miR-128-3p Regulates Follicular Granulosa Cell Proliferation and Apoptosis by Targeting the Growth Hormone Secretagogue Receptor
Abstract
:1. Introduction
2. Results
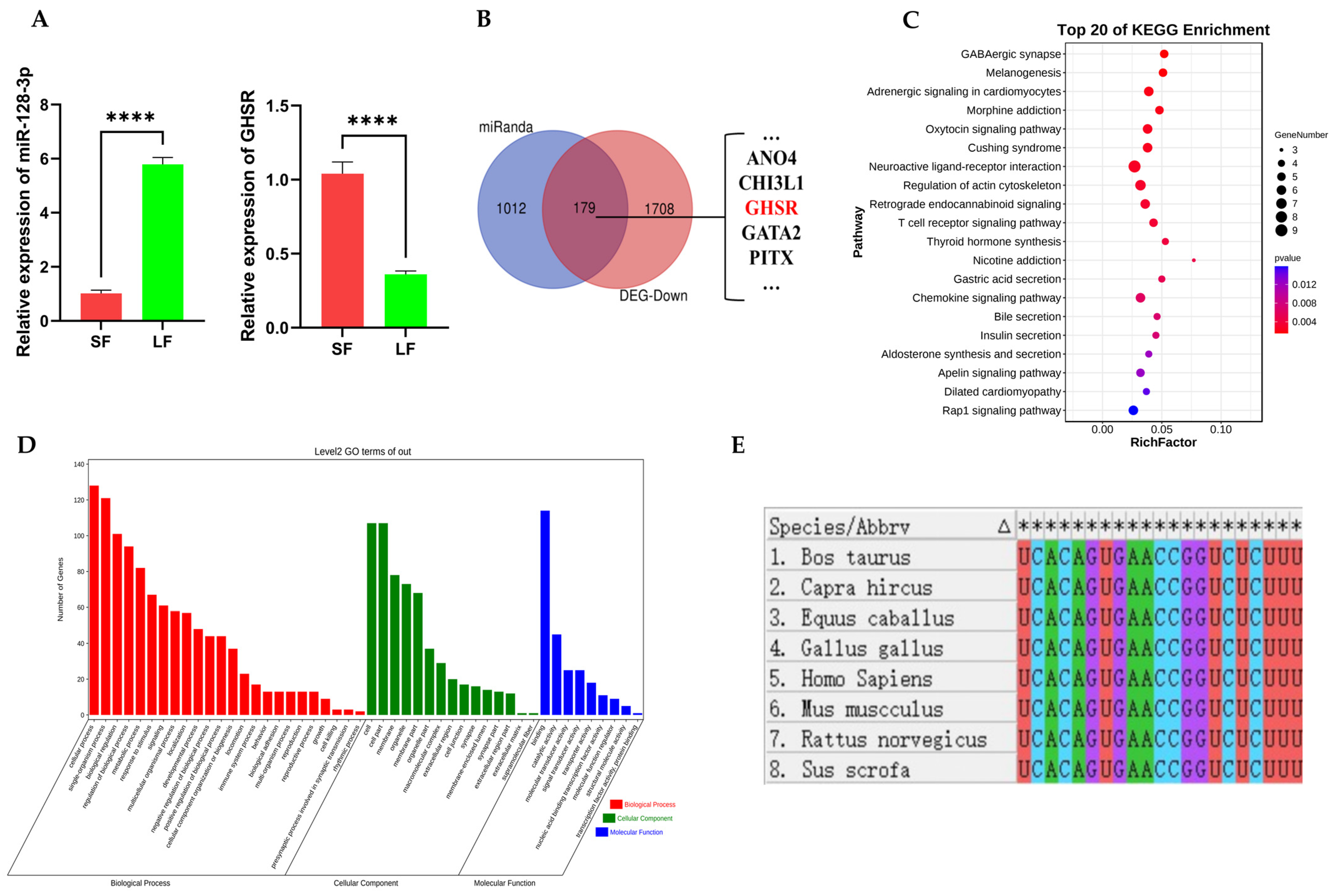
2.1. Prediction of miR-128-3p Target Genes and Functional Enrichment Analysis of Its Target Genes
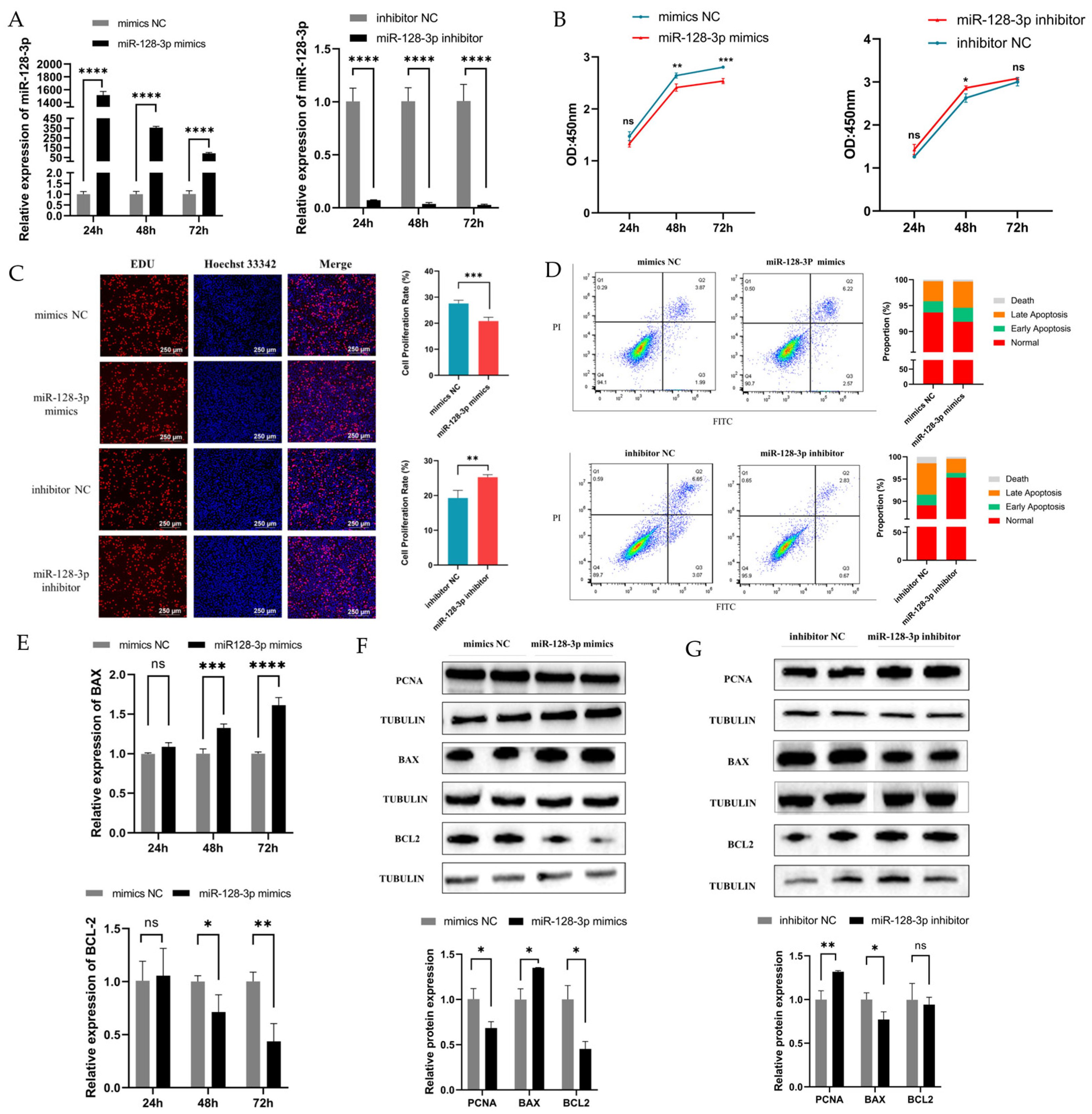
2.2. MiR-128-3p Is Involved in GC Apoptosis and Proliferation
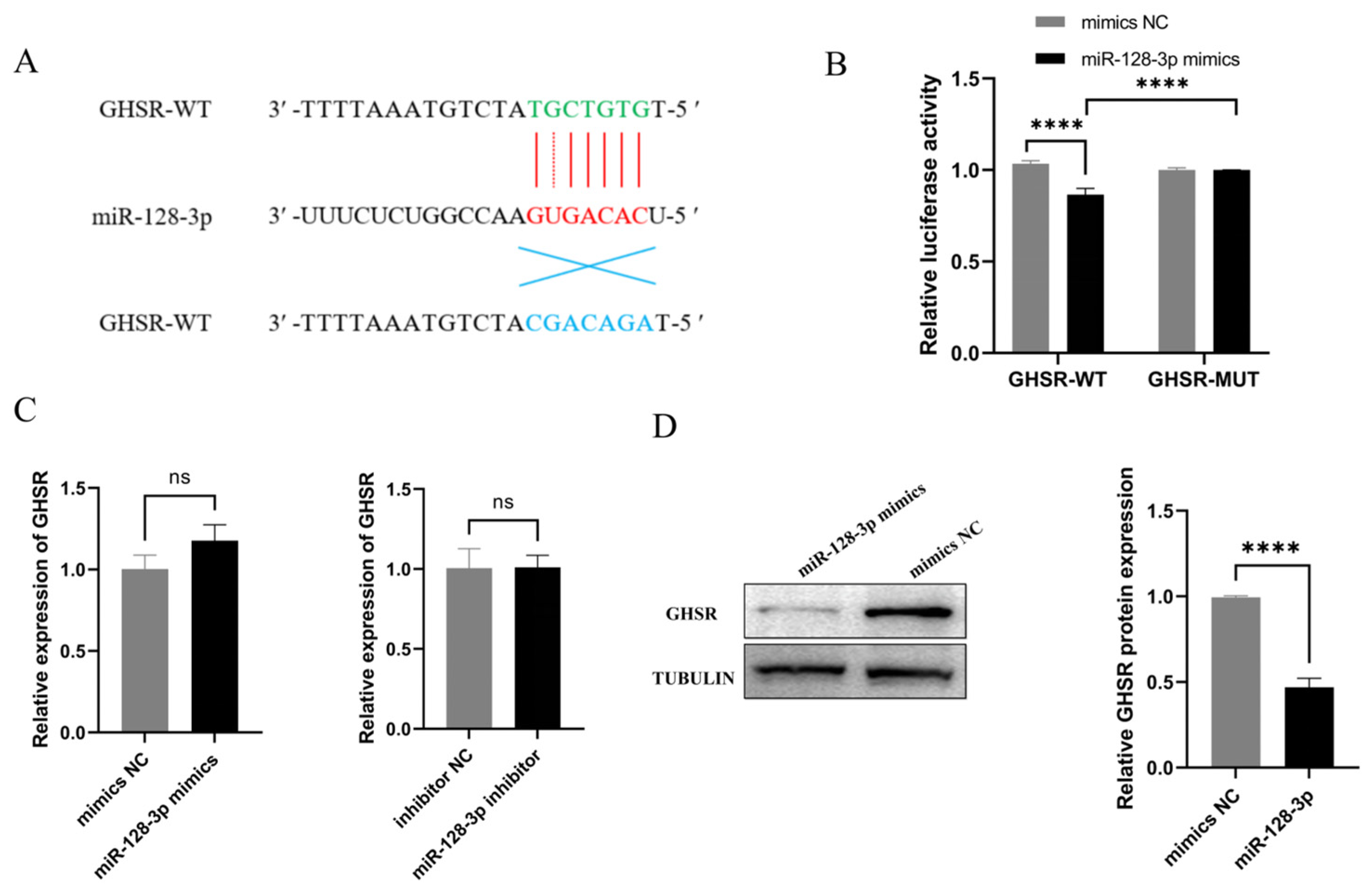
2.3. GHSR Is a Target Gene of miR-128-3p
2.4. GHSR Is Involved in GC Apoptosis and Proliferation
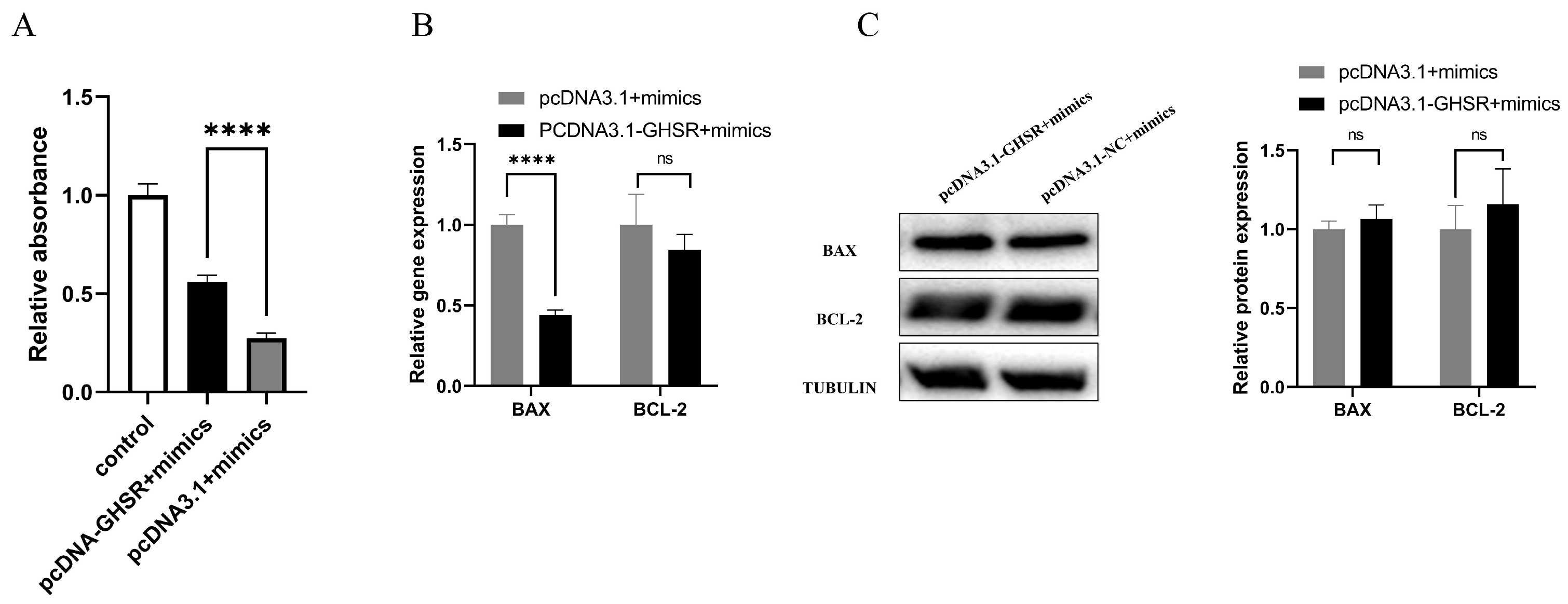
2.5. Cotransfection of GHSR and miR-128-3p Affects Apoptosis in GCs
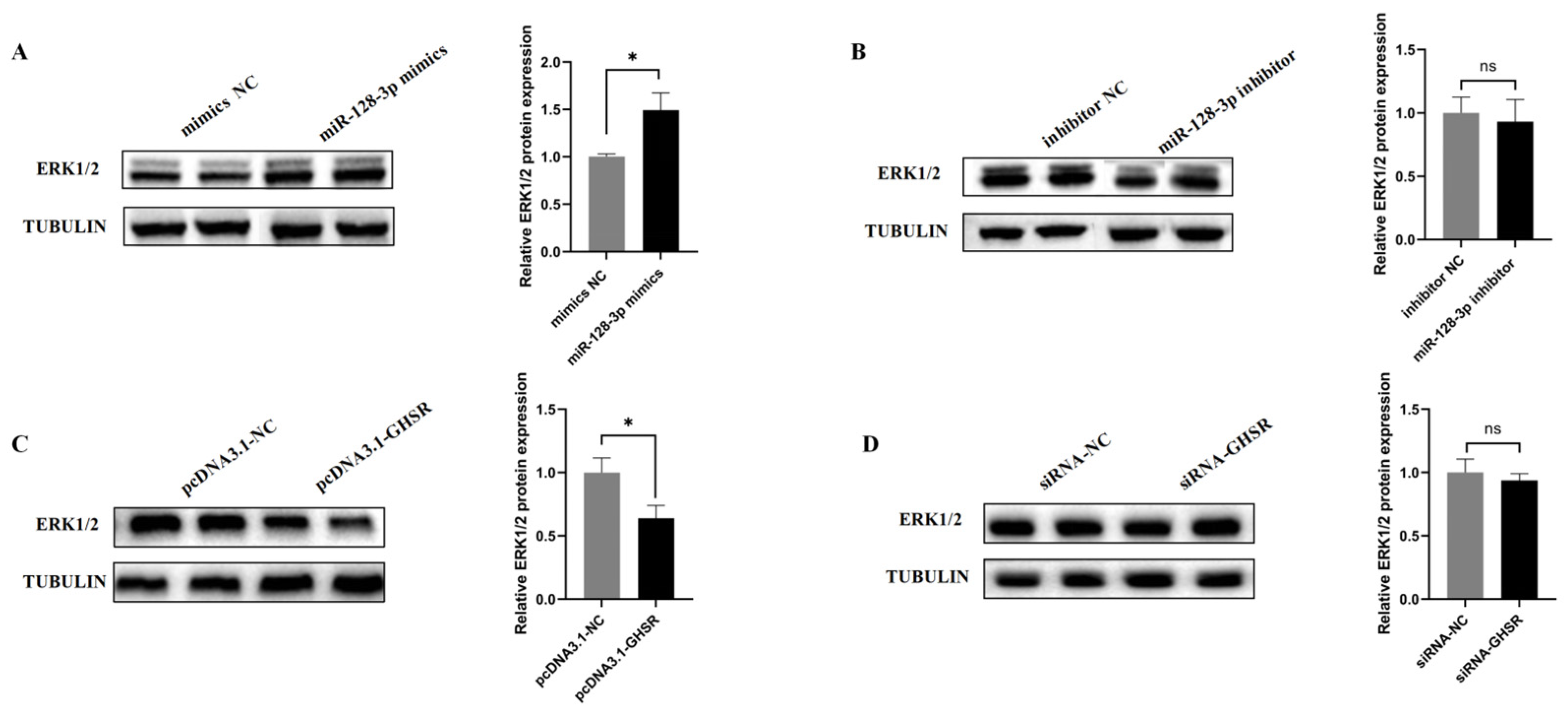
2.6. miR-128-3p and GHSR Regulate ERK1/2 Protein Expression
3. Discussion
4. Materials and Methods
4.1. Cell Line Selection and Culture
4.2. Vector Construction
4.3. Cell Transfection
4.4. RNA Extraction and qRT-PCR Analysis
4.5. Cell Counting Kit-8
4.6. EDU Proliferation Assay
4.7. Cell Apoptosis Analysis
4.8. Western Blot
4.9. Dual-Luciferase Reporter Assay
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sánchez, F.; Smitz, J. Molecular control of oogenesis. Biochim. Biophys. Acta 2012, 1822, 1896–1912. [Google Scholar] [CrossRef]
- Matsuda, F.; Inoue, N.; Manabe, N.; Ohkura, S. Follicular growth and atresia in mammalian ovaries: Regulation by survival and death of granulosa cells. J. Reprod. Dev. 2012, 58, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Regan, S.L.P.; Knight, P.G.; Yovich, J.L.; Leung, Y.; Arfuso, F.; Dharmarajan, A. Granulosa Cell Apoptosis in the Ovarian Follicle-A Changing View. Front. Endocrinol. 2018, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Duffy, D.M.; Ko, C.; Jo, M.; Brannstrom, M.; Curry, T.E. Ovulation: Parallels With Inflammatory Processes. Endocr. Rev. 2019, 40, 369–416. [Google Scholar] [CrossRef]
- Jaffe, L.A.; Egbert, J.R. Regulation of Mammalian Oocyte Meiosis by Intercellular Communication Within the Ovarian Follicle. Annu. Rev. Physiol. 2017, 79, 237–260. [Google Scholar] [CrossRef]
- D’Aurora, M.; Sperduti, S.; Di Emidio, G.; Stuppia, L.; Artini, P.G.; Gatta, V. Inside the granulosa transcriptome. Gynecol. Endocrinol. Off. J. Int. Soc. Gynecol. Endocrinol. 2016, 32, 951–956. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Peng, X.; Mei, S. Autophagy in Ovarian Follicular Development and Atresia. Int. J. Biol. Sci. 2019, 15, 726–737. [Google Scholar] [CrossRef]
- Tilly, J.L. Apoptosis and ovarian function. Rev. Reprod. 1996, 1, 162–172. [Google Scholar] [CrossRef]
- Han, S.; Wang, J.; Cui, C.; Yu, C.; Zhang, Y.; Li, D.; Ma, M.; Du, H.; Jiang, X.; Zhu, Q.; et al. Fibromodulin is involved in autophagy and apoptosis of granulosa cells affecting the follicular atresia in chicken. Poult. Sci. 2022, 101, 101524. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.; Dang, Y.; Li, D.; Lu, G.; Chan, W.Y.; Leung, P.C.K.; Zhao, S.; Qin, Y.; Chen, Z.J. Long noncoding RNA HCP5 participates in premature ovarian insufficiency by transcriptionally regulating MSH5 and DNA damage repair via YB1. Nucleic Acids Res. 2020, 48, 4480–4491. [Google Scholar] [CrossRef]
- Yu, Y.S.; Sui, H.S.; Han, Z.B.; Li, W.; Luo, M.J.; Tan, J.H. Apoptosis in granulosa cells during follicular atresia: Relationship with steroids and insulin-like growth factors. Cell Res. 2004, 14, 341–346. [Google Scholar] [CrossRef]
- Hu, J.; Jin, J.; Qu, Y.; Liu, W.; Ma, Z.; Zhang, J.; Chen, F. ERO1α inhibits cell apoptosis and regulates steroidogenesis in mouse granulosa cells. Mol. Cell. Endocrinol. 2020, 511, 110842. [Google Scholar] [CrossRef] [PubMed]
- Gershon, E.; Dekel, N. Newly Identified Regulators of Ovarian Folliculogenesis and Ovulation. Int. J. Mol. Sci. 2020, 21, 4565. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Samuels, D.C.; Zhao, S.; Xiang, Y.; Zhao, Y.Y.; Guo, Y. Current Research on Non-Coding Ribonucleic Acid (RNA). Genes 2017, 8, 366. [Google Scholar] [CrossRef] [PubMed]
- Bushati, N.; Cohen, S.M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, K.; LaPierre, M.P.; Gasser, E.; Denzler, R.; Yang, Y.; Rülicke, T.; Kero, J.; Latreille, M.; Stoffel, M. Loss of microRNA-7a2 induces hypogonadotropic hypogonadism and infertility. J. Clin. Investig. 2017, 127, 1061–1074. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xiao, H.; Zhang, X.; Qiukai, E.; Gong, X.; Li, T.; Han, Y.; Ying, X.; Cherrington, B.D.; Xu, B.; et al. Decreased microRNA-125b-5p disrupts follicle steroidogenesis through targeting PAK3/ERK1/2 signalling in mouse preantral follicles. Metab. Clin. Exp. 2020, 107, 154241. [Google Scholar] [CrossRef]
- Bartolucci, A.F.; Uliasz, T.; Peluso, J.J. MicroRNA-21 as a regulator of human cumulus cell viability and its potential influence on the developmental potential of the oocyte. Biol. Reprod. 2020, 103, 94–103. [Google Scholar] [CrossRef]
- Li, H.; Wang, X.; Mu, H.; Mei, Q.; Liu, Y.; Min, Z.; Zhang, L.; Su, P.; Xiang, W. Mir-484 contributes to diminished ovarian reserve by regulating granulosa cell function via YAP1-mediated mitochondrial function and apoptosis. Int. J. Biol. Sci. 2022, 18, 1008–1021. [Google Scholar] [CrossRef]
- Qu, C.; Liu, X.; Guo, Y.; Fo, Y.; Chen, X.; Zhou, J.; Yang, B. MiR-128-3p inhibits vascular smooth muscle cell proliferation and migration by repressing FOXO4/MMP9 signaling pathway. Mol. Med. 2020, 26, 116. [Google Scholar] [CrossRef]
- Zhao, J.; Li, D.; Fang, L. MiR-128-3p suppresses breast cancer cellular progression via targeting LIMK1. Biomed. Pharmacother.=Biomed. Pharmacother. 2019, 115, 108947. [Google Scholar] [CrossRef]
- Du, X.; Li, Y.; Lian, B.; Yin, X. microRNA-128-3p inhibits proliferation and accelerates apoptosis of gastric cancer cells via inhibition of TUFT1. World J. Surg. Oncol. 2023, 21, 47. [Google Scholar] [CrossRef]
- Feng, G.; Liu, J.; Lu, Z.; Li, Y.; Deng, M.; Liu, G.; Sun, B.; Guo, Y.; Zou, X.; Liu, D. miR-450-5p and miR-202-5p Synergistically Regulate Follicle Development in Black Goat. Int. J. Mol. Sci. 2022, 24, 401. [Google Scholar] [CrossRef]
- Woo, I.; Christenson, L.K.; Gunewardena, S.; Ingles, S.A.; Thomas, S.; Ahmady, A.; Chung, K.; Bendikson, K.; Paulson, R.; McGinnis, L.K. Micro-RNAs involved in cellular proliferation have altered expression profiles in granulosa of young women with diminished ovarian reserve. J. Assist. Reprod. Genet. 2018, 35, 1777–1786. [Google Scholar] [CrossRef] [PubMed]
- Gnanapavan, S.; Kola, B.; Bustin, S.A.; Morris, D.G.; McGee, P.; Fairclough, P.; Bhattacharya, S.; Carpenter, R.; Grossman, A.B.; Korbonits, M. The tissue distribution of the mRNA of ghrelin and subtypes of its receptor, GHS-R, in humans. J. Clin. Endocrinol. Metab. 2002, 87, 2988. [Google Scholar] [CrossRef] [PubMed]
- Davis, J. Hunger, ghrelin and the gut. Brain Res. 2018, 1693, 154–158. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.T.; Kola, B.; Korbonits, M. The ghrelin/GOAT/GHS-R system and energy metabolism. Rev. Endocr. Metab. Disord. 2011, 12, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Qian, L.; Li, J.; Ming, H.; Fang, L.; Li, Y.; Zhang, M.; Xu, Y.; Ban, Y.; Zhang, W.; et al. GHSR deficiency exacerbates cardiac fibrosis: Role in macrophage inflammasome activation and myofibroblast differentiation. Cardiovasc. Res. 2020, 116, 2091–2102. [Google Scholar] [CrossRef]
- Yuan, M.J.; Wang, T. The new mechanism of Ghrelin/GHSR-1a on autophagy regulation. Peptides 2020, 126, 170264. [Google Scholar] [CrossRef]
- Schalla, M.A.; Stengel, A. The Role of the Gastric Hormones Ghrelin and Nesfatin-1 in Reproduction. Int. J. Mol. Sci. 2021, 22, 11059. [Google Scholar] [CrossRef]
- Kheradmand, A.; Roshangar, L.; Taati, M.; Sirotkin, A.V. Morphometrical and intracellular changes in rat ovaries following chronic administration of ghrelin. Tissue Cell 2009, 41, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Rak, A.; Szczepankiewicz, D.; Gregoraszczuk, E. Expression of ghrelin receptor, GHSR-1a, and its functional role in the porcine ovarian follicles. Growth Horm. IGF Res. Off. J. Growth Horm. Res. Soc. Int. IGF Res. Soc. 2009, 19, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.H.; Chen, M.J. The Effect of Steroid Hormones on Ovarian Follicle Development. Vitam. Horm. 2018, 107, 155–175. [Google Scholar] [CrossRef]
- Andersen, C.Y.; Ezcurra, D. Human steroidogenesis: Implications for controlled ovarian stimulation with exogenous gonadotropins. Reprod. Biol. Endocrinol. RBE 2014, 12, 128. [Google Scholar] [CrossRef]
- Guo, I.C.; Hu, M.C.; Chung, B.C. Transcriptional regulation of CYP11A1. J. Biomed. Sci. 2003, 10, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Liu, T.; Shasaltaneh, M.D.; Wang, X.; Imani, S.; Wen, Q. Targeting Adenylate Cyclase Family: New Concept of Targeted Cancer Therapy. Front. Oncol. 2022, 12, 829212. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.; Li, J. The art of oocyte meiotic arrest regulation. Reprod. Biol. Endocrinol. RBE 2019, 17, 8. [Google Scholar] [CrossRef] [PubMed]
- Andrieu, T.; Pezzi, V.; Sirianni, R.; Le Bas, R.; Feral, C.; Benhaim, A.; Mittre, H. cAMP-dependent regulation of CYP19 gene in rabbit preovulatory granulosa cells and corpus luteum. J. Steroid Biochem. Mol. Biol. 2009, 116, 110–117. [Google Scholar] [CrossRef]
- Bhardwaj, J.K.; Paliwal, A.; Saraf, P.; Sachdeva, S.N. Role of autophagy in follicular development and maintenance of primordial follicular pool in the ovary. J. Cell. Physiol. 2022, 237, 1157–1170. [Google Scholar] [CrossRef]
- Chu, Y.L.; Xu, Y.R.; Yang, W.X.; Sun, Y. The role of FSH and TGF-β superfamily in follicle atresia. Aging 2018, 10, 305–321. [Google Scholar] [CrossRef]
- Meng, L.; Jan, S.Z.; Hamer, G.; van Pelt, A.M.; van der Stelt, I.; Keijer, J.; Teerds, K.J. Preantral follicular atresia occurs mainly through autophagy, while antral follicles degenerate mostly through apoptosis. Biol. Reprod. 2018, 99, 853–863. [Google Scholar] [CrossRef]
- Kumariya, S.; Ubba, V.; Jha, R.K.; Gayen, J.R. Autophagy in ovary and polycystic ovary syndrome: Role, dispute and future perspective. Autophagy 2021, 17, 2706–2733. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, Y.; Liu, H.; Pan, Z. MicroRNAs in ovarian follicular atresia and granulosa cell apoptosis. Reprod. Biol. Endocrinol. RBE 2019, 17, 9. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Z.; Wang, S.; Du, X.; Li, Q. miR-423 sponged by lncRNA NORHA inhibits granulosa cell apoptosis. J. Anim. Sci. Biotechnol. 2023, 14, 154. [Google Scholar] [CrossRef] [PubMed]
- Ning, Z.; Deng, X.; Li, L.; Feng, J.; Du, X.; Amevor, F.K.; Tian, Y.; Li, L.; Rao, Y.; Yi, Z.; et al. miR-128-3p regulates chicken granulosa cell function via 14-3-3β/FoxO and PPAR-γ/LPL signaling pathways. Int. J. Biol. Macromol. 2023, 241, 124654. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xue, L.; Xu, W.; Liu, P.; Li, F. rno-miR-128-3p promotes apoptosis in rat granulosa cells (GCs) induced by norepinephrine through Wilms tumor 1 (WT1). Vitr. Cell. Dev. Biol. Anim. 2021, 57, 775–785. [Google Scholar] [CrossRef] [PubMed]
- Stavast, C.J.; Erkeland, S.J. The Non-Canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells 2019, 8, 1465. [Google Scholar] [CrossRef] [PubMed]
- Rykova, E.; Ershov, N.; Damarov, I.; Merkulova, T. SNPs in 3′UTR miRNA Target Sequences Associated with Individual Drug Susceptibility. Int. J. Mol. Sci. 2022, 23, 13725. [Google Scholar] [CrossRef] [PubMed]
- Pu, M.; Chen, J.; Tao, Z.; Miao, L.; Qi, X.; Wang, Y.; Ren, J. Regulatory network of miRNA on its target: Coordination between transcriptional and post-transcriptional regulation of gene expression. Cell. Mol. Life Sci. CMLS 2019, 76, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Sirotkin, A.V.; Alexa, R.; Harrath, A.H. Puncturevine (Tribulus terrestris L.) affects the proliferation, apoptosis, and ghrelin response of ovarian cells. Reprod. Biol. 2020, 20, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Benco, A.; Sirotkin, A.V.; Vasícek, D.; Pavlová, S.; Zemanová, J.; Kotwica, J.; Darlak, K.; Valenzuela, F. Involvement of the transcription factor STAT1 in the regulation of porcine ovarian granulosa cell functions treated and not treated with ghrelin. Reproduction 2009, 138, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Sirotkin, A.V.; Grossmann, R. Effects of ghrelin and its analogues on chicken ovarian granulosa cells. Domest. Anim. Endocrinol. 2008, 34, 125–134. [Google Scholar] [CrossRef]
- Hsieh, M.; Lee, D.; Panigone, S.; Horner, K.; Chen, R.; Theologis, A.; Lee, D.C.; Threadgill, D.W.; Conti, M. Luteinizing hormone-dependent activation of the epidermal growth factor network is essential for ovulation. Mol. Cell. Biol. 2007, 27, 1914–1924. [Google Scholar] [CrossRef]
- Fan, H.Y.; Liu, Z.; Shimada, M.; Sterneck, E.; Johnson, P.F.; Hedrick, S.M.; Richards, J.S. MAPK3/1 (ERK1/2) in ovarian granulosa cells are essential for female fertility. Science 2009, 324, 938–941. [Google Scholar] [CrossRef]
- Liu, J.; Liu, D.; Zhang, X.; Li, Y.; Fu, X.; He, W.; Li, M.; Chen, P.; Zeng, G.; DiSanto, M.E.; et al. NELL2 modulates cell proliferation and apoptosis via ERK pathway in the development of benign prostatic hyperplasia. Clin. Sci. 2021, 135, 1591–1608. [Google Scholar] [CrossRef]
- Nishi, Y.; Yanase, T.; Mu, Y.; Oba, K.; Ichino, I.; Saito, M.; Nomura, M.; Mukasa, C.; Okabe, T.; Goto, K.; et al. Establishment and characterization of a steroidogenic human granulosa-like tumor cell line, KGN, that expresses functional follicle-stimulating hormone receptor. Endocrinology 2001, 142, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zheng, L.; Wang, Y.; Gao, Y.; Xu, Y. Arachidonic Acid in Follicular Fluid of PCOS Induces Oxidative Stress in a Human Ovarian Granulosa Tumor Cell Line (KGN) and Upregulates GDF15 Expression as a Response. Front. Endocrinol. 2022, 13, 865748. [Google Scholar] [CrossRef] [PubMed]
- Chao, Y.N.; Sun, D.; Peng, Y.C.; Wu, Y.L. Growth Hormone Releasing Peptide-2 Attenuation of Protein Kinase C-Induced Inflammation in Human Ovarian Granulosa Cells. Int. J. Mol. Sci. 2016, 17, 1359. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, S.; Jiang, S.; Hou, B.; Li, Y.; Sun, B.; Guo, Y.; Deng, M.; Liu, D.; Liu, G. miR-128-3p Regulates Follicular Granulosa Cell Proliferation and Apoptosis by Targeting the Growth Hormone Secretagogue Receptor. Int. J. Mol. Sci. 2024, 25, 2720. https://doi.org/10.3390/ijms25052720
Dong S, Jiang S, Hou B, Li Y, Sun B, Guo Y, Deng M, Liu D, Liu G. miR-128-3p Regulates Follicular Granulosa Cell Proliferation and Apoptosis by Targeting the Growth Hormone Secretagogue Receptor. International Journal of Molecular Sciences. 2024; 25(5):2720. https://doi.org/10.3390/ijms25052720
Chicago/Turabian StyleDong, Shucan, Shengwei Jiang, Biwei Hou, Yaokun Li, Baoli Sun, Yongqing Guo, Ming Deng, Dewu Liu, and Guangbin Liu. 2024. "miR-128-3p Regulates Follicular Granulosa Cell Proliferation and Apoptosis by Targeting the Growth Hormone Secretagogue Receptor" International Journal of Molecular Sciences 25, no. 5: 2720. https://doi.org/10.3390/ijms25052720
APA StyleDong, S., Jiang, S., Hou, B., Li, Y., Sun, B., Guo, Y., Deng, M., Liu, D., & Liu, G. (2024). miR-128-3p Regulates Follicular Granulosa Cell Proliferation and Apoptosis by Targeting the Growth Hormone Secretagogue Receptor. International Journal of Molecular Sciences, 25(5), 2720. https://doi.org/10.3390/ijms25052720





