New Biocides Based on N4-Alkylcytidines: Effects on Microorganisms and Application for the Protection of Cultural Heritage Objects of Painting
Abstract
1. Introduction
2. Results
2.1. Chemistry
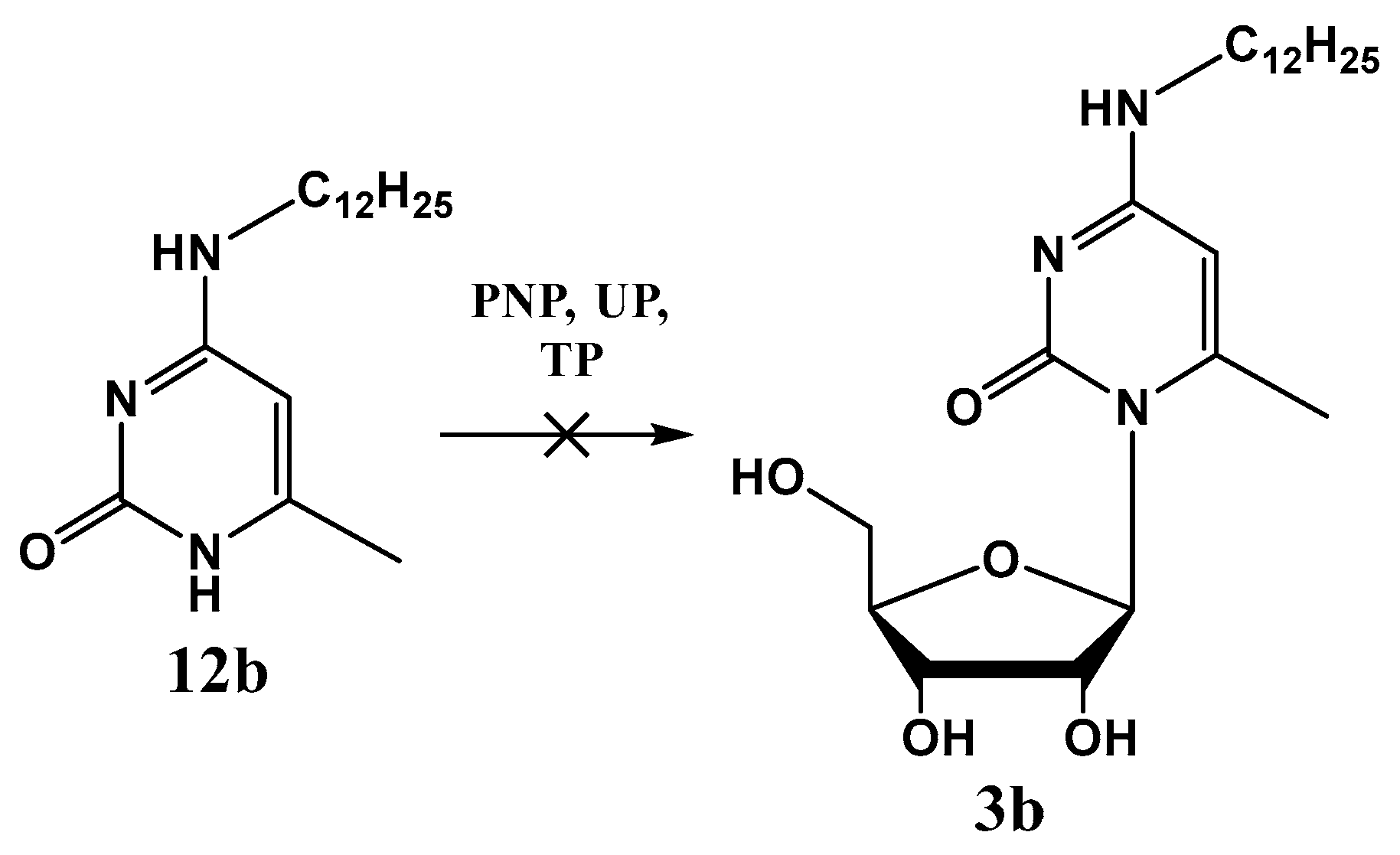
2.2. Enzymology
2.3. Cytotoxicity
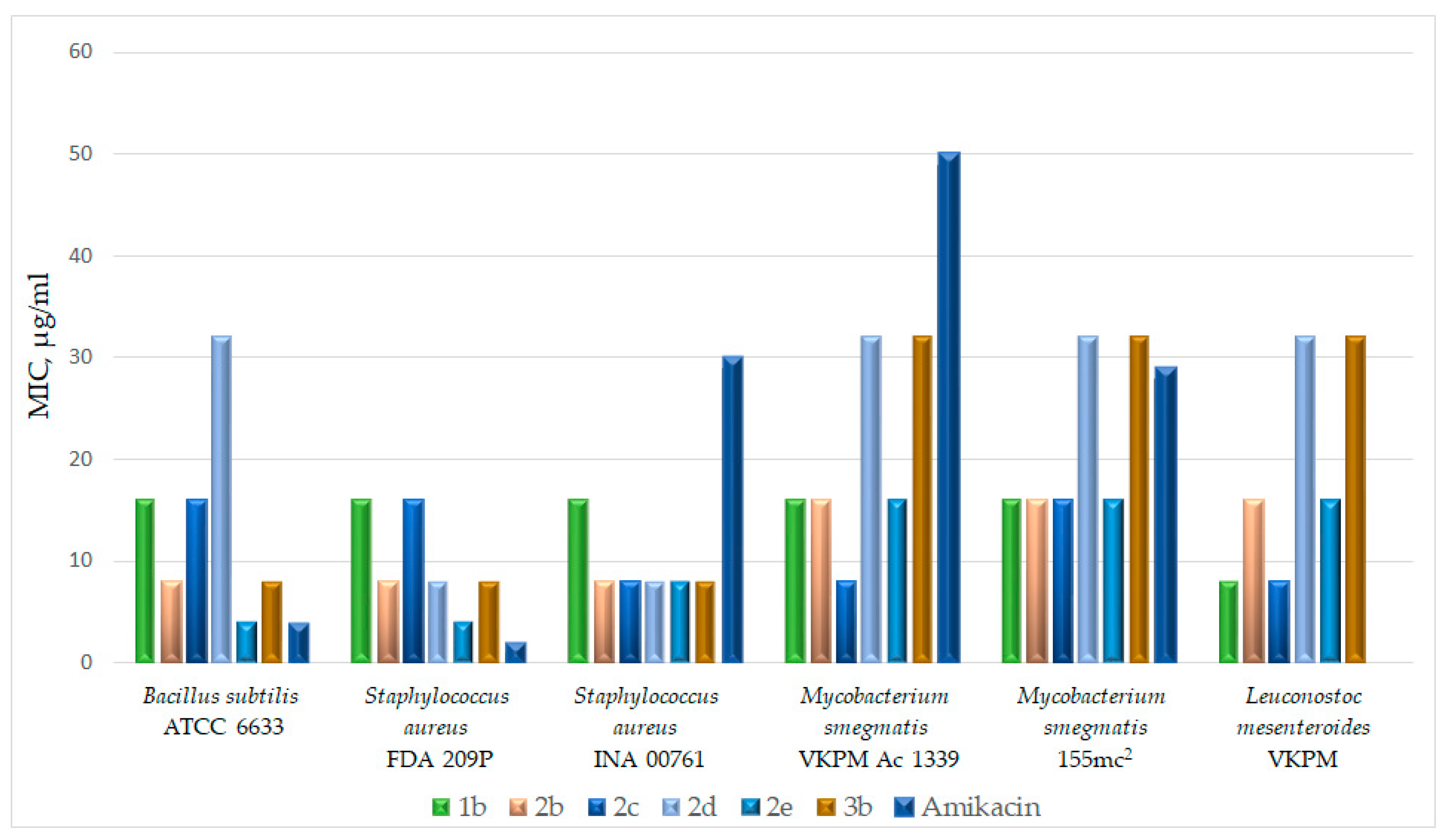
2.4. In Vitro Study of Antibacterial Activity of the Obtained Compounds
In Vitro Study of Antibacterial Activity of the Obtained Compounds
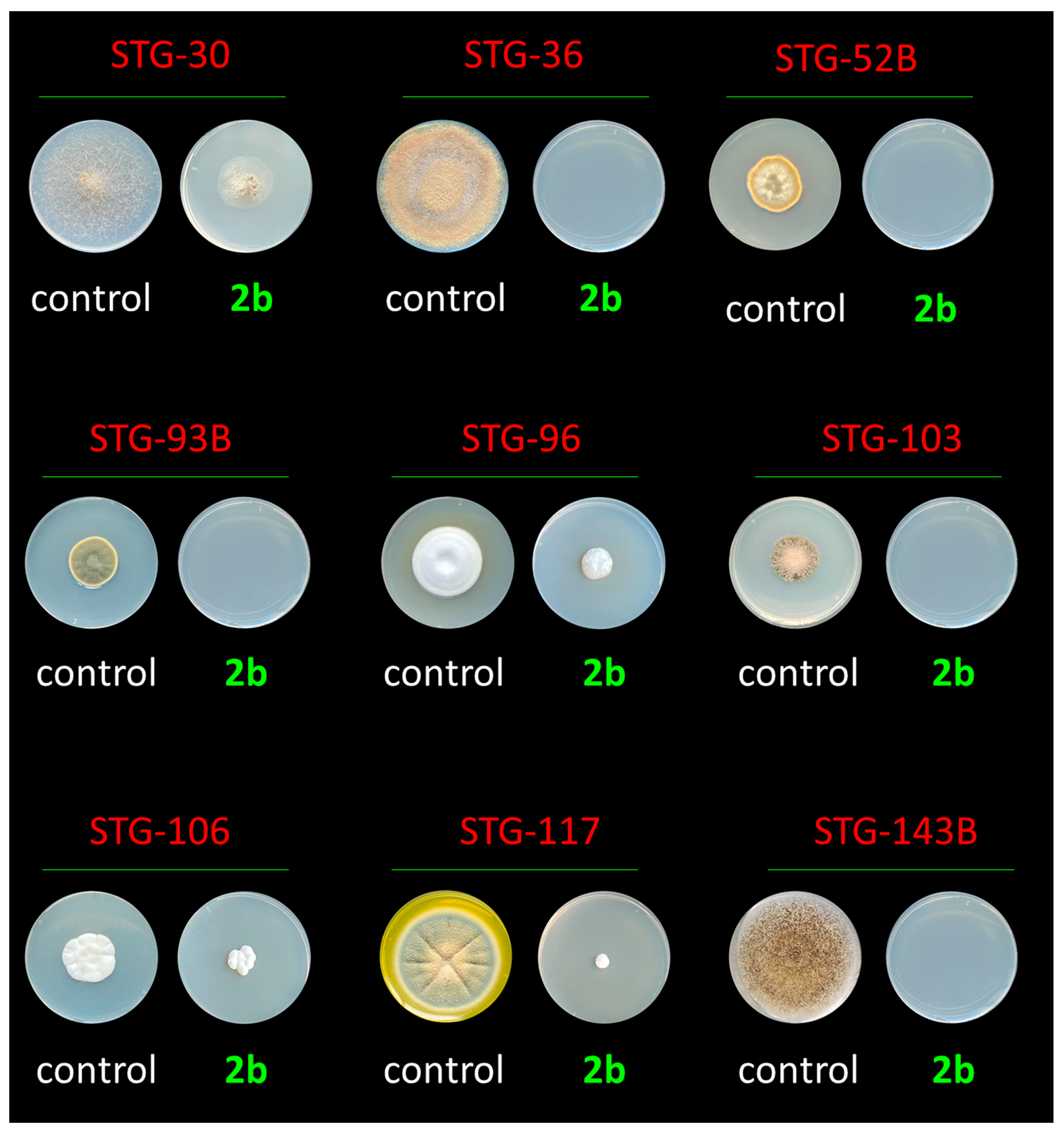
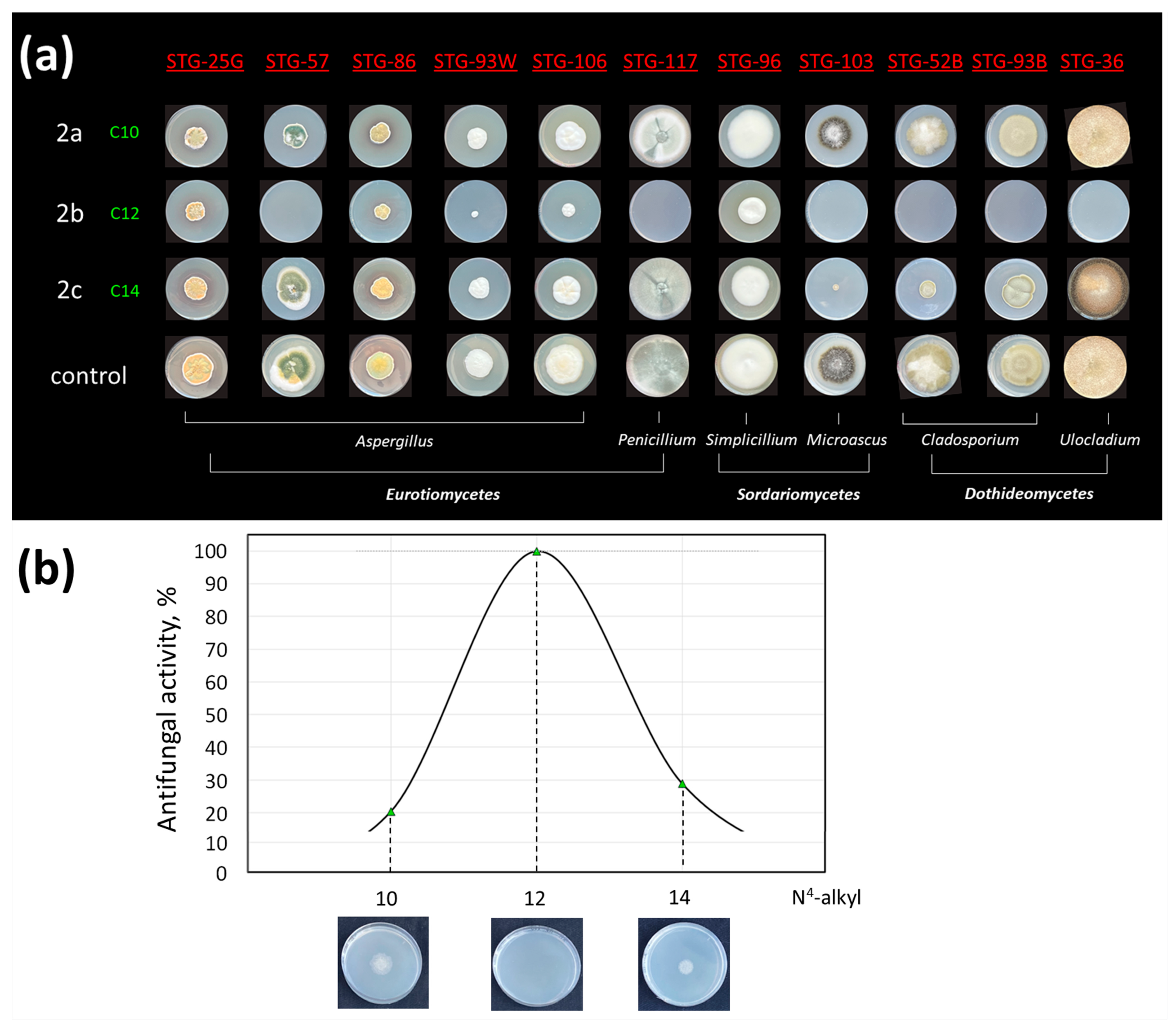
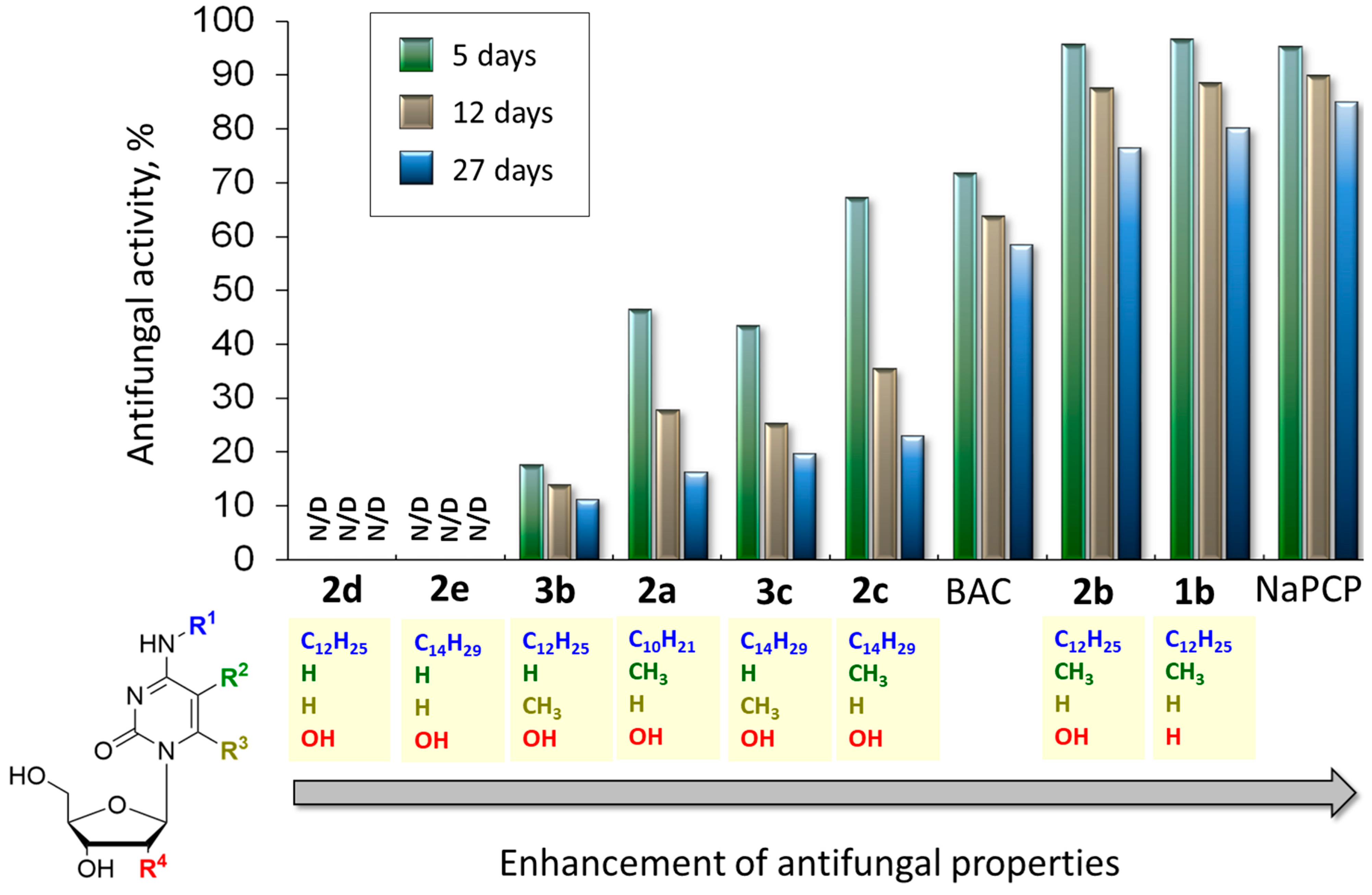
2.5. In Vitro Study of the Antifungal Activity of the Obtained Compounds
2.5.1. The Role of the Hydroxyl Group in the 2′-Position of Pentose
2.5.2. The Role of the Methyl Group in the Fifth or Sixth Position of Cytidine Derivatives
2.5.3. The Role of the Size of Alkyl Group in the N4 Position of Cytidine Derivatives
3. Discussion
4. Materials and Methods
4.1. General Information
General Method for the Synthesis of Compounds 2a–e
4.2. Enzymatic Synthesis of Nucleosides Using Nucleoside Phosphorylases
Enzymatic Reactions
4.3. Biological Evaluation
4.3.1. Antibacterial Effect
Bacterial Strains
In Vitro Study of the Antibacterial Effect
4.3.2. Fungal Growth Inhibition
Fungal Strains
Fungal Growth Inhibition
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aminov, R.I. A Brief History of the Antibiotic Era: Lessons Learned and Challenges for the Future. Front. Microbiol. 2010, 1, 134. [Google Scholar] [CrossRef]
- Butler, M.S.; Henderson, I.R.; Capon, R.J.; Blaskovich, M.A.T. Antibiotics in the clinical pipeline as of December 2022. J. Antibiot. 2023, 76, 431–473. [Google Scholar] [CrossRef]
- Cook, M.A.; Wright, G.D. The past, present, and future of antibiotics. Sci. Transl. Med. 2022, 14, eabo7793. [Google Scholar] [CrossRef]
- Zhgun, A.A. Industrial Production of Antibiotics in Fungi: Current State, Deciphering the Molecular Basis of Classical Strain Improvement and Increasing the Production of High-Yielding Strains by the Addition of Low-Molecular Weight Inducers. Fermentation 2023, 9, 1027. [Google Scholar] [CrossRef]
- Zhuang, M.; Achmon, Y.; Cao, Y.; Liang, X.; Chen, L.; Wang, H.; Siame, B.A.; Leung, K.Y. Distribution of antibiotic resistance genes in the environment. Environ. Pollut. 2021, 285, 117402. [Google Scholar] [CrossRef]
- Hutchings, M.; Truman, A.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef]
- Gould, K. Antibiotics: From prehistory to the present day. J. Antimicrob. Chemother. 2016, 71, 572–575. [Google Scholar] [CrossRef] [PubMed]
- Podolsky, S.H. The evolving response to antibiotic resistance (1945–2018). Palgrave Commun. 2018, 4, 124. [Google Scholar] [CrossRef]
- Upmanyu, N.; Malviya, V.N. Antibiotics: Mechanisms of action and modern challenges. In Microorganisms for Sustainable Environment and Health; Chowdhary, P., Raj, A., Verma, D., Akhter, Y., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; Volume 1, pp. 367–382. ISBN 9780128190012. [Google Scholar]
- Correia, J.; Borges, A.; Simões, M.; Simões, L.C. Beyond Penicillin: The Potential of Filamentous Fungi for Drug Discovery in the Age of Antibiotic Resistance. Antibiotics 2023, 12, 1250. [Google Scholar] [CrossRef]
- Prescott, T.A.K.; Hill, R.; Mas-Claret, E.; Gaya, E.; Burns, E. Fungal Drug Discovery for Chronic Disease: History, New Discoveries and New Approaches. Biomolecules 2023, 13, 986. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, P.S. Antibacterial Discovery: 21st Century Challenges. Antibiotics 2020, 9, 213. [Google Scholar] [CrossRef]
- World Health Organization. Global Antibiotic Resistance Report 2023; World Health Organization: Geneva, Switzerland, 2023. [Google Scholar]
- López-Miras, M.d.M.; Martín-Sánchez, I.; Yebra-Rodríguez, Á.; Romero-Noguera, J.; Bolívar-Galiano, F.; Ettenauer, J.; Sterflinger, K.; Piñar, G. Contribution of the Microbial Communities Detected on an Oil Painting on Canvas to Its Biodeterioration. PLoS ONE 2013, 8, e80198. [Google Scholar] [CrossRef]
- Zhgun, A.; Avdanina, D.; Shumikhin, K.; Simonenko, N.; Lyubavskaya, E.; Volkov, I.; Ivanov, V. Detection of potential biodeterioration risks for tempera painting in 16th century exhibits from State Tretyakov Gallery. PLoS ONE 2020, 15, e0230591. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, B.; He, Z.; Yang, X. Distribution and Diversity of Bacteria and Fungi Colonization in Stone Monuments Analyzed by High-Throughput Sequencing. PLoS ONE 2016, 11, e0163287. [Google Scholar] [CrossRef]
- Gadd, G.M.; Fomina, M.; Pinzari, F. Fungal biodeterioration and preservation of cultural heritage, artwork, and historical artifacts: Extremophily and adaptation. Microbiol. Mol. Biol. Rev. 2024, e00200-22. [Google Scholar] [CrossRef]
- Salvador, C.; Sandu, I.C.A.; Sandbakken, E.; Candeias, A.; Caldeira, A.T. Biodeterioration in art: A case study of Munch’s paintings. Eur. Phys. J. Plus 2022, 137, 11. [Google Scholar] [CrossRef]
- De Leo, F.; Isola, D. The Role of Fungi in Biodeterioration of Cultural Heritage: New Insights for Their Control. Appl. Sci. 2022, 12, 10490. [Google Scholar] [CrossRef]
- Zhgun, A.; Avdanina, D.; Shagdarova, B.; Nuraeva, G.; Shumikhin, K.; Zhuikova, Y.; Il’ina, A.; Troyan, E.; Shitov, M.; Varlamov, V. The Application of Chitosan for Protection of Cultural Heritage Objects of the 15–16th Centuries in the State Tretyakov Gallery. Materials 2022, 15, 7773. [Google Scholar] [CrossRef] [PubMed]
- Bastian, F.; Alabouvette, C.; Jurado, V.; Saiz-Jimenez, C. Impact of biocide treatments on the bacterial communities of the Lascaux Cave. Naturwissenschaften 2009, 96, 863–868. [Google Scholar] [CrossRef] [PubMed]
- Nagai, K.; Ohta, S.; Zenda, H.; Matsumoto, H.; Makino, M. Biochemical characterization of a Pseudomonas fluorescens strain isolated from a benzalkonium chloride solution. Biol. Pharm. Bull. 1996, 19, 873–875. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shim, H. Three Innovations of Next-Generation Antibiotics: Evolvability, Specificity, and Non-Immunogenicity. Antibiotics 2023, 12, 204. [Google Scholar] [CrossRef]
- Spagnolo, F.; Trujillo, M.; Dennehy, J.J. Why Do Antibiotics Exist? MBio 2021, 12, e0196621. [Google Scholar] [CrossRef]
- Alexandrova, L.A.; Khandazhinskaya, A.L.; Matyugina, E.S.; Makarov, D.A.; Kochetkov, S.N. Analogues of Pyrimidine Nucleosides as Mycobacteria Growth Inhibitors. Microorganisms 2022, 10, 1299. [Google Scholar] [CrossRef] [PubMed]
- Yssel, A.E.J.; Vanderleyden, J.; Steenackers, H.P. Repurposing of nucleoside- and nucleobase-derivative drugs as antibiotics and biofilm inhibitors. J. Antimicrob. Chemother. 2017, 72, 2326–2333. [Google Scholar] [CrossRef] [PubMed]
- Jordheim, L.P.; Durantel, D.; Zoulim, F.; Dumontet, C. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat. Rev. Drug Discov. 2013, 12, 447–464. [Google Scholar] [CrossRef] [PubMed]
- Osada, H. Discovery and applications of nucleoside antibiotics beyond polyoxin. J. Antibiot. 2019, 72, 855–864. [Google Scholar] [CrossRef] [PubMed]
- Bugg, T.D.H.; Kerr, R.V. Mechanism of action of nucleoside antibacterial natural product antibiotics. J. Antibiot. 2019, 72, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Alexandrova, L.A.; Jasko, M.V.; Negrya, S.D.; Solyev, P.N.; Shevchenko, O.V.; Solodinin, A.P.; Kolonitskaya, D.P.; Karpenko, I.L.; Efremenkova, O.V.; Glukhova, A.A.; et al. Discovery of novel N4−alkylcytidines as promising antimicrobial agents. Eur. J. Med. Chem. 2021, 215, 113212. [Google Scholar] [CrossRef] [PubMed]
- Alexandrova, L.A.; Shevchenko, O.V.; Jasko, M.V.; Solyev, P.N.; Karpenko, I.L.; Negrya, S.D.; Efremenkova, O.V.; Vasilieva, B.F.; Efimenko, T.A.; Avdanina, D.A.; et al. 3′-Amino modifications enhance the antifungal properties of N 4-alkyl-5-methylcytidines for potential biocides. New J. Chem. 2022, 46, 5614–5626. [Google Scholar] [CrossRef]
- Suck, D.; Saenger, W. Molecular and crystal structure of 6-methyluridine, a pyrimidine nucleoside in the syn conformation. J. Am. Chem. Soc. 1972, 94, 6520–6526. [Google Scholar] [CrossRef]
- Felczak, K.; Drabikowska, A.K.; Vilpo, J.A.; Kulikowski, T.; Shugar, D. 6-Substituted and 5,6-Disubstituted Derivatives of Uridine: Stereoselective Synthesis, Interaction with Uridine Phosphorylase, and in Vitro Antitumor Activity. J. Med. Chem. 1996, 39, 1720–1728. [Google Scholar] [CrossRef]
- Sochacka, E.; Czerwinska, G.; Guenther, R.; Cain, R.; Agris, P.F.; Malkiewicz, A. Synthesis and Properties of Uniquely Modified Oligoribonucleotides: Yeast Trna Phe Fragments with 6-Methyluridine and 5,6-Dimethyluridine at Site-Specific Positions. Nucleosides Nucleotides Nucleic Acids 2000, 19, 515–531. [Google Scholar] [CrossRef]
- Schweizer, M.P.; Witkowski, J.T.; Robins, R.K. Nuclear magnetic resonance determination of syn and anti conformations in pyrimidine nucleosides. J. Am. Chem. Soc. 1971, 93, 277–279. [Google Scholar] [CrossRef]
- Volov, A.N.; Volov, N.A.; Platonova, Y.B. Design and synthesis of novel 5-alkynyl pyrimidine nucleosides derivatives: Influence of C-6-substituent on antituberculosis activity. Bioorg. Med. Chem. Lett. 2021, 48, 128261. [Google Scholar] [CrossRef]
- Cody, V.; Kalman, T.I. Conformational Analysis of 6-Substituted Uridine Analogues: Crystal Structures of Uridine-6-Thiocarboxamide and 6-Cyanouridine. Nucleosides Nucleotides 1985, 4, 587–594. [Google Scholar] [CrossRef]
- Akula, H.K.; Kokatla, H.; Andrei, G.; Snoeck, R.; Schols, D.; Balzarini, J.; Yang, L.; Lakshman, M.K. Facile functionalization at the C4 position of pyrimidine nucleosides via amide group activation with (benzotriazol-1-yloxy)tris(dimethylamino)phosphonium hexafluorophosphate (BOP) and biological evaluations of the products. Org. Biomol. Chem. 2017, 15, 1130–1139. [Google Scholar] [CrossRef] [PubMed]
- Amblard, F.; LeCher, J.C.; De, R.; Goh, S.L.; Li, C.; Kasthuri, M.; Biteau, N.; Zhou, L.; Tber, Z.; Downs-Bowen, J.; et al. Synthesis of Novel N4-Hydrocytidine Analogs as Potential Anti-SARS-CoV-2 Agents. Pharmaceuticals 2022, 15, 1144. [Google Scholar] [CrossRef] [PubMed]
- Divakar, K.J.; Reese, C.B. 4-(1,2,4-Triazol-1-yl)-and 4-(3-nitro-1,2,4-triazol-1-yl)-1-(β-D-2,3,5-tri-O-acetylarabinofuranosyl)pyrimidin-2(1H)-ones. Valuable intermediates in the synthesis of derivatives of 1-(β-D-arabinofuranosyl)cytosine (ara-C). J. Chem. Soc. Perkin Trans. 1982, 1, 1171–1176. [Google Scholar] [CrossRef]
- Lin, T.S.; Gao, Y.S.; Mancini, W.R. Synthesis and biological activity of various 3′-azido and 3′-amino analogs of 5-substituted pyrimidine deoxyribonucleosides. J. Med. Chem. 1983, 26, 1691–1696. [Google Scholar] [CrossRef]
- Niedballa, U.; Vorbrüggen, H. A General Synthesis of Pyrimidine Nucleosides. Angew. Chemie Int. Ed. English 1970, 9, 461–462. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, K.; Katayama, H.; Wakabayashi, T.; Kumonaka, T. Pyrimidine Derivatives; 1. The Highly Regioselective 4-Thionation of Pyrimidine-2,4(1 H,3 H)-dione Derivatives with Lawesson Reagent. Synthesis 1988, 1988, 152–154. [Google Scholar] [CrossRef]
- Phillips, A.P. Some 5-Substituted Aminouracils. J. Am. Chem. Soc. 1951, 73, 1061–1062. [Google Scholar] [CrossRef]
- Kezin, V.A.; Matyugina, E.S.; Novikov, M.S.; Chizhov, A.O.; Snoeck, R.; Andrei, G.; Kochetkov, S.N.; Khandazhinskaya, A.L. New Derivatives of 5-Substituted Uracils: Potential Agents with a Wide Spectrum of Biological Activity. Molecules 2022, 27, 2866. [Google Scholar] [CrossRef]
- Alexeev, C.S.; Sivets, G.G.; Safonova, T.N.; Mikhailov, S.N. Substrate specificity of E. coli uridine phosphorylase. Further evidences of high- syn conformation of the substrate in uridine phosphorolysis. Nucleosides Nucleotides Nucleic Acids 2017, 36, 107–121. [Google Scholar] [CrossRef]
- Khandazhinskaya, A.; Eletskaya, B.; Fateev, I.; Kharitonova, M.; Konstantinova, I.; Barai, V.; Azhayev, A.; Hyvonen, M.T.; Keinanen, T.A.; Kochetkov, S.; et al. Novel fleximer pyrazole-containing adenosine analogues: Chemical, enzymatic and highly efficient biotechnological synthesis. Org. Biomol. Chem. 2021, 19, 7379–7389. [Google Scholar] [CrossRef] [PubMed]
- Niks, M.; Otto, M. Towards an optimized MTT assay. J. Immunol. Methods 1990, 130, 149–151. [Google Scholar] [CrossRef]
- Alexandrova, L.A.; Efremenkova, O.V.; Andronova, V.L.; Galegov, G.A.; Solyev, P.N.; Karpenko, I.L.; Kochetkov, S.N. 5-(4-alkyl-1,2,3-triazol-1-yl)methyl derivatives of 2′-deoxyuridine as inhibitors of viral and bacterial growth. Russ. J. Bioorganic Chem. 2016, 42, 677–684. [Google Scholar] [CrossRef]
- Negrya, S.D.; Jasko, M.V.; Solyev, P.N.; Karpenko, I.L.; Efremenkova, O.V.; Vasilyeva, B.F.; Sumarukova, I.G.; Kochetkov, S.N.; Alexandrova, L.A. Synthesis of water-soluble prodrugs of 5-modified 2′-deoxyuridines and their antibacterial activity. J. Antibiot. 2020, 73, 236–246. [Google Scholar] [CrossRef]
- Zhgun, A.A.; Avdanina, D.A.; Shagdarova, B.T.; Troyan, E.V.; Nuraeva, G.K.; Potapov, M.P.; Il’ina, A.V.; Shitov, M.V.; Varlamov, V.P. Search for Efficient Chitosan-Based Fungicides to Protect the 15th–16th Centuries Tempera Painting in Exhibits from the State Tretyakov Gallery. Microbiology 2020, 89, 750–755. [Google Scholar] [CrossRef]
- Alexandrova, L.A.; Karpenko, I.L.; Kochetkov, S.N.; Negrya, S.D.; Solyev, P.N.; Shevchenko, O.V.; Jasko, M.V.; Nuraeva, G.K.; Potapov, M.P.; Avdanina, D.A.; et al. Patent RU 2766333C1. New N4-modified 5-methyl-2′-deoxycytidines exhibiting antimicotic activity. Materials 2022, 15, 7773. [Google Scholar]
- Makarov, D.A.; Negrya, S.D.; Jasko, M.V.; Karpenko, I.L.; Solyev, P.N.; Chekhov, V.O.; Kaluzhny, D.N.; Efremenkova, O.V.; Vasilyeva, B.F.; Chizhov, A.O.; et al. 5-Substituted Uridines with Activity against Gram-Positive Bacteria. ChemMedChem 2023, 18, e202300366. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D. How dangerous is pentachlorphenol? Med. J. Aust. 1956, 2, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Esipov, R.S.; Gurevich, A.I.; Chuvikovsky, D.V.; Chupova, L.A.; Muravyova, T.I.; Miroshnikov, A.I. Overexpression of Escherichia coli genes encoding nucleoside phosphorylases in the pET/Bl21(DE3) system yields active recombinant enzymes. Protein Expr. Purif. 2002, 24, 56–60. [Google Scholar] [CrossRef] [PubMed]
- Dumina, M.V.; Zhgun, A.A.; Kerpichnikov, I.V.; Domracheva, A.G.; Novak, M.I.; Valiachmetov, A.Y.; Knorre, D.A.; Severin, F.F.; Eldarov, M.A.; Bartoshevich, Y.E. Functional analysis of MFS protein CefT involved in the transport of beta-lactam antibiotics in Acremonium chrysogenum and Saccharomyces cerevisiae. Appl. Biochem. Microbiol. 2013, 49, 368–377. [Google Scholar] [CrossRef]
- Zhgun, A.A.; Nuraeva, G.K.; Dumina, M.V.; Voinova, T.M.; Dzhavakhiya, V.V.; Eldarov, M.A. 1,3-Diaminopropane and Spermidine Upregulate Lovastatin Production and Expression of Lovastatin Biosynthetic Genes in Aspergillus terreus via LaeA Regulation. Appl. Biochem. Microbiol. 2019, 55, 243–254. [Google Scholar] [CrossRef]
- Hyvönen, M.T.; Keinänen, T.A.; Nuraeva, G.K.; Yanvarev, D.V.; Khomutov, M.; Khurs, E.N.; Kochetkov, S.N.; Vepsäläinen, J.; Zhgun, A.A.; Khomutov, A.R. Hydroxylamine analogue of agmatine: Magic bullet for arginine decarboxylase. Biomolecules 2020, 10, 406. [Google Scholar] [CrossRef]
- Zhgun, A.A.; Nuraeva, G.K.; Volkov, I.A. High-Yielding Lovastatin Producer Aspergillus terreus Shows Increased Resistance to Inhibitors of Polyamine Biosynthesis. Appl. Sci. 2020, 10, 8290. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alexandrova, L.A.; Oskolsky, I.A.; Makarov, D.A.; Jasko, M.V.; Karpenko, I.L.; Efremenkova, O.V.; Vasilyeva, B.F.; Avdanina, D.A.; Ermolyuk, A.A.; Benko, E.E.; et al. New Biocides Based on N4-Alkylcytidines: Effects on Microorganisms and Application for the Protection of Cultural Heritage Objects of Painting. Int. J. Mol. Sci. 2024, 25, 3053. https://doi.org/10.3390/ijms25053053
Alexandrova LA, Oskolsky IA, Makarov DA, Jasko MV, Karpenko IL, Efremenkova OV, Vasilyeva BF, Avdanina DA, Ermolyuk AA, Benko EE, et al. New Biocides Based on N4-Alkylcytidines: Effects on Microorganisms and Application for the Protection of Cultural Heritage Objects of Painting. International Journal of Molecular Sciences. 2024; 25(5):3053. https://doi.org/10.3390/ijms25053053
Chicago/Turabian StyleAlexandrova, Liudmila A., Ivan A. Oskolsky, Dmitry A. Makarov, Maxim V. Jasko, Inna L. Karpenko, Olga V. Efremenkova, Byazilya F. Vasilyeva, Darya A. Avdanina, Anna A. Ermolyuk, Elizaveta E. Benko, and et al. 2024. "New Biocides Based on N4-Alkylcytidines: Effects on Microorganisms and Application for the Protection of Cultural Heritage Objects of Painting" International Journal of Molecular Sciences 25, no. 5: 3053. https://doi.org/10.3390/ijms25053053
APA StyleAlexandrova, L. A., Oskolsky, I. A., Makarov, D. A., Jasko, M. V., Karpenko, I. L., Efremenkova, O. V., Vasilyeva, B. F., Avdanina, D. A., Ermolyuk, A. A., Benko, E. E., Kalinin, S. G., Kolganova, T. V., Berzina, M. Y., Konstantinova, I. D., Chizhov, A. O., Kochetkov, S. N., & Zhgun, A. A. (2024). New Biocides Based on N4-Alkylcytidines: Effects on Microorganisms and Application for the Protection of Cultural Heritage Objects of Painting. International Journal of Molecular Sciences, 25(5), 3053. https://doi.org/10.3390/ijms25053053










