Abstract
The BRI1 EMS suppressor 1(BES1) transcription factor is a crucial regulator in the signaling pathway of Brassinosteroid (BR) and plays an important role in plant growth and response to abiotic stress. Although the identification and functional validation of BES1 genes have been extensively explored in various plant species, the understanding of their role in woody plants—particularly the endangered species Phoebe bournei (Hemsl.) Yang—remains limited. In this study, we identified nine members of the BES1 gene family in the genome of P. bournei; these nine members were unevenly distributed across four chromosomes. In our further evolutionary analysis of PbBES1, we discovered that PbBES1 can be divided into three subfamilies (Class I, Class II, and Class IV) based on the evolutionary tree constructed with Arabidopsis thaliana, Oryza sativa, and Solanum lycopersicum. Each subfamily contains 2–5 PbBES1 genes. There were nine pairs of homologous BES1 genes in the synteny analysis of PbBES1 and AtBES1. Three segmental replication events and one pair of tandem duplication events were present among the PbBES1 family members. Additionally, we conducted promoter cis-acting element analysis and discovered that PbBES1 contains binding sites for plant growth and development, cell cycle regulation, and response to abiotic stress. PbBES1.2 is highly expressed in root bark, stem bark, root xylem, and stem xylem. PbBES1.3 was expressed in five tissues. Moreover, we examined the expression profiles of five representative PbBES1 genes under heat and drought stress. These experiments preliminarily verified their responsiveness and functional roles in mediating responses to abiotic stress. This study provides important clues to elucidate the functional characteristics of the BES1 gene family, and at the same time provides new insights and valuable information for the regulation of resistance in P. bournei.
1. Introduction
Plants often suffer from a series of biotic and abiotic stresses, leading to a decline in yield and quality. Transcription factors (TFs) play a crucial role in plant growth and development, as well as stress response regulation, by activating or inhibiting the transcription of target genes [1]. In plants subjected to biotic and abiotic stresses, TFs can also activate various defense mechanisms [2]. The BRI1 EMS suppressor 1(BES1) transcription factor is a crucial regulator in the signaling pathway of Brassinosteroid (BR), which is a plant steroid hormone engaged in plant growth and development as well as environmental stress responses [3,4,5]; the application of exogenous BR has been shown to promote plant growth under abiotic stress conditions [6]. BR is the sixth type of phytohormone after auxin and was discovered in Brassica napus, gibberellins, cytokinin, abscisic acid, and ethylene [7,8]. BR induces a local accumulation of hormones, which triggers signaling for the gradual transformation of cells in the meristematic zone into the elongation zone and promotes cell elongation. The correct balance of BR levels appears to be crucial for normal root growth and development [9,10]. In A. thaliana, the deletion mutants of BR exhibit dwarfism, reduced cell elongation, reduced apical dominance, delayed flowering and senescence, and male sterility [11,12]. The signaling pathway of BR can interact with multiple responses and cooperatively regulate plant growth, development, and response to abiotic stresses [7]; this pathway also cross-regulates with other pathways, including the ABA, light, and GA signaling pathways [13,14,15], through the BES1 family of transcription factors. In response to abiotic stresses, the BR signaling pathway negatively regulates the plant’s drought stress response by crosstalking with drought stress through RD26 [16,17]. The BR signaling pathway also interacts with ABA signaling to enhance plant resistance to abiotic stresses [15].
BZR1/BES1 is a plant-specific transcription factor that can bind to and regulate BR-responsive genes [18,19]. Research indicates that BZR1 and BES1 are two closely related nuclear proteins that share high homology in their amino acid sequences. They bind to DNA through a conserved N-terminal DNA-binding domain [20], and they associate with genes related to BR and regulate their expression [21,22,23]. Both BZR1 and BES1 play positive regulatory roles in the BR signaling pathway [24]. In plant cells, BR binds directly to the extracellular structural domain of Brassinosteroid insensitive 1 (BRI1) and activates its intracellular activating enzyme activity [25]. This promotes the dissociation of BRI1 kinase inhibitor 1 (BKI1) from the cell membrane, allowing it to bind to BRI1-associated kinase 1 (BAK1) and to signal BR into the cell [26]. In the cytoplasm, BR-signaling kinase 1 (BSK1) through a phosphorylation/dephosphorylation cascade, BSK, which in turn activates BRI1-suppressor 1 (BSU1), which dephosphorylates and degrades Brassinosteroid-insensitive 2 (BIN2) [27]. This rescues the inhibition of BES1 by BIN2 [18,28]. BES1 is dephosphorylated by protein phosphatase 2A (PP2A) and accumulates in the nucleus, where it acts on a large number of target genes downstream of BES1 [5,29]. For example, BES1 binds to ACO1 to increase ethylene production and promotes gravitropic responses in A. thaliana roots [30]. BES1 possesses an unusual bHLH structural domain, which is the largest family of transcription factors in the model plant A. thaliana [31,32]. This domain can regulate the expression of thousands of downstream genes in conjunction with the E-box of the promoter region of the target genes (CANNTG) and the oleoresin lactone response element (CGTGT/CG) [33,34]. Furthermore, the BES1 gene family interacts with PIF4, WRKY46, WRKY54, and WRKY70 to co-regulate plant cell elongation and plant stress tolerance [35,36]. Mecchia et al. demonstrated that the sporophyte geophyte BES1 plays a crucial role in controlling cell proliferation and differentiation [37]. In a study of the BES1 gene family in the angiosperm Populus trichocarpa, it was found that BES1 enhances the drought stress tolerance and scavenging of reactive oxygen species in plants [38].
Recently, it has been discovered that A. thaliana [23] has 8 members of the BES1 transcription factor family, rapeseed [39] has 28, cotton [40] has 22, corn [41] has 11, tomato [42] has 9, and cabbage [43] has 15. Previous research has demonstrated the involvement of BES1 transcription factors in plant growth, development, and stress responses, such as cell growth [44], pollen development [45], plant immune signaling [46], and resistance physiology [23]. Studies indicate that BES1 is involved in the formation of the primary periderm; it inhibits the genes AIB3 and AIB5, which are insensitive to abscisic acid (ABA), attenuates ABA signaling during seedling development, accelerates flowering in the reproductive stage, and regulates elongation of the hypocotyl [47]. In cotton research, the GhBES1 gene exhibits functional diversity, affecting fiber growth and consequently influencing cotton plant structure [41]. Additionally, research shows that the expression levels of most BES1 genes change significantly under hormone stimulation, indicating that BES1 transcription factors mediate plant responses to hormonal stress. The cotton variety ‘Xinluzao 17’ rapidly responds to drought stress under the regulation of BES1 transcription factors [48]. Furthermore, studies suggest that the majority of tomato BES1 genes are significantly up-regulated 6 and 24 h after salt treatment, emphasizing the crucial role of the BES1 transcription factor family in salt tolerance in tomatoes [42].
Phoebe bournei (Hemsl.) Yang, which belongs to the Lauraceae family and Phoebe genus, stands as a renowned arboreal species in the horticultural field. This species holds a prominent position among the flora within subtropical evergreen broad-leaved forests and has substantial economic worth and ecological relevance [23]. Due to commercial and development needs, P. bournei has been heavily cut down and destroyed in recent years, leading to a gradual decrease in its population. Its slow growth has resulted in its classification as an endangered plant. P. bournei grows in monsoon climates with abundant precipitation. However, its growth is affected by droughts and prolonged summers [49]. Studies have shown that global warming, caused by the rising global carbon dioxide concentrations due to the combustion of industrial fuels, significantly affects the incidence of forest fires and the efficiency of photosynthesis in forest trees [50,51]. Additionally, global warming contributes to the dryness of the climate [49]. Drought has a significant impact on the dry weight of stems and roots, as well as chlorophyll synthesis and photosynthesis in P. bournei [52]. Abiotic stresses can also affect chlorophyll biosynthesis, leading to reduced photosynthesis [53]. Therefore, it is highly desirable to improve the plants’ ability to respond to abiotic stresses.
This study analyzed the physicochemical properties, evolutionary relationships, gene structure, conserved structural domains, and intraspecies and interspecies covariance of the BES1 gene family of P. bournei. It also investigated and determined the expression of the PbBES1 family of genes under different abiotic stresses, providing novel insights and information for future research on the selection and regulation of stress tolerance. Furthermore, it offers valuable insights and information for the further understanding of the functional characteristics of the BES1 gene family.
2. Results
2.1. Analysis of Physicochemical Properties of the BES1 Gene Family in Phoebe bournei
Predictive analysis of the physicochemical properties of the amino acid sequences of nine members of the PbBES1 family was conducted; these nine members were named PbBES1.1~PbBES1.9 (Table 1). They spanned a wide range of amino acids, from 134 (PbBES1.8) to 696 (PbBES1.6). The protein had a molecular weight ranging from 15,094.98 kDa (PbBES1.8) ~ 78,314.37 kDa (PbBES1.6), and the theoretical isoelectric point ranged from 5.59 (PbBES1.9) to 10.21 (PbBES1.3), with a mean value of 7.44. In addition, four of the genes had larger isoelectric points; the differences in isoelectric points indicated that PbBES1s function in different microenvironments. Additionally, all nine PbBES1s had negative hydrophilicity, and the lipid solubility index was located between 50.56 (PbBES1.2) and 75.66 (PbBES1.9), making them hydrophilic proteins. Four genes had a lipid solubility index greater than seventy and were thermally stable. The proteins with instability coefficients of less than 40 are generally considered to be stable proteins, and the proteins with coefficients greater than 40 are considered to be unstable proteins. Among the nine members of the PbBES1 family, only PbBES1.9 had an instability coefficient lower than 40, indicating that it belongs to the stable proteins. The remaining members of the PbBES1 family were unstable proteins.

Table 1.
Physicochemical characterization of 9 PbBES1 genes and their encoded proteins.
2.2. Protein Evolution and Collinearity Analysis of PbBES1 Genes
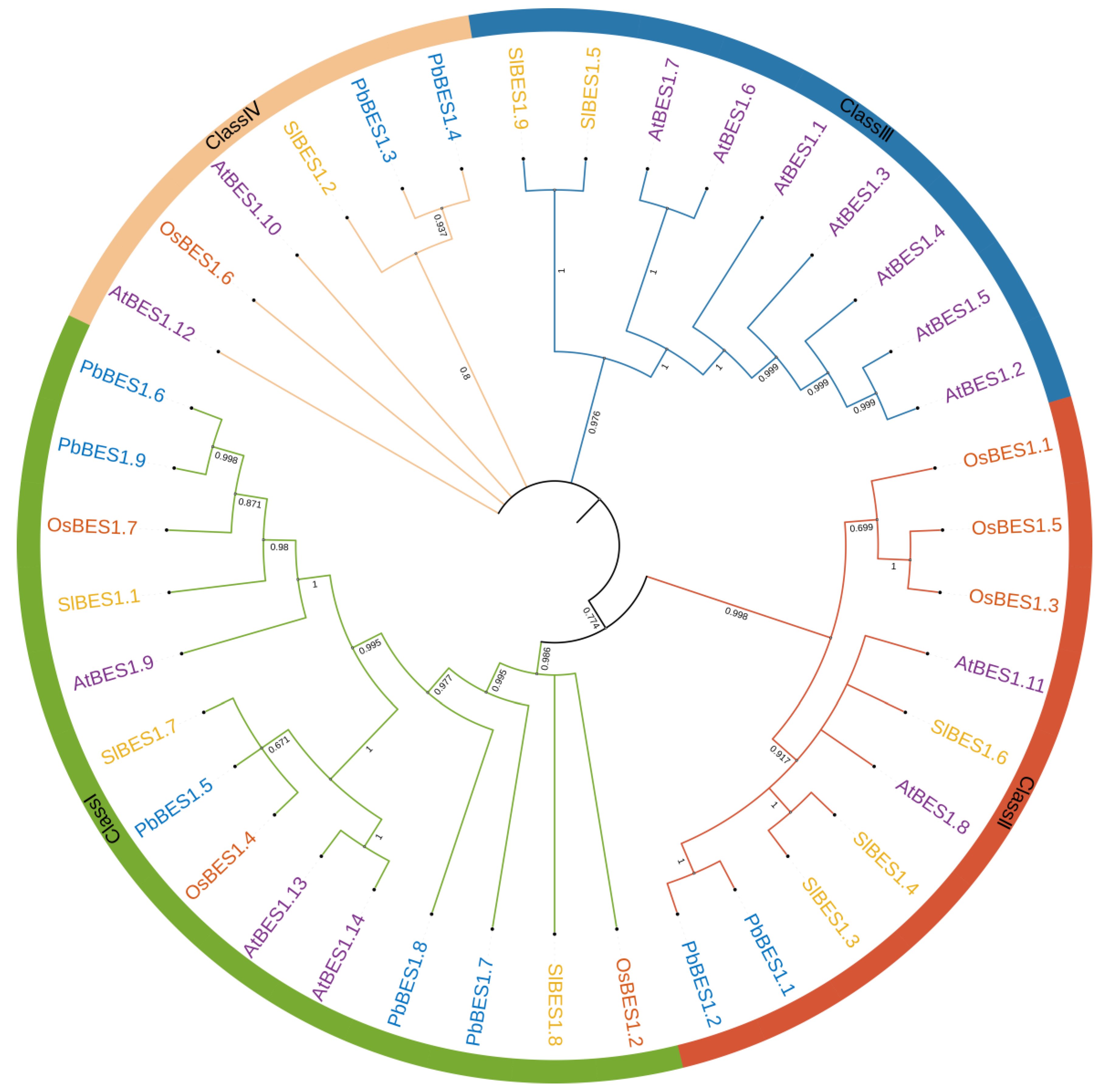
Phylogenetic trees display the affinities and evolutionary relationships between individual species and genes. To investigate the evolutionary relationship between the PbBES1 gene family and other species, we performed multiple sequence comparisons and constructed maximum likelihood phylogenetic trees with the PbBES1s using three representative plants: Arabidopsis thaliana (14), Solanum lycopersicum (9), and the monocotyledonous plant Oryza sativa (7) (Figure 1). The distribution of PbBES1 genes among the different fractions was uneven, with Class I having a maximum of five PbBES1s. Class II contained two PbBES1s, followed by two PbBES1s in Class IV. The phylogenetic tree revealed that PbBES1.6 was closely related to PbBES1.9, while PbBES1.1 was closely related to PbBES1.2 and PbBES1.3 was closely related to PbBES1.4. Notably, there were no PbBES1 genes in Class III, and the BES1 genes were only distributed in dicotyledonous A. thaliana and S. lycopersicum.
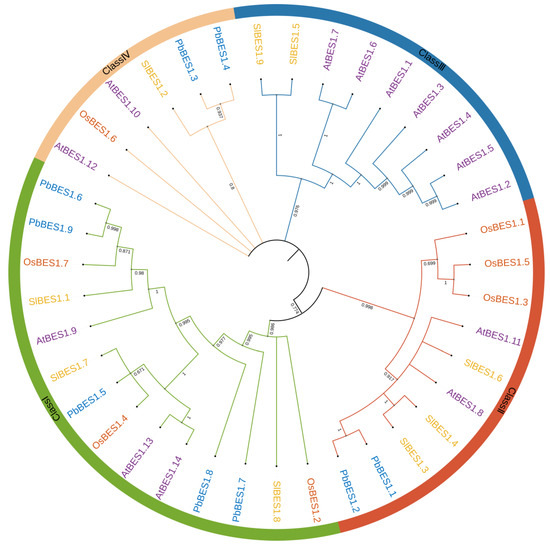
Figure 1.
The phylogenetic tree of BES1 proteins in Phoebe bournei, Arabidopsis thaliana, Solanum lycopersicum, and Oryza sativa. Classes I–IV referred to the phylogenetic tree clusters.
The evolutionary tree analysis revealed that the BES1 family members were interspersed in four plants, but Class III contained only the BES1 genes of A. thaliana and S. lycopersicum. This suggests that the BES1 of P. bournei may have amplified after the monocotyledonous divergence, which was more conserved. Martin analyzed the evolutionary tree of BES1 and suggested that it first appeared in bryophytes and may have developed after phytoplankton [37]. Furthermore, the evolutionary origins of PbBES1.6 and PbBES1.9 are homologous to those of PbBES1.5 from the consensus branch.
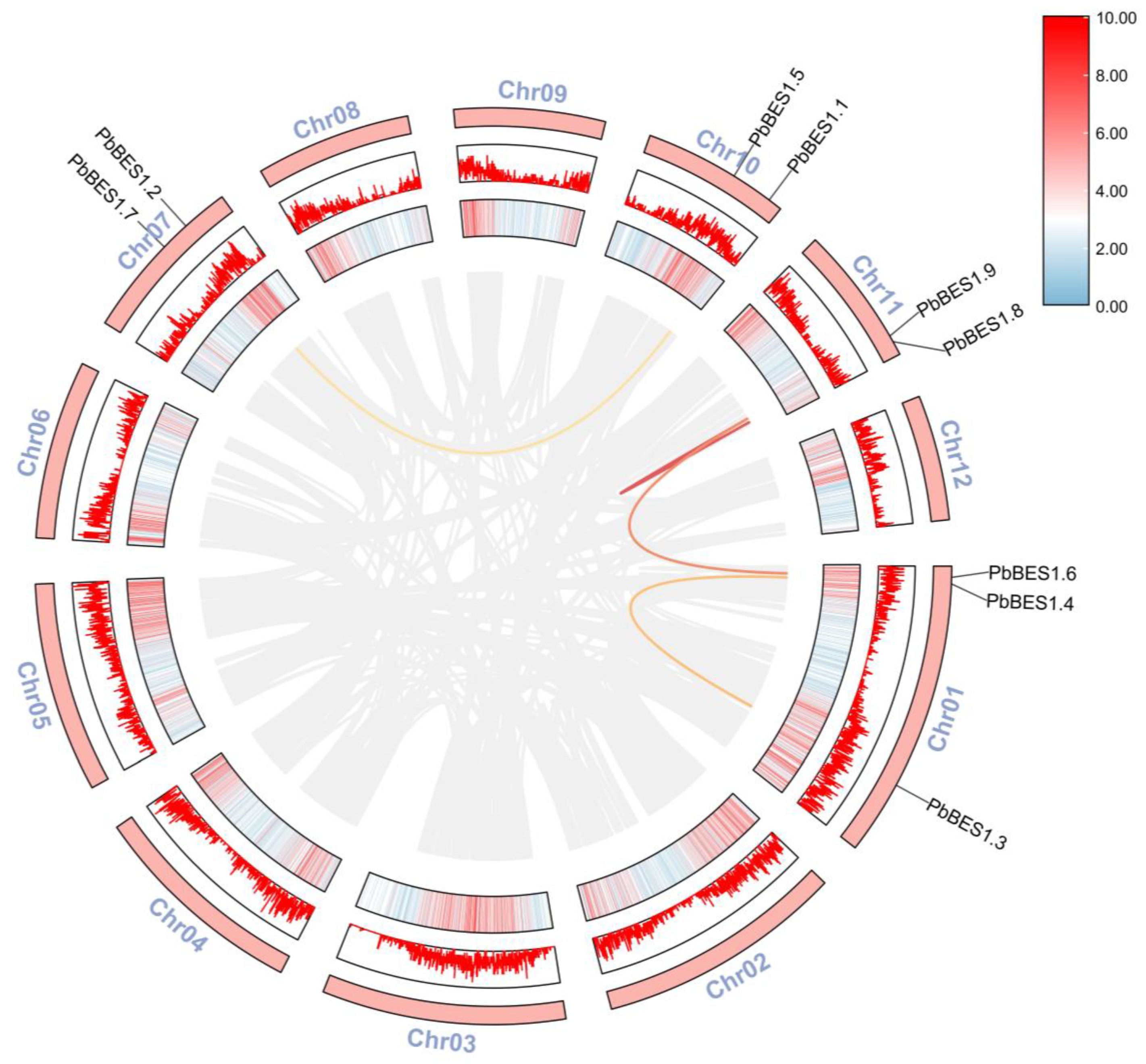
The collinearity model provides an historical insight into the genome and enables downstream analyses [54]. Following the analysis of the PbBES1 gene family (Figure 2), its intraspecies covariance showed that the 9 PbBES1 genes were mainly located on 4 of the 12 chromosomes of P. bournei. The chromosome Chr01 contained three PbBES1s and was the chromosome with the highest number of PbBES1 genes, with two genes distributed on each of the remaining three chromosomes. The PbBES1 genes had a total of four replication events: PbBES1.3 on Chr01 and PbBES1.4 were tandem replication events; PbBES1.8/PbBES1.9/PbBES1.6 and PbBES1.2/PbBES1.1 were fragment replication events.
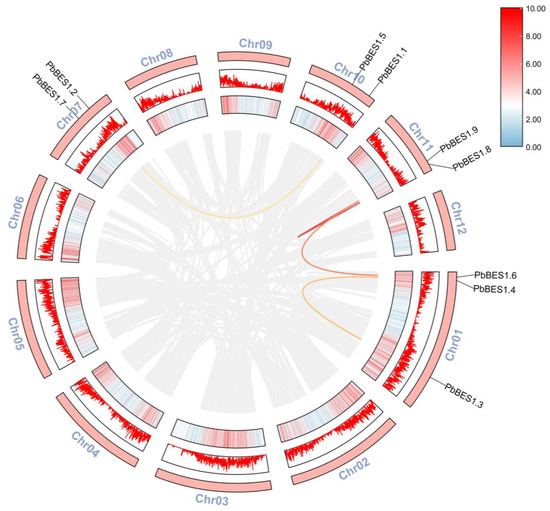
Figure 2.
Chromosomal distribution and inter-chromosomal relationship of PbBES1 genes. The outer red bar indicates the Phoebe bournei chromosome, the middle red box indicates the corresponding GC relationship of each chromosome, and the colored line represents the covariance of PbBES1; the gray line indicates the covariance of the whole Phoebe bournei genes.
The abundance of duplicate genes in plant genomes contributes to the evolution of new functions [38,55]. Tandem duplication events are genes that contain two or more copies within a region smaller than 200 kb. Fragment duplication events, on the other hand, are homologous gene pairs found on different chromosomes [56].
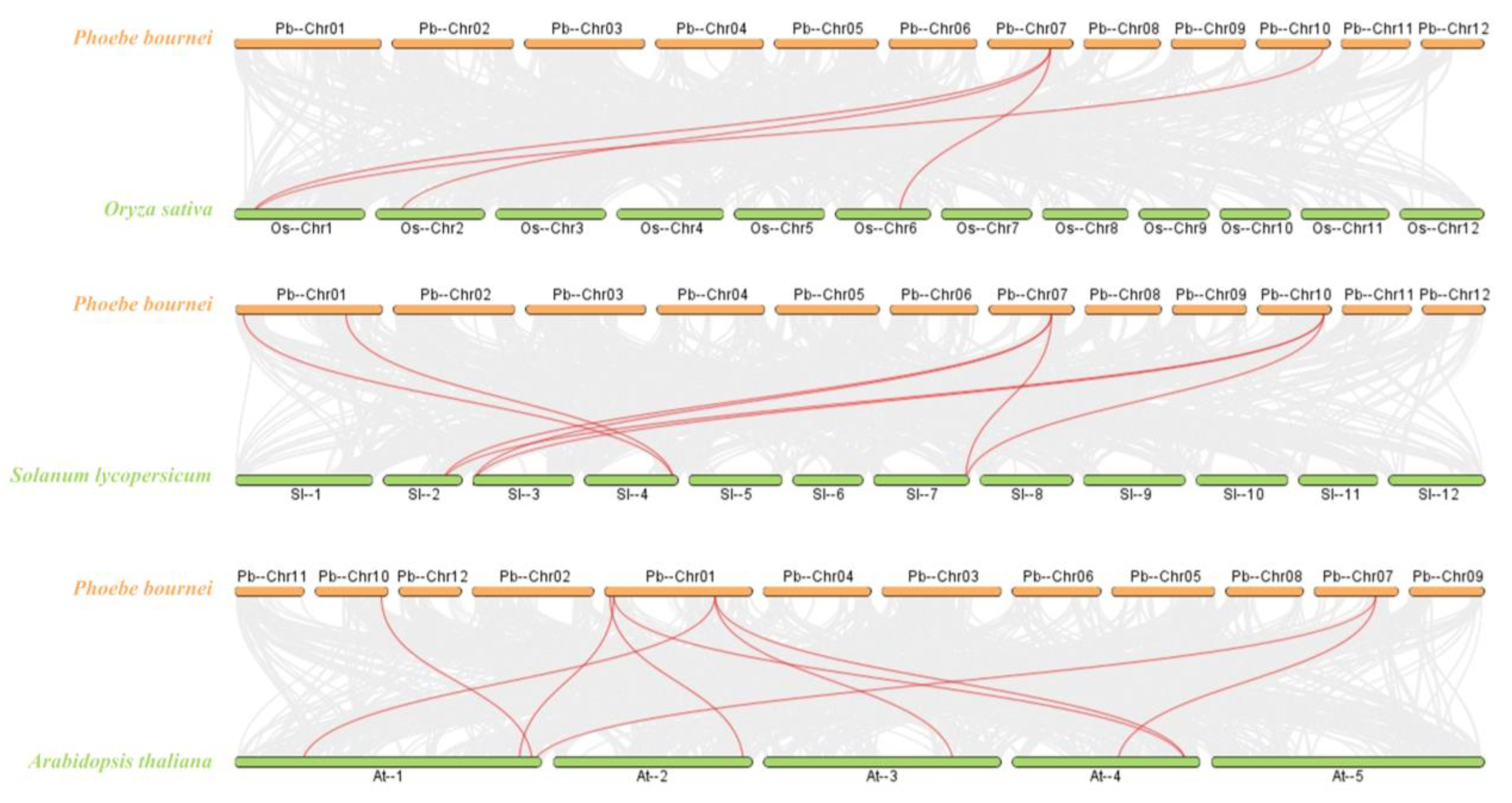
To gain a deeper understanding of the evolutionary mechanism of P. bournei, interspecific collinearity analyses were performed between P. bournei and the three representative species used to construct the evolutionary tree (Figure 3). Four homologous gene pairs were identified between PbBES1 and OsBES1. Additionally, eight homologous gene pairs were identified between PbBES1 and SlBES1, and PbBES1 had nine homologous gene pairs with AtBES1. Analyzing the collinearity between species can provide an insight into the timing of gene family amplification.
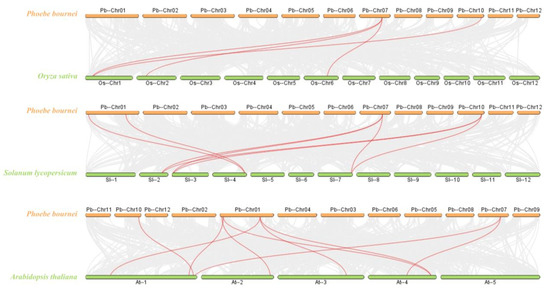
Figure 3.
Collinearity analysis plot of Phoebe bournei with Oryza sativa, Solanum lycopersicum, and Arabidopsis thaliana. The red line represents the collinearity comparison of the BES1 gene family, and the gray portion represents the collinearity comparison of the other gene families.
2.3. Analysis of Gene Structure and Conserved Motifs in Phoebe bournei
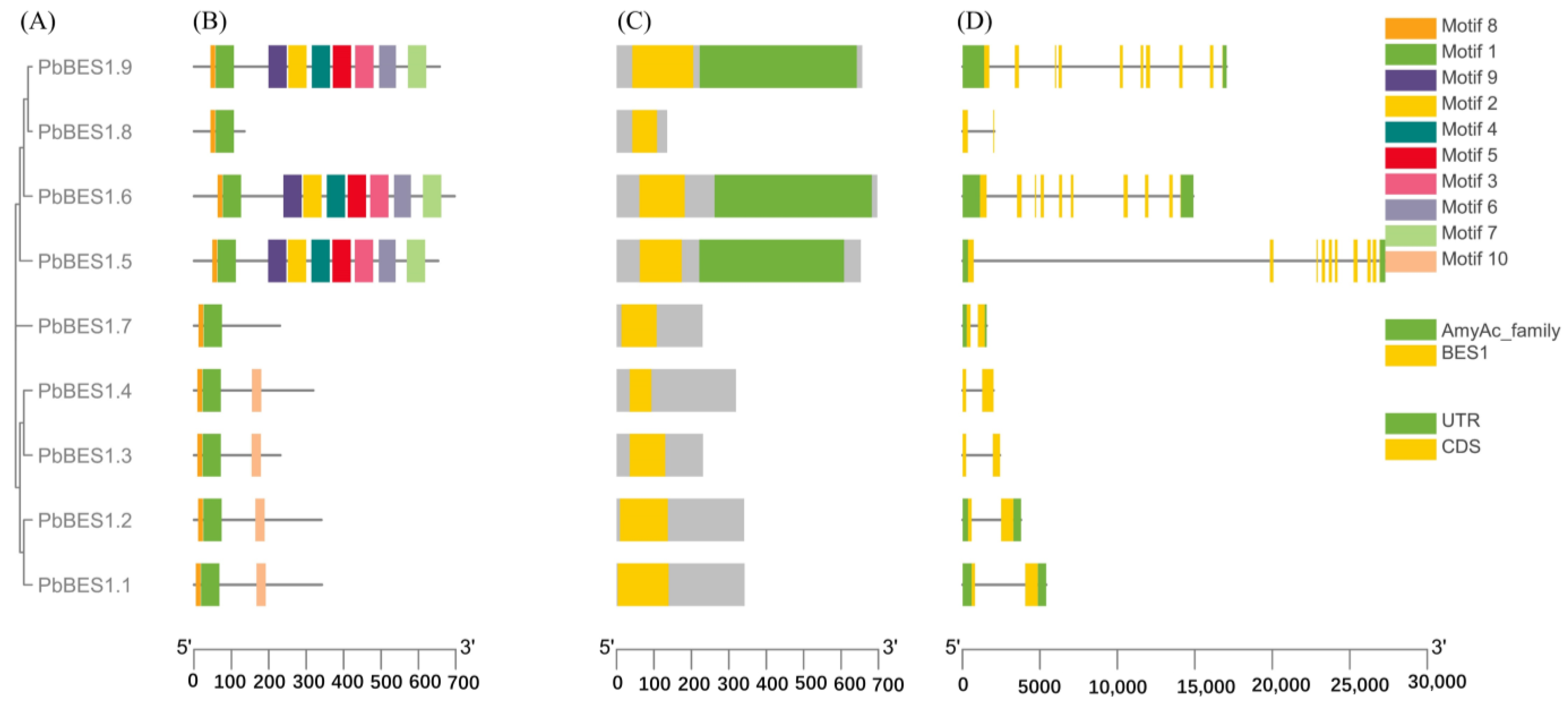
The analysis of the 9 identified PbBES1 family members revealed that they had 10 conserved motifs (Figure 4A). The members of the same subfamily shared the same motif composition and arrangement order. All the genes contained motif 1 and motif 8, indicating that these are the most conserved motifs. PbBES1.7 and PbBES1.8 only contained motif 8 and motif1, while PbBES1.1, PbBES1.2, PbBES1.3, and PbBES1.4 contained motif8, motif1, and motif2. This suggests that they may have similar functions. The differences in conserved motifs among the members of the PbBES1 family suggest that gene loss or deletion may have occurred during the evolutionary process. On the conservative structural domains (Figure 4B), PbBES1.9 and PbBES1.8 had two conserved structural domains, AmyAc family and BES1. The remaining family members possessed the BES1 structural domain, suggesting a higher degree of conservation.
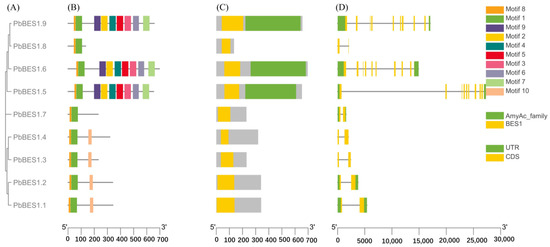
Figure 4.
Schematic diagram of the conserved motif of the PbBES1 gene. (A) Phylogenetic tree of 9 PbBES1 proteins. (B) Different colors correspond to different types of motifs with the numbers 1–10. (C) Indicates a conserved structural domain of PbBES1. (D) Exon/intron structure of the PbBES1 gene. The exon is represented by the yellow box, while the intron is represented by the black line. The green box indicates the PbBES1 gene’s UTR region.
The analysis of the gene structure revealed that there were nine genes with exons ranging from two to nine and introns ranging from one to nine. The number of exons and introns in PbBES1.9, PbBES1.6, and PbBES1.5 was nine. The numbers of exons and introns in PbBES1.8, PbBES1.7, PbBES1.4, PbBES1.3, PbBES1.2, and PbBES1.1 were two and nine, respectively. Furthermore, it is worth noting that PbBES1.8, PbBES1.4, and PbBES1.3 lacked non-coding regions. These regions play a crucial role in regulating the gene expression of mRNA stability.
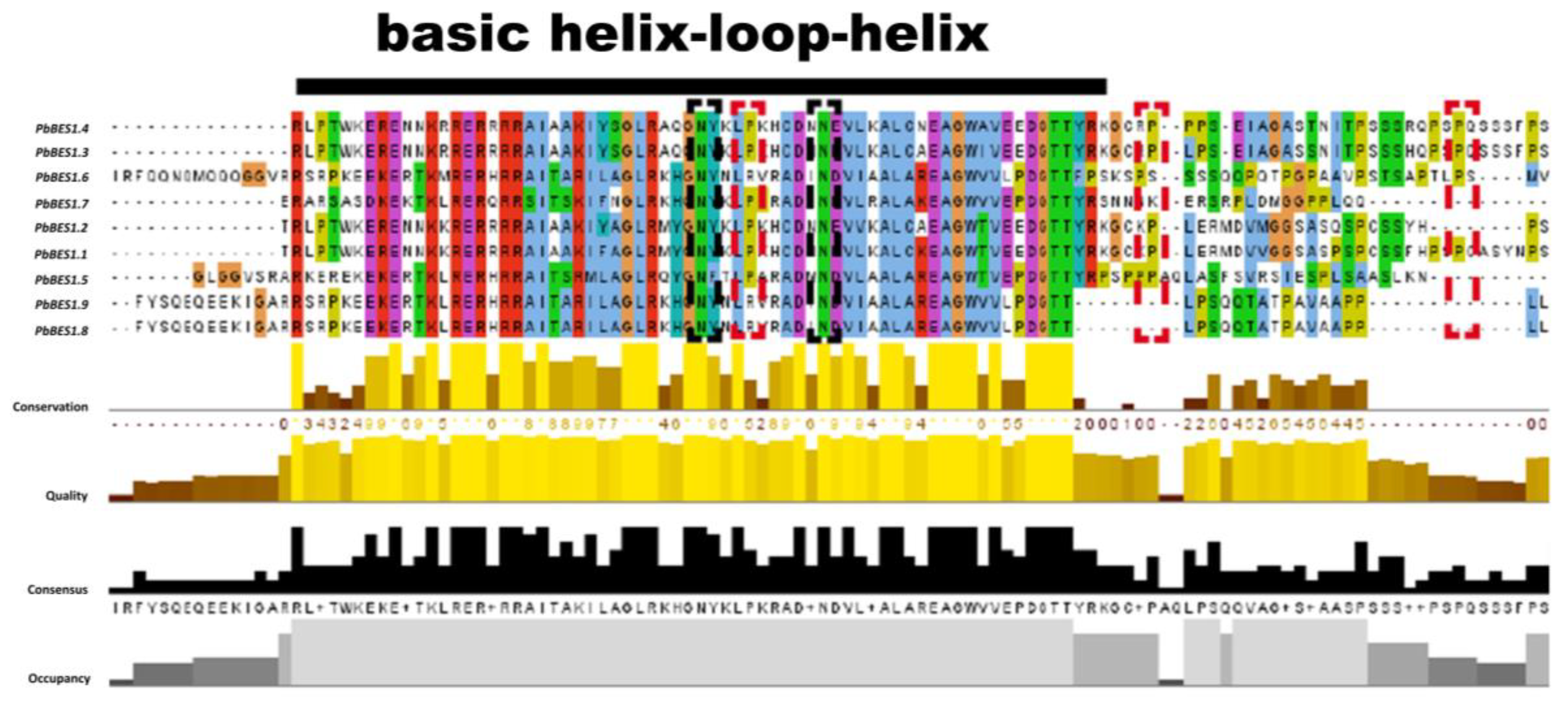
The protein conserved structural domains are highly conserved, and protein sequence similarity can provide valuable clues to evolutionary processes [57]. To further investigate the evolution and function of the PbBES1 gene family, we performed multiple sequence comparisons of its conserved structural protein domains (Figure 5). With reference to previous studies [19,58], comparisons were made with the structural domains of the AtBES1 protein. The conserved structural domains of the PbBES1 family proteins were found to have atypical basic helix–loop–helix (bHLH) structural domains and highly conserved amino-terminal structural domains (N) and BIN2 phosphorylation binding sites (P). This suggests that the PbBES1 gene family is more conserved in the evolutionary processes.
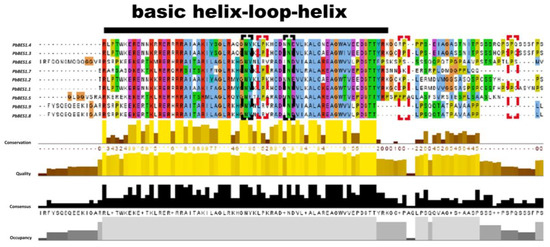
Figure 5.
Multiple sequence comparison of PbBES1 protein using Jalview software (v2.11. 3.0). Different amino acids are labeled with different colors, and the possible functional sites or elements are encircled by a box. Black line shows portion of BHLH conserved sequence; black dashed box shows conserved amino-conserved structural domains; red dashed box shows conserved BIN2 phosphorylation binding site.
2.4. Cis-Acting Analysis and Structure of the PbBES1 Promoter
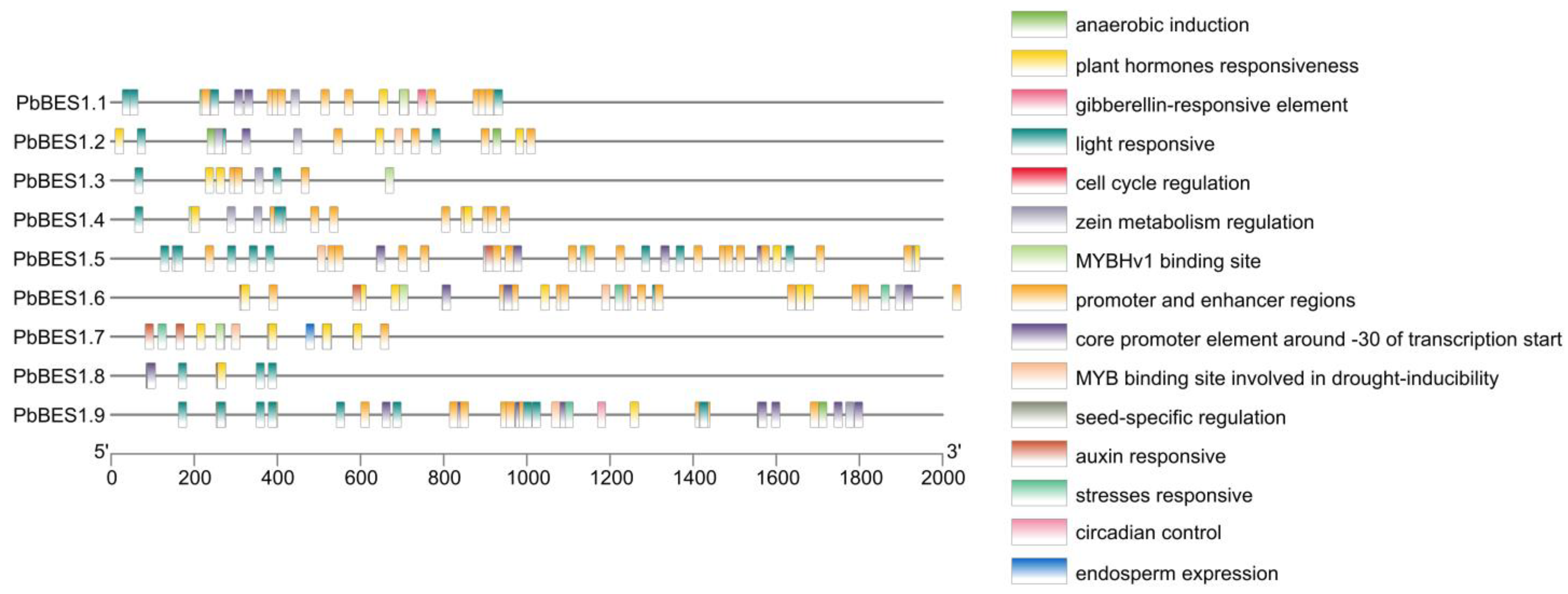
To enhance comprehension of the function of the PbBES1 genes, we predicted the cis-acting elements in their promoter regions (Figure 6). The results revealed that the PbBES1 gene family contained a total of 15 cis-acting elements that are related to environmental stress and hormone response. Each cis-element represented a distinct function, with the highest number of homeostatic components being related to light response; there were 34 of these components, and they were contained in the promoter and enhancer regions. All the members, except PbBES1.6 and PbBES1.7, contained light-responsive cis-acting elements. This suggests that the PbBES1 gene family may play a role in regulating plant light response. The PbBES1.7 genes had the highest number of cis-acting elements, with 37 each, indicating their potential involvement in transcriptional regulation. All the genes had phytohormone response elements, and PbBES1.2, PbBES1.5, PbBES1.6, PbBES1.7, and PbBES1.9 also had MYB transcription factor binding sites that were involved in drought induction. The involvement of MYB transcription factors in various abiotic stress responses, including drought stress [59], suggests that the PbBES1 gene family may also regulate plant resistance to drought stress.
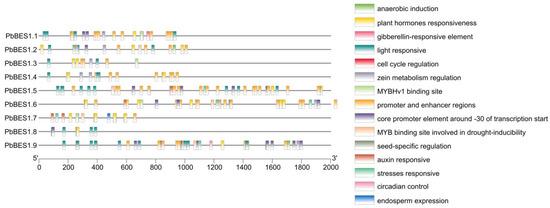
Figure 6.
Analysis of cis-acting elements of the PbBES1 gene family, with different colored squares representing different cis-elements; the scale bar at the bottom indicates the position of different cis-elements in the promoter region.
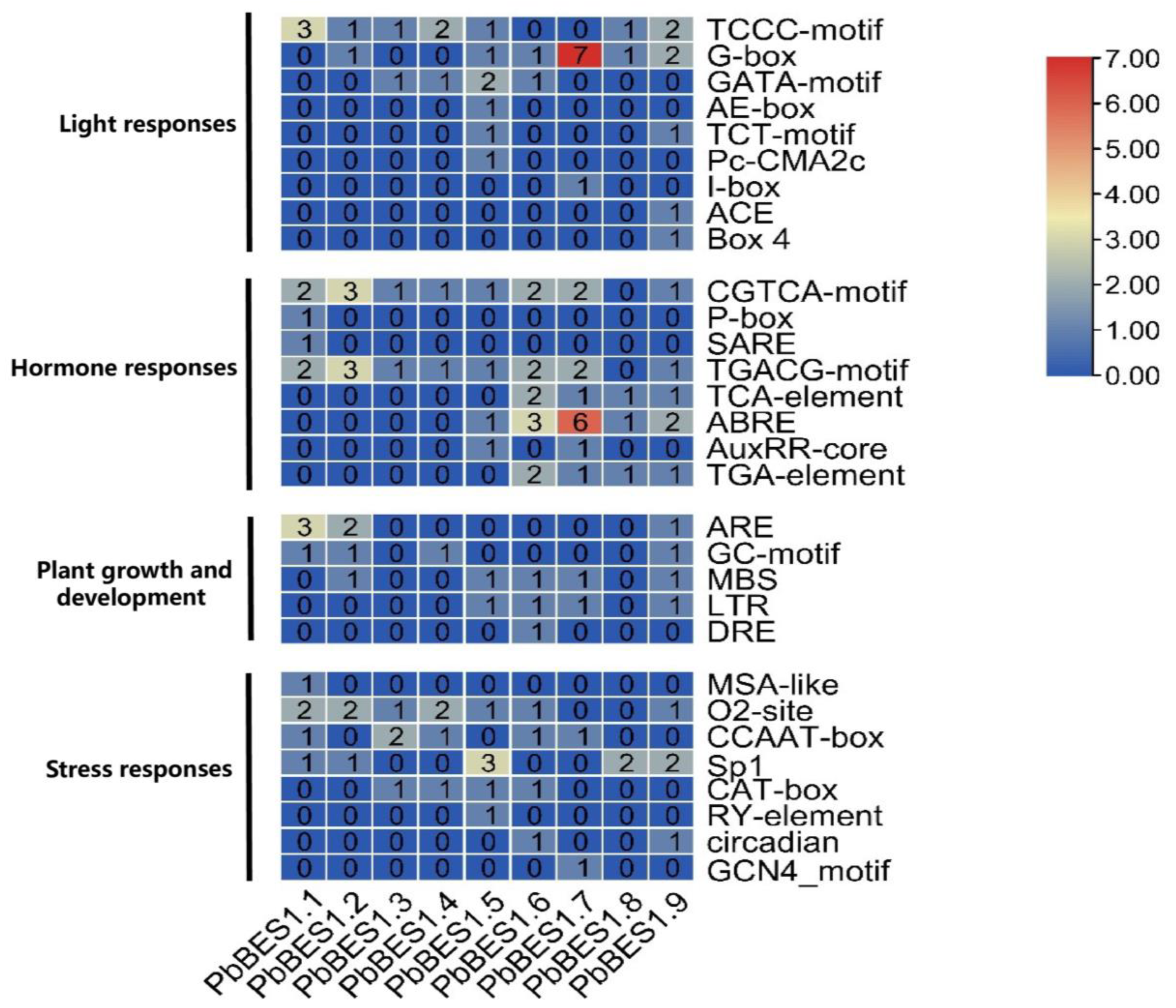
Further analysis of the cis-acting progenitor of the PbBES1 promoter region (Figure 7) was conducted. In the PbBES1 gene family, there were 36 cis-elements related to stress; PbBES1.7 contained up to 7 stress-responsive cis-elements and 52 cis-elements related to growth and development; PbBES1.7 contained up to 13 growth-responsive cis-elements and 20 hormone-responsive cis-elements; PbBES1.1, PbBES1.2, and PbBES1.9 contained 4 hormone-responsive cis-elements each and 34 cis-elements related to light response; and PbBES1.6 was the most numerous, with 6 light-responsive cis-elements. The PbBES1 gene family contained hormone-responsive homeotic elements, including abscisic acid-responsive elements and growth hormone-responsive elements. This suggests a potential link between the PbBES1 gene family and the abscisic acid and growth hormone signaling pathways, which is consistent with previous research [13,60]. Among the nine members of the PbBES1 family, PbBES1.7 contained the most stress-corresponding and hormone-responsive homeotic elements. This suggests that PbBES1.7 may play a major role in the family and that it warrants further study.
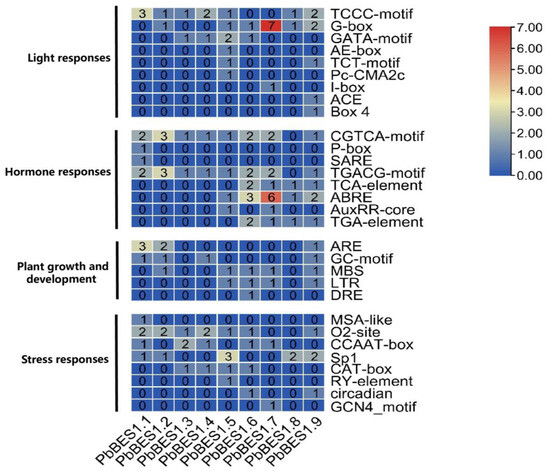
Figure 7.
The numbers of the 30 cis-elements of the 9 PbBES1 genes.
2.5. Heat Map of PbBES1 Gene Expression in Different Tissues
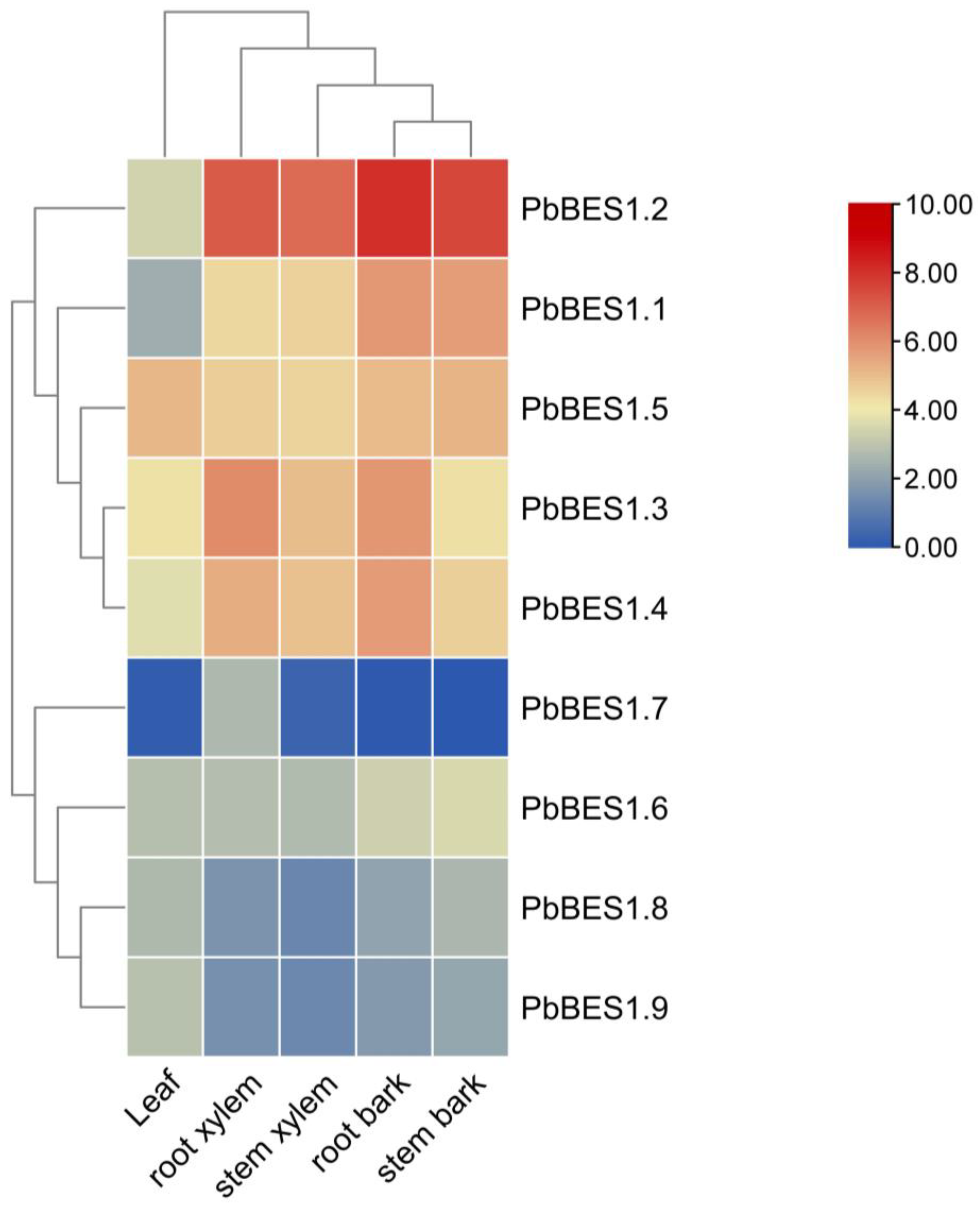
To enhance comprehension of PbBES1′s role in plant growth and development, we analyzed the expression of the PbBES1 gene family in various organs (Figure 8). We classified the nine genes into three subfamilies. With the exception of PbBES1.7, which was not expressed in the leaves, root bark, or stem bark, all the genes were expressed to varying degrees in all five organs. The expression of the PbBES1.5 gene was highest in the leaves, indicating its involvement in the growth and development of leaf organs. PbBES1.2, a separate subfamily, had the highest expression in the root xylem, stem xylem, root bark, and stem bark. Its expression was higher in the root bark than in the other organs, suggesting a strong correlation between PbBES1.2 and root growth and development. The expression of each gene in the third subfamily was low in all five organs, and PbBES1.7 only had low expression in the root xylem and stem xylem. This suggests that PbBES1.7 may be involved in plant growth and development with fewer corresponding functions. The genes in the third subfamily may play other roles.
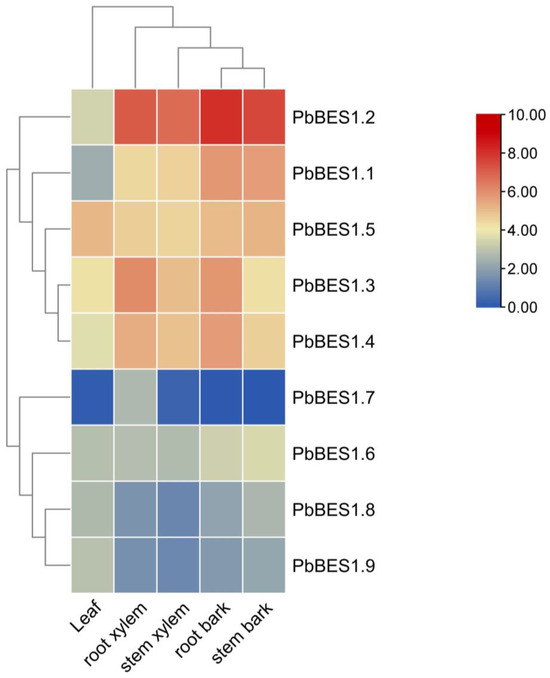
Figure 8.
Heat map of PbBES1 gene family expression in leaves, root xylem, stem xylem, root bark, and stem bark; different colors represent different levels of expression, with blue representing the lowest expression and red representing the highest.
2.6. Abiotic Stress Experiments on the BES1 Gene Family of Phoebe bournei
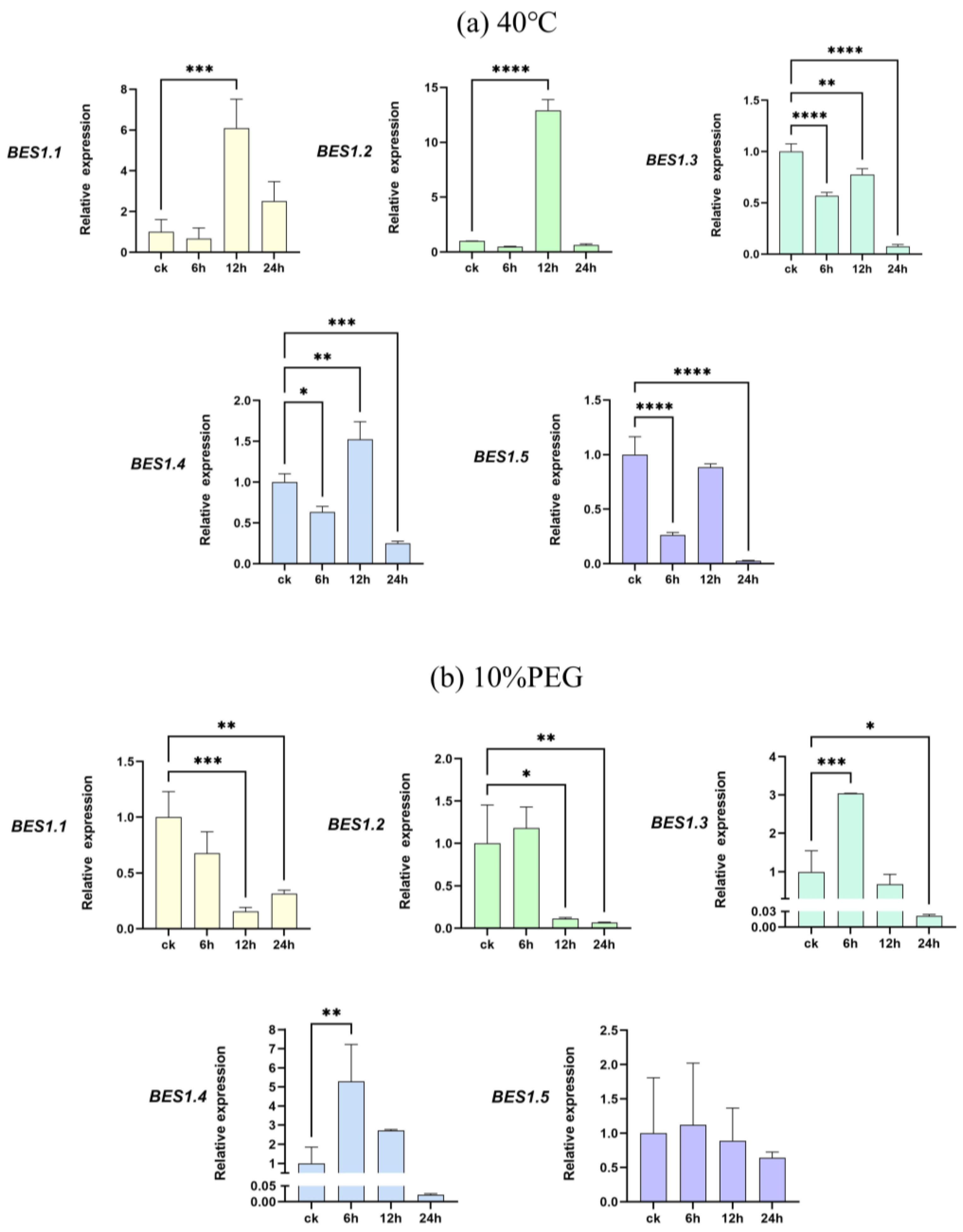
To further investigate the response of the BES1 gene family of P. bournei to abiotic stresses, we subjected its leaves to high-temperature and drought stress treatments. The leaves of P. bournei were collected at 6 h, 12 h, and 24 h after treatment using qRT-PCR to compare their gene expressions with that of leaves untreated with heat and drought under the same culture conditions (Figure 6). We observed changes in the up- and down-regulation of the different genes. The experimental results indicate that the expression of PbBES1 in the leaves was decreased to varying degrees after 6h under high-temperature stress treatment (Figure 9a). The PbBES1.3, PbBES1.4, and PbBES1.5 expression was significantly decreased, with PbBES1.5 being approximately one-third of the control (ck). The expression of PbBES1.1, PbBES1.2, and PbBES1.4 was significantly increased after 12 h of high-temperature stress. Among them, PbBES1.2, which was highly expressed in the root bark, showed a 12-fold increase compared to the control group. However, after 24 h of treatment, the expression of all the genes decreased, except for PbBES1.1; PbBES1.3 was only 1/14 of the control.
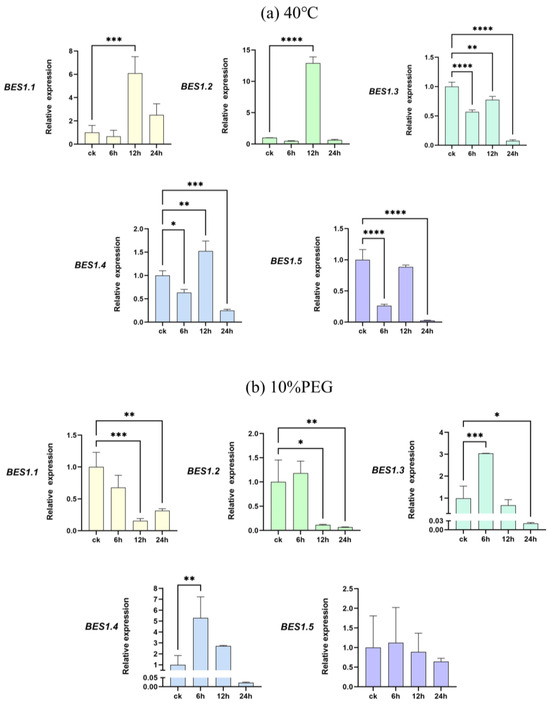
Figure 9.
The expression profiles of five representative PbBES1 genes in response to high-temperature (a) and drought (b) stresses. The relative expression levels of PbBES1 genes in response to abiotic stresses, assessed using RT-qPCR. The error bars indicate the standard deviations of the three independent RT-qPCR biological replicates. (X: process time; Y: relative expression) * represents a significant difference relative to the 0 h group (* p < 0.05, ** p < 0.01, *** p < 0.0005, **** p < 0.0001).
Under drought stress treatment (Figure 9b), all the genes except PbBES1.1 were up-regulated after 6 h, and PbBES1.3 had an expression that was five times higher than that of the control group. After 12 h, all the genes except PbBES1.3 decreased in expression. After 24 h, the expression of the PbBES1 family genes was suppressed, and the expression of PbBES1.3 tended to be absent. This result indicates that the expression of PbBES1.3 was significantly altered after 6 h of exposure to both drought and high-temperature treatments. This suggests that PbBES1.3 may play a more significant role in the response to abiotic stresses.
3. Discussion
BR signaling enters cells through membrane receptor BRI1 and acts on a series of transcription factors to finally activate BES1, which binds to a large number of downstream target genes and regulates plant growth and development. According to the analysis, there were nine BES1 genes (BES1.1~1.9) in P. bournei. The nine BES1 genes were identified through bioinformatics; this number of genes was lower than that in P. trichocarpa (14) [38] and higher than in rice (6) [61]; it was similar to the number in grape (8) [62], and PbBES1s share the same number of genes and similar amino acid counts with tomato BES1s [63]. The physicochemical properties of PbBES1 showed that PbBES1 was an unstable hydrophilic protein that could adapt to the slightly acidic and slightly alkaline environment. The evolutionary tree analysis revealed that the BES1 family members were interspersed in four plants, but Class III contained only the BES1 genes of A. thaliana and S. lycopersicum. This suggests that the BES1 of P. bournei may have been amplified after the monocotyledonous divergence, which was more conserved. Martin analyzed the evolutionary tree of BES1 and suggested that it first appeared in bryophytes and may have developed after phytoplankton [37]. Furthermore, the evolutionary origins of PbBES1.6 and PbBES1.9 are homologous to those of PbBES1.5 from the consensus branch. The expansion of the PbBES1 gene family mainly relies on segmental duplication events, which is the primary mode of expansion for this gene family. Similarly, in wheat, the expansion of the BES1 gene number also occurs through segmental duplication, while in gramineous plants, it occurs mainly through segmental duplication [64]. In addition, the collinearity analysis showed that PbBES1 was more closely related to dicots. Furthermore, introns play a crucial role in the evolution of various plant species [61]. The members of the PbBES1 family have different exon and intron positions, and they can be divided into two main categories based on their number, indicating changes in the structure of the PbBES1 gene family members. The variations in the gene structure and conserved protein domains of the PbBES1 family members may imply differences in their gene functions. The PbBES1 gene family is highly conserved. PbBES1.7 and PbBES1.8 have fewer motifs compared to the other three genes in the same subfamily. Additionally, their conserved structural domains and gene structures differ, suggesting that PbBES1.7 and PbBES1.8 may have been lost or deleted during evolution and that their structures may have changed accordingly. PbBES1.5, PbBES1.6, and PbBES1.9 exhibit similar conserved motifs and gene structures; this is supported by the results of the phylogenetic analyses (Figure 1). In contrast to the PbBES1 gene family, the grape BES1 [62] gene family contains at least two exons in total, but VvBES1–3 contains only one exon and no introns. In an analysis that made a comparison with the wheat and foxtail millet BES1/BZR1 gene family [65], it was shown that P. bournei contained fewer homeotic-acting elements (15), but the wheat and foxtail millet BES1/BZR1 families contained 66 homeotic response elements. This difference may be due to the fact that P. bournei has fewer BES1 genes. Additionally, all three species contained phytohormone response elements, light response elements, and stress response-related elements. An analysis of BES1 gene transcription factor expression during citrus senescence in a study by Izadi et al. [66] also noted the presence of high-temperature stress-related genes among the targets of the CsBES1 transcription factor.
The gene expression analysis showed that the expression of PbBES1.1~1.5 was down-regulated 24 h after drought treatment. In contrast to the PbBES1 gene expression results, the expression analysis of BES1 in P. trichocarpa showed that most of the PtrBES1 expression was up-regulated under drought stress for 24 h. This suggests that it plays a positive role in the drought stress response in P. trichocarpa. This may be due to the influence of external factors such as the environment during the evolutionary process. In a study by Mahesh et al. [67], the aim was to determine whether BR could promote the growth of Raphanus sativus seedlings under drought stress by regulating the increase in nucleic acid and soluble protein content and reducing ribonuclease activity; the plants alleviated drought stress by increasing the activities of superoxide dismutase, catalase, and ascorbate peroxidase, but the activities of superoxide dismutase, catalase, and ascorbate catalase in R. sativus seedlings under drought treatment need to be further studied [14].
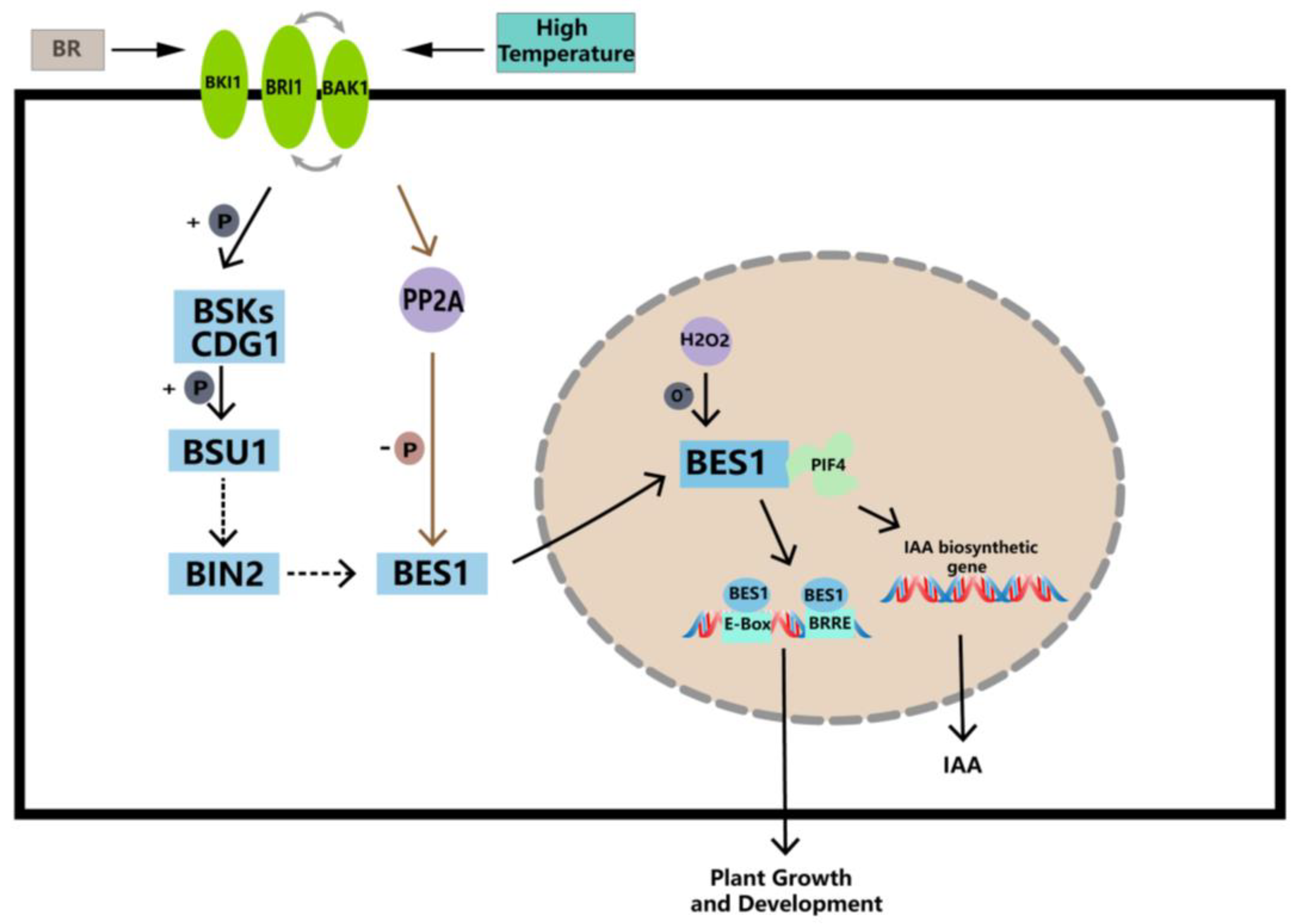
BR mitigates high-temperature stress by enhancing enzymatic and non-enzymatic antioxidant defense and glyoxalase systems [68,69]. EBR, which is a type of BR, significantly improved the survival rate of A. thaliana seedlings when exposed to 43 °C for a period of time [70]. EBR treatment was applied to Ficus concinna seedlings at 40 °C, and the reduced glutathione (GSH) and the ratio of reduced glutathione to oxidative glutathione (GSH/GSSG) of Ficus melisa were significantly increased. EBR treatment reduced 40 °C-induced increases in O2−, H2O2, and MG (Methylglyoxal) levels, and this process was likely associated with a decrease in lipid peroxidation. EBR attenuated the 40 °C-induced oxidative stress by simultaneously increasing non-enzymatic and enzymatic antioxidant responses, as well as MG detoxification systems [67]. Based on the qRT-PCR results, significant differences in the gene expression of the PbBES1 gene environments were observed under high-temperature conditions. Martins et al. demonstrated [71] that BR regulates thousands of genes in roots that are regulated by high temperatures. In addition, in a model that simulates the BR signaling pathway and downstream target genes of BES1 (Figure 10), we found that BES1 was able to bind to the PIF4, which is involved in auxin synthesis, thereby promoting the synthesis of IAA [36]. Therefore, we speculate that PbBES1 promotes auxin synthesis by activating PIF4, thereby alleviating the impact of auxin synthesis and transport at high temperatures and regulating plant growth; however, this needs to be further verified. The expression levels of PbBES1.1~1.5 under high-temperature stress showed that only PbBES1.1 was up-regulated after 24 h of high-temperature treatment, and the expression of the other four genes was down-regulated; thus, PbBES1.1 may be the key gene of the high-temperature stress. Further research, such as that which considers the knockout of PbBES1.1, may provide a more accurate understanding of the function of the gene.
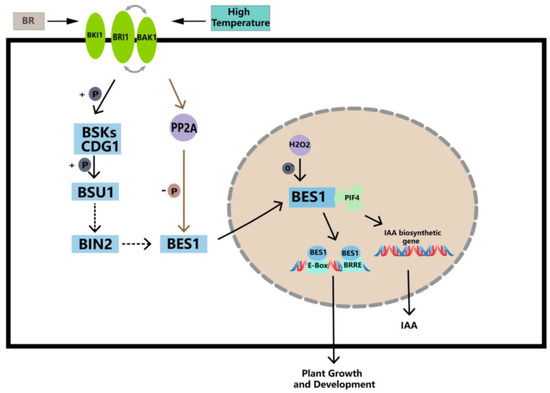
Figure 10.
Working model of BES1 transcription factor under high-temperature stresses. Activated BES1 interacts with PIF4, which promotes the expression of IAA synthesis genes and increases IAA content in high-temperature environments, thereby increasing the expression of BES1 genes and enhancing the ability of plants to cope with high temperatures (modified from Li et al. [47]; Choudhary et al. [72]).
4. Materials and Methods
4.1. Genomic Data
The genomic data and annotation information of P. bournei were downloaded from the sequence library of the Chinese National Genebank Database (CNSA), search number CNP0002030 [73]. The genomic data and annotation information for the A. thaliana, tomato, and rice were obtained from EnsemblPlents (https://plants.ensembl.org/index.html, accessed on 24 June 2023) and Phytozome v13 (https://phytozome-next.jgi.doe.gov, accessed on 24 June 2023), respectively. The RNA-seq data of the different tissues of P. bournei were downloaded from BioProject (https://www.ncbi.nlm.nih.gov/bioproject/, accessed on 24 June 2023) under the accession number PRJNA628065.
4.2. Plant Material Sources and Abiotic Stress Treatment
4.2.1. Plant Material Sources
The plant material was obtained from 1-year-old P. bournei seedlings that were cultured in an artificial climatic chamber with various stress treatments. The P. bournei samples were collected and stored in liquid nitrogen at −80 °C for RNA extraction.
4.2.2. Drought Stress Treatment
Six seedlings from the control group were washed root and soaked in distilled water. Meanwhile, the treatment group was soaked in a nutrient solution containing 10% Polyethylene glycol 6000 (PEG6000) [38,67] and incubated in an artificial climate chamber at a temperature of 25 °C and a humidity of 75%. The treatment group was sampled at 6 h, 12 h, and 24 h time periods with 6 plants in each time period, and the control group was sampled at 0 h.
4.2.3. Temperature Handling
For the high-temperature treatment, 6 plants from the control group were kept at room temperature, while the treatment group was incubated at 40 °C in a warm box. The treatment group was sampled at 6 h, 12 h, and 24 h time periods, with 6 plants sampled at each time point. The control group was sampled at 0 h.
4.3. Identification and Physical and Chemical Property Analysis
The protein sequences of the AtBES1 gene family were obtained from plantTFDB (http://planttfdb.gao-lab.org/ (accessed on 24 June 2023)), and the members of the BES1 family of P. bournei were initially screened by removing duplicates with local BLASTp against the P. bournei protein sequences. The BES1 structural domain number PF05687 was then obtained by searching the pfam database (http://pfam.xfam.org/ (accessed on 24 June 2023)), and the initial screened protein sequences were further examined using the default parameters of HMMER-3.2.1 (http://hmmer.org/download.html (accessed on 24 June 2023)) with an e-value of <105 to obtain the final P. bournei BES1 family members. Then, their physicochemical properties were analyzed using ExPASy (https://web.expasy.org/prot-param/ (accessed on 24 June 2023)).
4.4. Phylogenetic Tree Construction
P. bournei, A. thaliana, tomato, and rice were compared using the Muscle program of MEGA(version 7.0.26 (7170509-x86_64)), and the maximum likelihood trees were constructed using the default settings (bootstrap:1000). iTOL (https://itol.embl.de/, accessed on 26 June 2023) was used to improve and beautify the phylogenetic trees.
4.5. Chromosome Distribution and Covariance Analysis
The positional information of the PbBES1 gene was extracted from the genome (FASTA) file and annotation (GFF) file of P. bournei using TBtools(version 1.108). The covariate relationships between A. thaliana and P. bournei, rice and P. bournei, and tomato and P. bournei were analyzed using MCScanX (https://github.com/wyp1125/MCScanX/ (accessed on 18 July 2023)) software, respectively, and later visualized using TBtools (version 1.108).
4.6. Gene Family Conserved Motifs, Gene Structure Analysis
The PbBES1 protein sequence was characterized using the online software MEME (http://meme-suite.org/ (accessed on 24 June 2023)) with a motif number prediction of 10. A Batch CD-search search with default parameters (https://www.ncbi.nlm.nih.gov/Structure/index.shtml (accessed on 24 June 2023)) was used to detect the conserved structural domains of the PbBES1 proteins.
4.7. Multiple Sequence Comparison and Promoter Cis-Element Analysis of the PbBES1 Gene
Jalview software (v2.11. 3.0) was used to perform the multiple sequence comparison of the 9 PbBES1 genes. To explore the cis-acting elements in the sequences, we analyzed the cis-regulatory elements in the promoter region of the PbBES1 gene using the online software PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 25 June 2023)). After screening and classification, the data were visualized using TBtools (version 1.108) software.
4.8. The Expression Profiles of PbBES1 Genes
The expression data for the BES1 genes in P. bournei across the various tissues were obtained from the BioProject database (Table S1). TBtools (version 1.108) software was employed to analyze these expression data and to construct a gene expression heat map, offering a visual representation of the patterns and levels of gene expression.
4.9. RNA Extraction and qRT-PCR Analysis
An RNA extraction kit (HiPure Plant RNA Mini Kit from Magen, Shanghai, China) was used to extract RNA, while cDNA was synthesized using the Prime Script RT reagent Kit (Perfect Real Time from Takara, Dalian, China). TBtools (version 1.108) software was used to design specific primers in the non-conserved region of the target gene (Table S2); the primers were synthesized by Fuzhou Qingbaiwang Biotechnology Company. Real-time fluorescence quantitative analysis was used with cDNA template (1 µL), cDNA template SYBR Premix Ex TaqTM II (10 µL), specific primers (2 µL), and a ddH2O reaction program (7 µL): 95 °C for 30 s; 95 °C for 5 s; 60 °C for 30 s; 95 °C for 5 s; 60 °C for 60 s; and 50 °C for 30 s, with 40 cycles in total. The internal reference gene was PbEF1α (GenBank No. KX682032) [74]. The expression level of the target gene was calculated using the 2−∆∆Ct method, and the quantitative data were analyzed via a test using GraphPad Prism 9.5 software. Finally, GraphPad Prism 9.5 was used to construct the graphs.
5. Conclusions
In conclusion, the PbBES1 genes play a crucial role in the growth of P. bournei, which has great commercial value and development potential. This study identified nine BES1 genes in P. bournei and performed a comprehensive bioinformatics analysis. The phylogenetic analysis classified the nine genes into three subgroups. The evolutionary analysis revealed that PbBES1 is highly conserved and primarily amplified through fragment duplication. PbBES1 contains numerous cis-acting elements related to environmental stress and hormonal responses, particularly with regard to high-temperature and drought stress. These findings are also supported by the qRT-PCR results. After the stress treatments, PbBES1.1–1.5 exhibited varying degrees of expression effects, with PbBES1.1 and PbBES1.3 showing more significant responses to both stresses. They were significantly expressed after 12 h of high-temperature stress. The study offers a supplementary investigation of the BES1 gene family in P. bournei and provides novel insights and information for future research on the selection and regulation of stress tolerance. Furthermore, it offers valuable insights and information for the further understanding of the functional characteristics of the BES1 gene family.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25053072/s1.
Author Contributions
Conceptualization, S.C., H.S. and J.L.; methodology, W.C. and H.S.; software, D.F. and Y.W.; data curation, K.Z. and Q.Z.; formal analysis, Y.W., J.L. and J.Z.; writing—original draft preparation, Y.W. and J.L.; writing—review and editing, J.L., H.Y. and H.S.; investigation, K.Z.; supervision, W.C. and S.C.; project administration, W.C.; visualization, H.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was jointly funded by National Key Research and Development Program of China (grant no. 2021YFD2201303-03 to H.S.) and Laboratory of Virtual Teaching and Research on Forest Therapy Specialty of Taiwan Strait (111TD2104 to W.C.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The genome sequence data and annotation information of Phoebe bournei were downloaded from the Sequence Archive of China National GeneBank Database (CNSA) with accession number CNP0002030.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Guo, J.T.; Lofgren, S.; Farrel, A. Structure-based prediction of transcription factor binding sites. Tsinghua Sci. Technol. 2014, 19, 568–577. [Google Scholar]
- Century, K.; Reuber, T.L.; Ratcliffe, O.J. Regulating the regulators: The future prospects for transcription-factor-based agricultural biotechnology products. Plant Physiol. 2008, 147, 20–29. [Google Scholar] [CrossRef]
- Belkhadir, Y.; Jaillais, Y. The molecular circuitry of brassinosteroid signaling. New Phytol. 2015, 206, 522–540. [Google Scholar] [CrossRef]
- Kono, A.; Yin, Y. Updates on BES1/BZR1 Regulatory Networks Coordinating Plant Growth and Stress Responses. Front. Plant Sci. 2020, 11, 617162. [Google Scholar] [CrossRef]
- Wang, R.; Wang, R.; Liu, M.; Yuan, W.; Zhao, Z.; Liu, X.; Peng, Y.; Yang, X.; Sun, Y.; Tang, W. Nucleocytoplasmic trafficking and turnover mechanisms of BRASSINAZOLE RESISTANT1 in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2021, 118, e2101838118. [Google Scholar] [CrossRef]
- Guedes, F.R.C.M.; Maia, C.F.; da Silva, B.R.S.; Batista, B.L.; Alyemeni, M.N.; Ahmad, P.; Lobato, A.K.d.S. Exogenous 24-Epibrassinolide stimulates root protection, and leaf antioxidant enzymes in lead stressed rice plants: Central roles to minimize Pb content and oxidative stress. Environ. Pollut. 2021, 280, 116992. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.W.; Mandava, N.; Worley, J.F.; Plimmer, J.R.; Smith, M.V. Brassins—A New Family of Plant Hormones from Rape Pollen. Nature 1970, 225, 1065–1066. [Google Scholar] [CrossRef] [PubMed]
- Peres, A.L.G.L.; Soares, J.S.; Tavares, R.G.; Righetto, G.; Zullo, M.A.T.; Mandava, N.B.; Menossi, M. Brassinosteroids, the Sixth Class of Phytohormones: A Molecular View from the Discovery to Hormonal Interactions in Plant Development and Stress Adaptation. Int. J. Mol. Sci. 2019, 20, 331. [Google Scholar] [CrossRef] [PubMed]
- González-García, M.-P.; Vilarrasa-Blasi, J.; Zhiponova, M.; Divol, F.; Mora-García, S.; Russinova, E.; Caño-Delgado, A.I. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 2011, 138, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Planas-Riverola, A.; Gupta, A.; Betegón-Putze, I.; Bosch, N.; Ibañes, M.; Caño-Delgado, A.I. Brassinosteroid signaling in plant development and adaptation to stress. Development 2019, 146, dev151894. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Russinova, E. Brassinosteroid signalling. Curr. Biol. 2020, 30, R294–R298. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.M.; Vukašinović, N.; Hsu, C.-W.; Zhang, J.; Vanhoutte, I.; Shahan, R.; Taylor, I.W.; Greenstreet, L.; Heitz, M.; Afanassiev, A.; et al. Brassinosteroid gene regulatory networks at cellular resolution in the Arabidopsis root. Science 2023, 379, eadf4721. [Google Scholar] [CrossRef] [PubMed]
- Bulgakov, V.P.; Avramenko, T.V. Linking Brassinosteroid and ABA Signaling in the Context of Stress Acclimation. Int. J. Mol. Sci. 2020, 21, 5108. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Bartolomé, J.; Minguet, E.G.; Grau-Enguix, F.; Abbas, M.; Locascio, A.; Thomas, S.G.; Alabadí, D.; Blázquez, M.A. Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 13446–13451. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Bai, M.Y.; Oh, E.; Zhu, J.Y. Brassinosteroid signaling network and regulation of photo-morphogenesis. Annu. Rev. Genet. 2012, 46, 701–724. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Tang, B.; Xie, Z.; Nolan, T.; Ye, H.; Song, G.; Walley, J.; Yin, Y. GSK3-like kinase BIN2 phosphorylates RD26 to potentiate drought signaling in Arabidopsis. Plant J. 2019, 100, 923–937. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Liu, S.; Tang, B.; Chen, J.; Xie, Z.; Nolan, T.M.; Jiang, H.; Guo, H.; Lin, H.Y.; Li, L.; et al. RD26 mediates crosstalk be-tween drought and brassinosteroid signalling pathways. Nat. Commun. 2017, 8, 14573. [Google Scholar] [CrossRef]
- He, J.X.; Gendron, J.M.; Yang, Y.; Li, J.; Wang, Z.Y. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 10185–10190. [Google Scholar] [CrossRef]
- Yin, Y.; Vafeados, D.; Tao, Y.; Yoshida, S.; Asami, T.; Chory, J. A new class of transcription factors mediates brassinoster-oid-regulated gene expression in Arabidopsis. Cell 2005, 120, 249–259. [Google Scholar] [CrossRef]
- He, J.X.; Gendron, J.M.; Sun, Y.; Gampala, S.S.; Gendron, N.; Sun, C.Q.; Wang, Z.Y. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 2005, 307, 1634–1638. [Google Scholar] [CrossRef]
- Guo, R.; Qian, H.; Shen, W.; Liu, L.; Zhang, M.; Cai, C.; Zhao, Y.; Qiao, J.; Wang, Q. BZR1 and BES1 participate in regulation of glucosinolate biosynthesis by brassinosteroids in Arabidopsis. J. Exp. Bot. 2013, 64, 2401–2412. [Google Scholar] [CrossRef]
- Ryu, H.; Cho, H.; Bae, W.; Hwang, I. Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat. Commun. 2014, 5, 4138. [Google Scholar] [CrossRef]
- Jiang, J.; Zhang, C.; Wang, X. A recently evolved isoform of the transcription factor BES1 pro-motes brassinosteroid signaling and development in Arabidopsis thaliana. Plant Cell 2015, 27, 361–374. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, Z.Y.; Mora-Garcia, S.; Li, J.; Yoshida, S.; Asami, T.; Chory, J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Mueller, A.K.; Labaied, M.; Kappe, S.H.; Matuschewski, K. Genetically modified Plasmodium para-sites as a protective experimental malaria vaccine. Nature 2005, 433, 164–167. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wen, J.; Lease, K.A.; Doke, J.T.; Tax, F.E.; Walker, J.C. BAK1, an Arabidopsis LRR Receptor-like Protein Kinase, Interacts with BRI1 and Modulates Brassinosteroid Signaling. Cell 2002, 110, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-W.; Guan, S.; Burlingame, A.L.; Wang, Z.-Y. The CDG1 Kinase Mediates Brassinosteroid Signal Transduction from BRI1 Receptor Kinase to BSU1 Phosphatase and GSK3-like Kinase BIN2. Mol. Cell 2011, 43, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Nam, K.H. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 2002, 295, 1299–1301. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Park, Y.J.; Son, S.-H.; Roh, J.; Youn, J.H.; Kim, S.Y.; Kim, S.-K. Brassinosteroids signaling via BZR1 down-regulates expression of ACC oxidase 4 to control growth of Arabidopsis thaliana seedlings. Plant Signal. Behav. 2020, 15, 1734333. [Google Scholar] [CrossRef]
- Yu, Z.; Ma, J.; Zhang, M.; Li, X.; Sun, Y.; Zhang, M.; Ding, Z. Auxin promotes hypocotyl elongation by enhancing BZR1 nuclear accumulation in Arabidopsis. Sci. Adv. 2023, 9, eade2493. [Google Scholar] [CrossRef] [PubMed]
- Bernardo-Garcia, S.; de Lucas, M.; Martinez, C.; Espinosa-Ruiz, A.; Daviere, J.M.; Prat, S. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes. Dev. 2014, 28, 1681–1694. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, X.; Lai, Y.; He, G.; Wen, S.; He, H.; Li, Z.; Zhang, B.; Zhang, D. Transcriptomic analysis reveals the significant effects of fertilization on the biosynthesis of sesquiterpenes in Phoebe bournei. Genomics 2022, 114, 110375. [Google Scholar] [CrossRef]
- Nolan, T.M.; Vukasinovic, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional Regulators of Plant Growth, Development, and Stress Responses. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Nolan, T.M.; Ye, H.; Zhang, M.; Tong, H.; Xin, P.; Chu, J.; Chu, C.; Li, Z.; Yin, Y. Arabidopsis WRKY46, WRKY54, and WRKY70 Transcription Factors Are Involved in Brassinosteroid-Regulated Plant Growth and Drought Responses. Plant Cell 2017, 29, 1425–1439. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A.; Lee, S.H.; Patel, D.; Kumar, S.V.; Spartz, A.K.; Gu, C.; Ye, S.; Yu, P.; Breen, G.; Cohen, J.D.; et al. Phytochrome-Interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA 2011, 108, 20231–20235. [Google Scholar] [CrossRef]
- Mecchia, M.A.; García-Hourquet, M.; Lozano-Elena, F.; Planas-Riverola, A.; Blasco-Escamez, D.; Marquès-Bueno, M.; Mora-García, S.; Caño-Delgado, A.I. The BES1/BZR1-family transcription factor MpBES1 regulates cell division and differentiation in Marchantia polymorpha. Curr. Biol. 2021, 31, 4860–4869.e8. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wu, Y.; Li, L.; Li, C. Comprehensive analysis of the BES1 gene family and its expression under abiotic stress and hormone treatment in Populus trichocarpa. Plant Physiol. Biochem. 2022, 173, 1–13. [Google Scholar] [CrossRef]
- Song, X.; Ma, X.; Li, C.; Hu, J.; Yang, Q.; Wang, T.; Wang, L.; Wang, J.; Guo, D.; Ge, W.; et al. Comprehensive analyses of the BES1 gene family in Brassica napus and examination of their evolutionary pattern in representative species. BMC Genom. 2018, 19, 346. [Google Scholar] [CrossRef]
- Liu, Z.; Qanmber, G.; Lu, L.; Qin, W.; Liu, J.; Li, J.; Ma, S.; Yang, Z.; Yang, Z. Genome-wide analysis of BES1 genes in Gossypium revealed their evolutionary conserved roles in brassinosteroid signaling. Sci. China Life Sci. 2018, 61, 1566–1582. [Google Scholar] [CrossRef]
- Yu, H.; Feng, W.; Sun, F.; Zhang, Y.; Qu, J.; Liu, B.; Lu, F.; Yang, L.; Fu, F.; Li, W. Cloning and characterization of BES1/BZR1 transcription factor genes in maize. Plant Growth Regul. 2018, 86, 235–249. [Google Scholar] [CrossRef]
- Gao, Y.; Hu, J.; Zhao, T.; Xu, X.; Jiang, J.; Li, J. Genome-wide Identification and Expression Pattern Analysis of BRI1-EMS–suppressor Transcription Factors in Tomato under Abiotic Stresses. J. Am. Soc. Hortic. Sci. 2018, 143, 84–90. [Google Scholar] [CrossRef]
- Song, X.; Li, Y.; Hou, X. Genome-wide analysis of the AP2/ERF transcription factor super-family in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genom. 2013, 14, 573. [Google Scholar] [CrossRef]
- Xie, L.; Yang, C.; Wang, X. Brassinosteroids can regulate cellulose biosynthesis by con-trolling the expression of CESA genes in Arabidopsis. J. Exp. Bot. 2011, 62, 4495–4506. [Google Scholar] [CrossRef]
- Ye, Q.; Zhu, W.; Li, L.; Zhang, S.; Yin, Y.; Ma, H.; Wang, X. Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc. Natl. Acad. Sci. USA 2010, 107, 6100–6105. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Durán, R.; Macho, A.P.; Boutrot, F.; Segonzac, C.; Somssich, I.E.; Zipfel, C. The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. eLife 2013, 2, e00983. [Google Scholar] [CrossRef]
- Li, Q.-F.; Lu, J.; Yu, J.-W.; Zhang, C.-Q.; He, J.-X.; Liu, Q.-Q. The brassinosteroid-regulated transcription factors BZR1/BES1 function as a coordinator in multisignal-regulated plant growth. Biochim. Biophys. Acta (BBA) 2018, 1861, 561–571. [Google Scholar] [CrossRef]
- Wenkai, A.N.; Dan, C.; Fuchun, Z. Expression Characteristics of Transcription Factor BES1/BRI1 of Cotton Seedling in Response to Brassinosteroid under Drought Stress. Acta Bot. Boreal.-Occident. Sin. 2015, 35, 1311–1316. [Google Scholar]
- Li, X.; Liu, L.; Sun, S.; Li, Y.; Jia, L.; Ye, S.; Yu, Y.; Dossa, K.; Luan, Y. Leaf-transcriptome profiles of Phoebe bournei provide insights into temporal drought stress responses. Front. Plant Sci. 2022, 13, 1010314. [Google Scholar] [CrossRef]
- Carnicer, J.; Alegria, A.; Giannakopoulos, C.; Di Giuseppe, F.; Karali, A.; Koutsias, N.; Lionello, P.; Parrington, M.; Vitolo, C. Global warming is shifting the relationships between fire weather and realized fire-induced CO2 emissions in Europe. Sci. Rep. 2022, 12, 10365. [Google Scholar] [CrossRef]
- Dai, A. Drought under global warming: A review. WIREs Clim. Chang. 2010, 2, 45–65. [Google Scholar] [CrossRef]
- Mansoor, S.; Kour, N.; Manhas, S.; Zahid, S.; Wani, O.A.; Sharma, V.; Wijaya, L.; Alyemeni, M.N.; Alsahli, A.A.; El-Serehy, H.A.; et al. Biochar as a tool for effective management of drought and heavy metal toxicity. Chemosphere 2020, 271, 129458. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; He, X.; Wang, J.; Jiang, B.; Ye, R.; Lin, X. Physiological and biochemical responses of Phoebe bournei seedlings to water stress and recovery. Acta Physiol. Plant. 2014, 36, 1241–1250. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [PubMed]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef]
- Kolodny, R. Searching protein space for ancient sub-domain segments. Curr. Opin. Struct. Biol. 2021, 68, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Nosaki, S.; Miyakawa, T.; Xu, Y.; Nakamura, A.; Hirabayashi, K.; Asami, T.; Nakano, T.; Tanokura, M. Structural basis for brassinosteroid response by BIL1/BZR1. Nat. Plants 2018, 4, 771–776. [Google Scholar] [CrossRef]
- Wang, X.; Niu, Y.; Zheng, Y. Multiple Functions of MYB Transcription Factors in Abiotic Stress Responses. Int. J. Mol. Sci. 2021, 22, 6125. [Google Scholar] [CrossRef]
- Gendron, J.M.; Wang, Z.Y. Multiple mechanisms modulate Brassinosteroid signaling. Curr. Opin. Plant Biol. 2007, 10, 436–441. [Google Scholar] [CrossRef]
- Cheng, M.; Yuan, H.; Wang, R.; Wang, W.; Zhang, L.; Fan, F.; Li, S. Identification and characterization of BES1 genes involved in grain size development of Oryza sativa L. Int. J. Biol. Macromol. 2023, 253, 127327. [Google Scholar] [CrossRef]
- Li, J.; Wang, L.; Leng, F.; Ma, C.; Zhang, C.; Wang, S. Genome-wide identification, characterization and gene expression of BES1 transcription factor family in grapevine (Vitis vinifera L.). Sci. Rep. 2023, 13, 240. [Google Scholar] [CrossRef] [PubMed]
- Su, D.; Xiang, W.; Wen, L.; Lu, W.; Shi, Y.; Liu, Y.; Li, Z. Genome-wide identification, characterization and expression analysis of BES1 gene family in tomato. BMC Plant Biol. 2021, 21, 161. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.W.; Gilbert, W. The evolution of spliceosomal introns: Patterns, puzzles and progress. Nat. Rev. Genet. 2006, 7, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Cui, Y.; Zhao, Z.; Li, S.; Liang, D.; Wang, C.; Feng, G.; Wang, J.; Liu, Z. Genome-wide identification and characterization of the BES/BZR gene family in wheat and foxtail millet. BMC Genom. 2021, 22, 682. [Google Scholar] [CrossRef]
- Izadi, F.; Zarrini, H.N.; Jelodar, N.B. In silico analysis of BES1 transcription factor in citrus senescence. Res. J. Life Sci. Bioinform. Pharm. Chem. Sci. 2016, 2, 29. [Google Scholar]
- Mahesh, K.; Balaraju, P.; Ramakrishna, B.; Rao, S.S.R. Effect of Brassinosteroids on Germination and Seedling Growth of Radish (Raphanus sativus L.) under PEG-6000 Induced Water Stress. Am. J. Plant Sci. 2013, 4, 2305–2313. [Google Scholar] [CrossRef]
- Anwar, A.; Liu, Y.; Dong, R.; Bai, L.; Yu, X.; Li, Y. The physiological and molecular mechanism of brassinosteroid in response to stress: A review. Biol. Res. 2018, 51, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.H.; Li, X.Q.; Wang, G.G.; Zhu, X.T. Brassinosteroids alleviate high-temperature injury in Ficus concinna seedlings via maintaining higher antioxidant defence and glyoxalase systems. AoB Plants 2015, 7, plv009. [Google Scholar] [CrossRef]
- Kagale, S.; Divi, U.K.; Krochko, J.E.; Keller, W.A.; Krishna, P. Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 2007, 225, 353–364. [Google Scholar] [CrossRef]
- Martins, S.; Montiel-Jorda, A.; Cayrel, A.; Huguet, S.; Roux, C.P.-L.; Ljung, K.; Vert, G. Brassinosteroid signaling-dependent root responses to prolonged elevated ambient temperature. Nat. Commun. 2017, 8, 309. [Google Scholar] [CrossRef]
- Choudhary, S.P.; Yu, J.Q.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S. Benefits of brassinosteroid crosstalk. Trends Plant Sci. 2012, 17, 594–605. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Zhang, J.; Han, S.; Chong, S.L.; Meng, G.; Song, M.; Wang, Y.; Zhou, S.; Liu, C.; Lou, L.; et al. The chromosome-scale genome of Phoebe bournei reveals contrasting fates of terpene synthase (TPS)-a and TPS-b subfamilies. Plant Commun. 2022, 3, 100410. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhu, Y.; Pan, Y.; Huang, H.; Li, C.; Li, G.; Tong, Z. Transcriptomic profiling and identification of candidate genes in two Phoebe bournei ecotypes with contrasting cold stress responses. Trees 2018, 32, 1315–1333. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).