M.L.; Urrea, V.; Malats, N.; Van Steen, K. Mbmdr: An R package for exploring gene-gene interactions associated with binary or quantitative traits. Bioinformatics 2010, 26, 2198–2199. [Google Scholar] [CrossRef][Green Version]
- Calle, M.L.; Urrea, V.; Vellalta, G.; Malats, N.; Steen, K.V. Improving strategies for detecting genetic patterns of disease susceptibility in association studies. Stat. Med. 2008, 27, 6532–6546. [Google Scholar] [CrossRef]
- Moore, J.H.; Gilbert, J.C.; Tsai, C.T.; Chiang, F.T.; Holden, T.; Barney, N.; White, B.C. A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J. Theor. Biol. 2006, 241, 252–261. [Google Scholar] [CrossRef]
- Gola, D.; Mahachie John, J.M.; van Steen, K.; König, I.R. A roadmap to multifactor dimensionality reduction methods. Brief Bioinform. 2016, 7, 293–308. [Google Scholar] [CrossRef]
- Che, R.; Jack, J.R.; Motsinger-Reif, A.A.; Brown, C.C. An adaptive permutation approach for genome-wide association study: Evaluation and recommendations for use. BioData Min. 2014, 7, 9. [Google Scholar] [CrossRef]
- Guo, Y.F.; Li, J.; Chen, Y.; Zhang, L.S.; Deng, H.W. A new permutation strategy of pathway-based approach for genome-wide association study. BMC Bioinform. 2009, 10, 429. [Google Scholar] [CrossRef] [PubMed]
- Gauderman, W.; Morrison, J. QUANTO 1.1: A Computer Program for Power and Sample Size Calculations Genetic–Epidemiology Studies. 2006. Available online: http://hydra.usc.edu/gxe (accessed on 17 April 2023).
- Ivanova, T.; Churnosova, M.; Abramova, M.; Plotnikov, D.; Ponomarenko, I.; Reshetnikov, E.; Aristova, I.; Sorokina, I.; Churnosov, M. Sex-Specific Features of the Correlation between GWAS-Noticeable Polymorphisms and Hypertension in Europeans of Russia. Int. J. Mol. Sci. 2023, 24, 7799. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.; Demin, S.; Churnosov, M.; Reshetnikov, E.; Aristova, I.; Churnosova, M.; Ponomarenko, I. Matrix Metalloproteinase Gene Polymorphisms Are Associated with Breast Cancer in the Caucasian Women of Russia. Int. J. Mol. Sci. 2022, 23, 12638. [Google Scholar] [CrossRef]
- Golovchenko, I.; Aizikovich, B.; Golovchenko, O.; Reshetnikov, E.; Churnosova, M.; Aristova, I.; Ponomarenko, I.; Churnosov, M. Sex Hormone Candidate Gene Polymorphisms Are Associated with Endometriosis. Int. J. Mol. Sci. 2022, 23, 13691. [Google Scholar] [CrossRef]
- Ivanova, T.; Churnosova, M.; Abramova, M.; Ponomarenko, I.; Reshetnikov, E.; Aristova, I.; Sorokina, I.; Churnosov, M. Risk Effects of rs1799945 Polymorphism of the HFE Gene and Intergenic Interactions of GWAS-Significant Loci for Arterial Hypertension in the Caucasian Population of Central Russia. Int. J. Mol. Sci. 2023, 24, 8309. [Google Scholar] [CrossRef]
- Li, M.J.; Yan, B.; Sham, P.C.; Wang, J. Exploring the function of genetic variants in the non-coding genomic regions: Approaches for identifying human regulatory variants affecting gene expression. Brief Bioinform. 2015, 16, 393–412. [Google Scholar] [CrossRef] [PubMed]
- Butkiewicz, M.; Bush, W.S. In Silico Functional Annotation of Genomic Variation. Curr. Protoc. Hum. Genet. 2016, 88, 6.15.1–6.15.17. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Taskesen, E.; van Bochoven, A.; Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017, 8, 1826. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.J.; Jiang, S.; Shi, F.Y.; Ding, Y.; Gao, G. Systematic identification and annotation of multiple-variant compound effects at transcription factor binding sites in human genome. J. Genet. Genom. 2018, 45, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Škunca, N.; Roberts, R.J.; Steffen, M. Evaluating Computational Gene Ontology Annotations. Methods Mol. Biol. 2017, 1446, 97–109. [Google Scholar] [CrossRef] [PubMed]
- GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Westra, H.J.; Peters, M.J.; Esko, T.; Yaghootkar, H.; Schurmann, C.; Kettunen, J.; Christiansen, M.W.; Fairfax, B.P.; Schramm, K.; Powell, J.E.; et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013, 45, 1238–1243. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009, 7, 1073–1081. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reshetnikov, E.; Churnosova, M.; Reshetnikova, Y.; Stepanov, V.; Bocharova, A.; Serebrova, V.; Trifonova, E.; Ponomarenko, I.; Sorokina, I.; Efremova, O.; et al. Maternal Age at Menarche Genes Determines Fetal Growth Restriction Risk. Int. J. Mol. Sci. 2024, 25, 2647. https://doi.org/10.3390/ijms25052647
Reshetnikov E, Churnosova M, Reshetnikova Y, Stepanov V, Bocharova A, Serebrova V, Trifonova E, Ponomarenko I, Sorokina I, Efremova O, et al. Maternal Age at Menarche Genes Determines Fetal Growth Restriction Risk. International Journal of Molecular Sciences. 2024; 25(5):2647. https://doi.org/10.3390/ijms25052647
Chicago/Turabian StyleReshetnikov, Evgeny, Maria Churnosova, Yuliya Reshetnikova, Vadim Stepanov, Anna Bocharova, Victoria Serebrova, Ekaterina Trifonova, Irina Ponomarenko, Inna Sorokina, Olga Efremova, and et al. 2024. "Maternal Age at Menarche Genes Determines Fetal Growth Restriction Risk" International Journal of Molecular Sciences 25, no. 5: 2647. https://doi.org/10.3390/ijms25052647
APA StyleReshetnikov, E., Churnosova, M., Reshetnikova, Y., Stepanov, V., Bocharova, A., Serebrova, V., Trifonova, E., Ponomarenko, I., Sorokina, I., Efremova, O., Orlova, V., Batlutskaya, I., Ponomarenko, M., Churnosov, V., Aristova, I., Polonikov, A., & Churnosov, M. (2024). Maternal Age at Menarche Genes Determines Fetal Growth Restriction Risk. International Journal of Molecular Sciences, 25(5), 2647. https://doi.org/10.3390/ijms25052647








