The Effect of HMGB1 and HMGB2 on Transcriptional Regulation Differs in Neuroendocrine and Adenocarcinoma Models of Prostate Cancer
Abstract
1. Introduction
2. Results
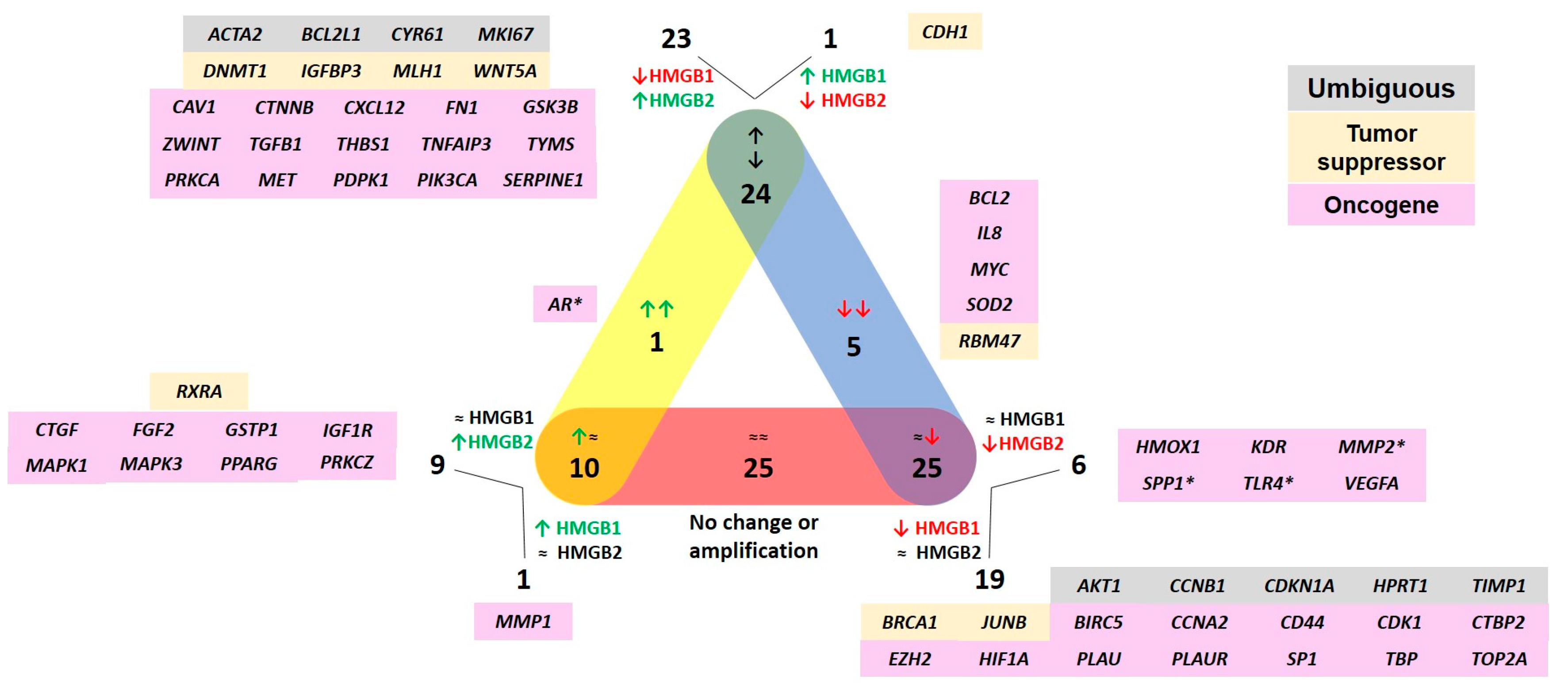
2.1. Transcription of Genes Related to PCa after HMGB1 or HMGB2 Silencing in PC-3
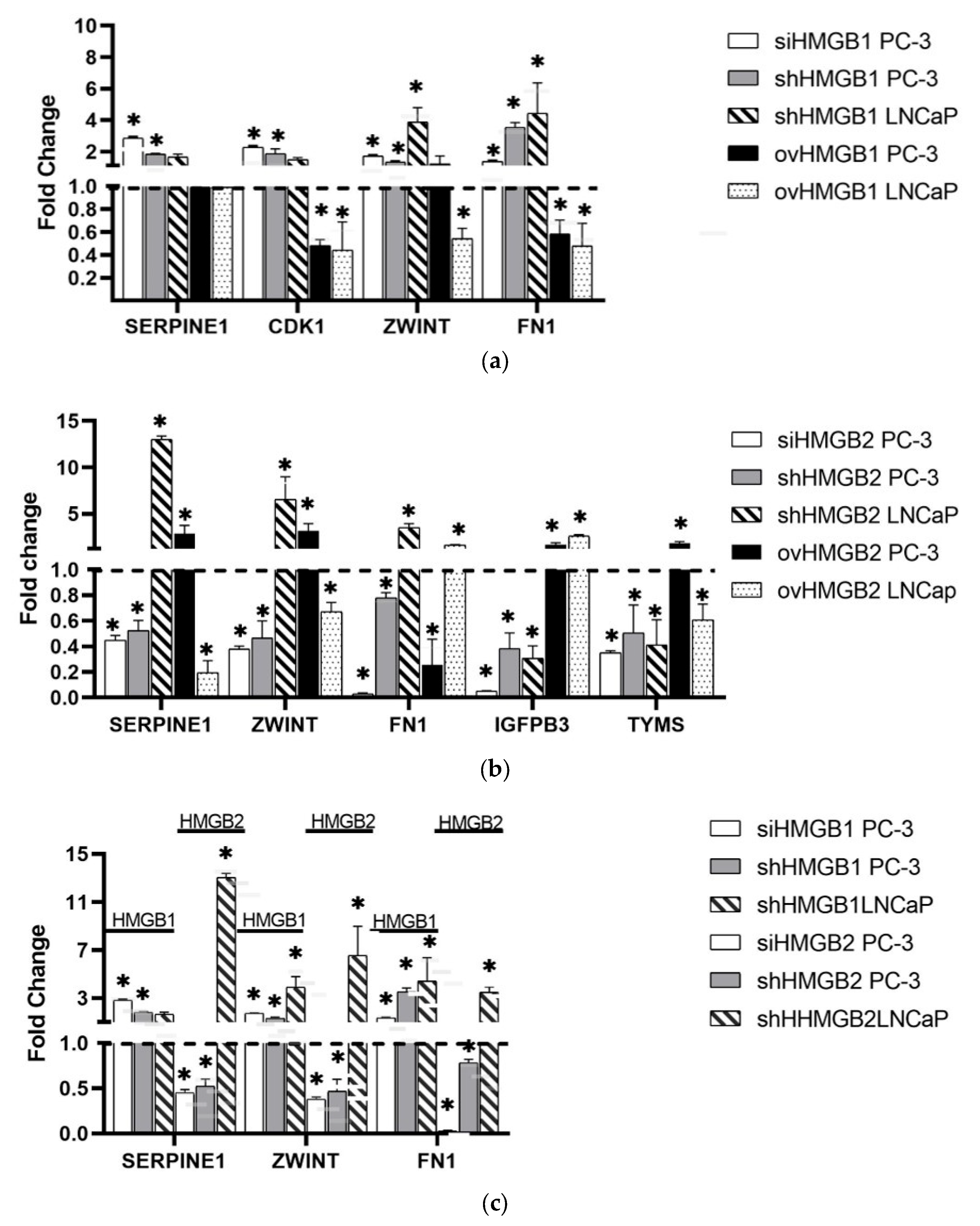
2.2. Validation of Data Array and Differential Regulation in PCa Cell Lines
2.3. Correlation between the Expression of HMGB2 and Genes Involved in PCa in Patients Classified as AR-Negative-Neuroendocrine-Positive or AR-Positive-Neuroendocrine-Negative
3. Discussion
4. Materials and Methods
4.1. Biological Materials
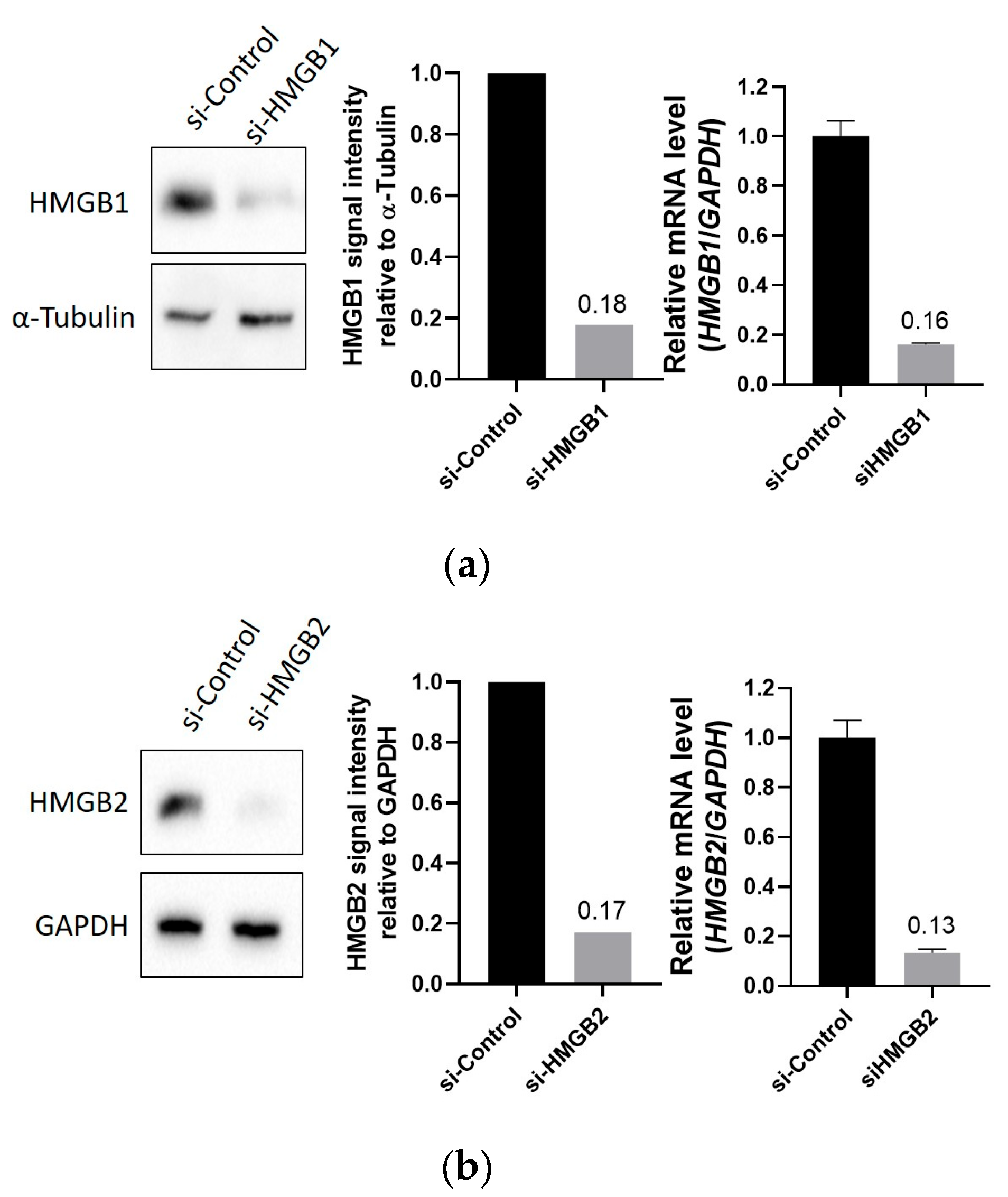
4.2. HMGB1 and HMGB2 Silencing by siRNA in PC-3
4.3. Analysis of Gene Expression in siHMGB1 and siHMGB2 PC-3 Cells Using Prime-PCR Panels
4.4. HMGB1 and HMGB2 Silencing by shRNA in PCa Cell Lines
4.5. HMGB1 and HMGB2 Overexpression in PCa Cell Lines
4.6. Relative Expression Analyses
4.7. Correlation Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Rubin, M.A.; Maher, C.A.; Chinnaiyan, A.M. Common Gene Rearrangements in Prostate Cancer. J. Clin. Oncol. 2011, 29, 3659–3668. [Google Scholar] [CrossRef] [PubMed]
- Urbanucci, A.; Sahu, B.; Seppala, J.; Larjo, A.; Latonen, L.M.; Waltering, K.K.; Tammela, T.L.J.; Vessella, R.L.; Lahdesmaki, H.; Janne, O.A.; et al. Overexpression of Androgen Receptor Enhances the Binding of the Receptor to the Chromatin in Prostate Cancer. Oncogene 2012, 31, 2153–2163. [Google Scholar] [CrossRef] [PubMed]
- Tai, S.; Sun, Y.; Squires, J.M.; Zhang, H.; Oh, W.K.; Liang, C.; Huang, J. PC3 is a Cell Line Characteristic of Prostatic Small Cell Carcinoma. Prostate 2011, 71, 1668–1679. [Google Scholar] [CrossRef] [PubMed]
- Beltran, H.; Prandi, D.; Mosquera, J.M.; Benelli, M.; Puca, L.; Cyrta, J.; Marotz, C.; Giannopoulou, E.; Chakravarthi, B.V.S.K.; Varambally, S.; et al. Divergent Clonal Evolution of Castration-Resistant Neuroendocrine Prostate Cancer. Nat. Med. 2016, 22, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Ostano, P.; Mello-Grand, M.; Sesia, D.; Gregnanin, I.; Peraldo-Neia, C.; Guana, F.; Jachetti, E.; Farsetti, A.; Chiorino, G. Gene Expression Signature Predictive of Neuroendocrine Transformation in Prostate Adenocarcinoma. Int. J. Mol. Sci. 2020, 21, 1078. [Google Scholar] [CrossRef] [PubMed]
- Slominski, R.M.; Raman, C.; Chen, J.Y.; Slominski, A.T. How Cancer Hijacks the Body’s Homeostasis through the Neuroendocrine System. Trends Neurosci. 2023, 46, 263–275. [Google Scholar] [CrossRef]
- Agresti, A.; Bianchi, M.E. HMGB Proteins and Gene Expression. Curr. Opin. Genet. Dev. 2003, 13, 170–178. [Google Scholar] [CrossRef]
- Ishiguro, H.; Nakaigawa, N.; Miyoshi, Y.; Fujinami, K.; Kubota, Y.; Uemura, H. Receptor for Advanced Glycation End Products (RAGE) and its Ligand, Amphoterin are Overexpressed and Associated with Prostate Cancer Development. Prostate 2005, 64, 92–100. [Google Scholar] [CrossRef]
- Wu, T.; Zhang, W.; Yang, G.; Li, H.; Chen, Q.; Song, R.; Zhao, L. HMGB1 Overexpression as a Prognostic Factor for Survival in Cancer: A Meta-Analysis and Systematic Review. Oncotarget 2016, 7, 50417–50427. [Google Scholar] [CrossRef]
- Gnanasekar, M.; Thirugnanam, S.; Ramaswamy, K. Short Hairpin RNA (shRNA) Constructs Targeting High Mobility Group Box-1 (HMGB1) Expression Leads to Inhibition of Prostate Cancer Cell Survival and Apoptosis. Int. J. Oncol. 2009, 34, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Elangovan, I.; Thirugnanam, S.; Chen, A.; Zheng, G.; Bosland, M.C.; Kajdacsy-Balla, A.; Gnanasekar, M. Targeting Receptor for Advanced Glycation End Products (RAGE) Expression Induces Apoptosis and Inhibits Prostate Tumor Growth. Biochem. Biophys. Res. Commun. 2012, 417, 1133–1138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lv, F.; Qiao, L.; Zhao, Q. MiR-505 Inhibits Prostate Cancer Cell Invasion, Metastasis and Epithelial-to-Mesenchymal Transition through Targeting HMGB-1. J. BUON 2020, 25, 2036–2044. [Google Scholar] [PubMed]
- Lv, D.; Song, X.; Huang, B.; Yu, Y.; Shu, F.; Wang, C.; Chen, H.; Zhang, H.; Zhao, S. HMGB1 Promotes Prostate Cancer Development and Metastasis by Interacting with Brahma-Related Gene 1 and Activating the Akt Signaling Pathway. Theranostics 2019, 9, 5166–5182. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shao, S.; Han, D.; Xu, Y.; Jiao, D.; Wu, J.; Yang, F.; Ge, Y.; Shi, S.; Li, Y.; et al. High Mobility Group Box 1 Promotes the Epithelial-to-Mesenchymal Transition in Prostate Cancer PC3 Cells Via the RAGE/NF-kappaB Signaling Pathway. Int. J. Oncol. 2018, 53, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Kato, H.; Fuji, S.; Naiki, T.; Naiki-Ito, A.; Yamashita, Y.; Takahashi, S. Early Detection of Prostate Carcinogens by Immunohistochemistry of HMGB2. J. Toxicol. Sci. 2018, 43, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Hu, X.; Zhang, T.; Zhang, J.; Wu, C.; Hong, W.; Jiang, Y.; Wang, Q.; Xie, Y.; Zhao, Y.; et al. HMGB1 Release Promotes Paclitaxel Resistance in Castration-Resistant Prostate Cancer Cells Via Activating C-Myc Expression. Cell. Signal. 2020, 72, 109631. [Google Scholar] [CrossRef]
- Barreiro-Alonso, A.; Camara-Quilez, M.; Salamini-Montemurri, M.; Lamas-Maceiras, M.; Vizoso-Vazquez, A.; Rodriguez-Belmonte, E.; Quindos-Varela, M.; Martinez-Iglesias, O.; Figueroa, A.; Cerdan, M. Characterization of HMGB1/2 Interactome in Prostate Cancer by Yeast Two Hybrid Approach: Potential Pathobiological Implications. Cancers 2019, 11, 1729. [Google Scholar] [CrossRef]
- Chen, J.; Xu, D.; Wang, T.; Yang, Z.; Yang, Y.; He, K.; Zhao, L. HMGB1 Promotes the Development of Castration-resistant Prostate Cancer by Regulating Androgen Receptor Activation. Oncol. Rep. 2022, 48, 197. [Google Scholar] [CrossRef]
- Horoszewicz, J.S.; Leong, S.S.; Chu, T.M.; Wajsman, Z.L.; Friedman, M.; Papsidero, L.; Kim, U.; Chai, L.S.; Kaka, S.; Arya, S.K.; et al. The LNCaP Cell Line—A New Model for Studies on Human Prostatic Carcinoma. Prog. Clin. Biol. Res. 1980, 37, 115–132. [Google Scholar]
- Zhou, J.; Chen, X.; Gilvary, D.L.; Tejera, M.M.; Eksioglu, E.A.; Wei, S.; Djeu, J.Y. HMGB1 Induction of Clusterin Creates a Chemoresistant Niche in Human Prostate Tumor Cells. Sci. Rep. 2015, 5, 15085. [Google Scholar] [CrossRef]
- Vizoso-Vázquez, Á.; Barreiro-Alonso, A.; Rico-Díaz, A.; Lamas-Maceiras, M.; Rodríguez-Belmonte, E.; Becerra, M.; González-Siso, M.I.; Cerdán, M.E. Gene Regulation, DNA Repair and Beyond. In Old Yeasts-New Questions; Lucas, C., Pais, C., Eds.; Intech: Berlin, Germany, 2017; Chapter 7. [Google Scholar]
- Mao, Y.; Li, W.; Hua, B.; Gu, X.; Pan, W.; Chen, Q.; Xu, B.; Wang, Z.; Lu, C. Silencing of ELK3 Induces S-M Phase Arrest and Apoptosis and Upregulates SERPINE1 Expression Reducing Migration in Prostate Cancer Cells. Biomed. Res. Int. 2020, 2020, 2406159. [Google Scholar] [CrossRef]
- Kuo, Y.; Huo, C.; Lin, C.; Lin, H.; Liu, J.; Wang, W.; Chang, C.; Chuu, C. Caffeic Acid Phenethyl Ester Suppresses Androgen Receptor Signaling and Stability Via Inhibition of Phosphorylation on Ser81 and Ser213. Cell. Commun. Signal. 2019, 17, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Koryakina, Y.; Knudsen, K.E.; Gioeli, D. Cell-Cycle-Dependent Regulation of Androgen Receptor Function. Endocr. Relat. Cancer 2015, 22, 249–264. [Google Scholar] [CrossRef]
- Song, Z.; Chao, F.; Zhuo, Z.; Ma, Z.; Li, W.; Chen, G. Identification of Hub Genes in Prostate Cancer using Robust Rank Aggregation and Weighted Gene Co-Expression Network Analysis. Aging 2019, 11, 4736–4756. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Liu, Y.; Zhang, Y.; Ouyang, J.; Feng, Y.; Li, S.; Wang, J.; Zhang, C.; Tan, L.; Zhong, J.; et al. MicroRNA-142-3P Suppresses the Progression of Papillary Thyroid Carcinoma by Targeting FN1 and Inactivating FAK/ERK/PI3K Signaling. Cell. Signal. 2023, 109, 110792. [Google Scholar] [CrossRef] [PubMed]
- Carreras, C.W.; Santi, D.V. The Catalytic Mechanism and Structure of Thymidylate Synthase. Annu. Rev. Biochem. 1995, 64, 721–762. [Google Scholar] [CrossRef]
- Rustum, Y.M. Thymidylate Synthase: A Critical Target in Cancer Therapy? Front. Biosci. 2004, 9, 2467–2473. [Google Scholar] [CrossRef]
- Burdelski, C.; Strauss, C.; Tsourlakis, M.C.; Kluth, M.; Hube-Magg, C.; Melling, N.; Lebok, P.; Minner, S.; Koop, C.; Graefen, M.; et al. Overexpression of Thymidylate Synthase (TYMS) is Associated with Aggressive Tumor Features and Early PSA Recurrence in Prostate Cancer. Oncotarget 2015, 6, 8377–8387. [Google Scholar] [CrossRef]
- Chan, J.M.; Stampfer, M.J.; Ma, J.; Gann, P.; Gaziano, J.M.; Pollak, M.; Giovannucci, E. Insulin-Like Growth Factor-I (IGF-I) and IGF Binding Protein-3 as Predictors of Advanced-Stage Prostate Cancer. J. Natl. Cancer Inst. 2002, 94, 1099–1106. [Google Scholar] [CrossRef]
- Boyle, B.J.; Zhao, X.Y.; Cohen, P.; Feldman, D. Insulin-Like Growth Factor Binding Protein-3 Mediates 1 Alpha,25-Dihydroxyvitamin D3 Growth Inhibition in the LNCaP Prostate Cancer Cell Line through p21/WAF1. J. Urol. 2001, 165, 1319–1324. [Google Scholar] [CrossRef] [PubMed]
- Mayo, J.C.; Hevia, D.; Quiros-Gonzalez, I.; Rodriguez-Garcia, A.; Gonzalez-Menendez, P.; Cepas, V.; Gonzalez-Pola, I.; Sainz, R.M. IGFBP3 and MAPK/ERK Signaling Mediates Melatonin-Induced Antitumor Activity in Prostate Cancer. J. Pineal Res. 2017, 62, e12373. [Google Scholar] [CrossRef] [PubMed]
- Natsuizaka, M.; Ohashi, S.; Wong, G.S.; Ahmadi, A.; Kalman, R.A.; Budo, D.; Klein-Szanto, A.J.; Herlyn, M.; Diehl, J.A.; Nakagawa, H. Insulin-Like Growth Factor-Binding Protein-3 Promotes Transforming Growth Factor-Beta1-Mediated Epithelial-to-Mesenchymal Transition and Motility in Transformed Human Esophageal Cells. Carcinogenesis 2010, 31, 1344–1353. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Ma, Z.; Liu, B.; Qian, H.; Shao, X.; Liu, J.; Wang, Q.; Xue, W. Identification of Cancer-Associated Fibroblasts Subtypes in Prostate Cancer. Front. Immunol. 2023, 14, 1133160. [Google Scholar] [CrossRef]
- Ochman, H.; Gerber, A.S.; Hartl, D.L. Genetic Applications of an Inverse Polymerase Chain Reaction. Genetics 1988, 120, 621–623. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

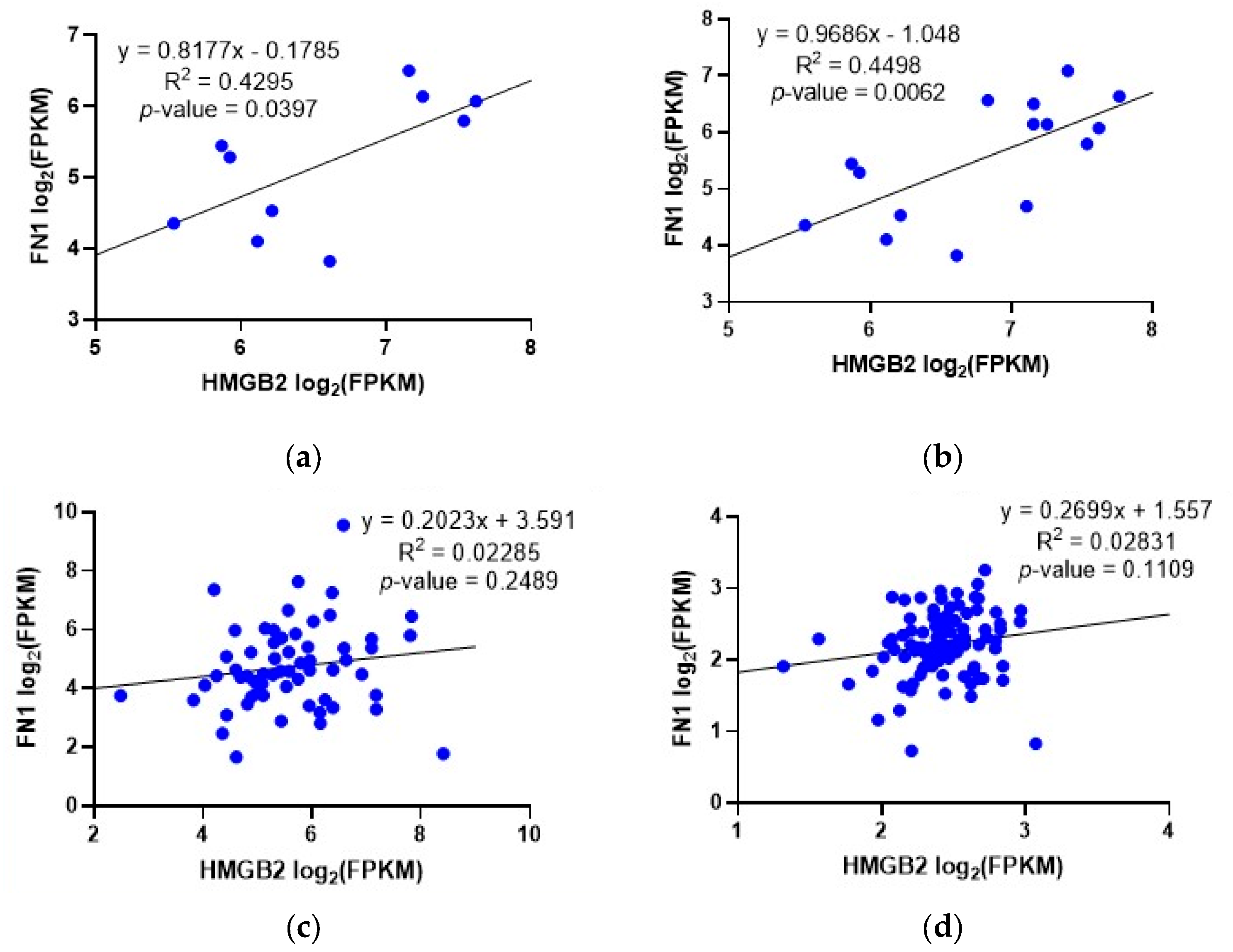
| GENE | cBioportal Study | Spearman Coefficient SC | p-Value SC | Pearson Coefficient PC | p-Value PC |
|---|---|---|---|---|---|
| CDK1 | MSK, Cancer Cell 2010 | −0.32 | 3.449 × 10−4 | −0.22 | 0.013 |
| FN1 | DKFZ, Cancer Cell 2018 | −0.32 | 3.712 × 10−4 | −0.27 | 3.282 × 10−3 |
| FN1 | Fred Hutchinson CRC, Nat Med 2016 | −0.21 | 0.016 | −0.23 | 6.517 × 10−3 |
| FN1 | TCGA, Firehose Legacy | −0.12 | 6.638 × 10−3 | −0.08 | ns |
| SERPINE1 | Fred Hutchinson CRC, Nat Med 2016 | −0.30 | 5.118 × 10−4 | −0.35 | 4.322 × 10−5 |
| SERPINE1 | TCGA, Cell 2015 | −0.12 | 0.0488 | −0.09 | ns |
| ZWINT | DKFZ, Cancer Cell 2018 | −0.43 | 7.12 × 10−7 | −0.37 | 3.14 × 10−5 |
| Gene | cBioportal Study | Spearman Coefficient SC | p-Value SC | Pearson Coefficient PC | p-Value SC |
|---|---|---|---|---|---|
| FN1 | MSK, Cancer Cell 2010 | 0.36 | 5.22 × 10−5 | 0.40 | 3.72 × 10−6 |
| FN1 | TCGA, Cell 2015 | 0.21 | 3.43 × 10−4 | 0.20 | 7.86 × 10−4 |
| FN1 | TCGA, Firehose Legacy | 0.21 | 2.95 × 10−6 | 0.20 | 4.13 × 10−6 |
| FN1 | TCGA, PanCancer Atlas | 0.22 | 5.62 × 10−7 | 0.21 | 2.83 × 10−6 |
| IGFBP3 | DKFZ, Cancer Cell 2018 | 0.25 | 5.78 × 10−3 | 0.38 | 1.72 × 10−5 |
| IGFBP3 | MSK, Cancer Cell 2010 | 0.16 | ns | 0.28 | 1.65 × 10−3 |
| IGFBP3 | TCGA, Cell 2015 | 0.40 | 8.72 × 10−13 | 0.35 | 1.37 × 10−9 |
| IGFBP3 | TCGA, Firehose Legacy | 0.41 | 4.78 × 10−22 | 0.36 | 1.41 × 10−16 |
| IGFBP3 | TCGA, PanCancer Atlas | 0.42 | 1.27 × 10−22 | 0.37 | 2.86 × 10−17 |
| TYMS | DKFZ, Cancer Cell 2018 | 0.45 | 3.11 × 10−7 | 0.56 | 5.99 × 10−11 |
| TYMS | Fred Hutchinson CRC, Nat Med 2016 | 0.60 | 3.75 × 10−14 | 0.59 | 6.71 × 10−14 |
| TYMS | MSK, Cancer Cell 2010 | 0.06 | ns | 0.40 | 4.58 × 10−6 |
| TYMS | SUC2C PCF Dream Team Cell 2015 | 0.69 | 9.03 × 10−18 | 0.69 | 6.81 × 10−18 |
| TYMS | SUC2C PCF Dream Team PNAS 2019 | 0.59 | 1.02 × 10−20 | 0.61 | 2.54 × 10−22 |
| TYMS | TCGA, Cell 2015 | 0.51 | 3.51 × 10−20 | 0.60 | 1.40 × 10−29 |
| TYMS | TCGA, Firehose Legacy | 0.48 | 1.99 × 10−30 | 0.59 | 3.05 × 10−47 |
| TYMS | TCGA, PanCancer Atlas | 0.47 | 1.65 × 10−28 | 0.58 | 3.01 × 10−45 |
| ZWINT | DKFZ, Cancer Cell 2018 | 0.47 | 1.10 × 10−7 | 0.60 | 4.17 × 10−13 |
| ZWINT | Fred Hutchinson CRC, Nat Med 2016 | 0.67 | 2.33 × 10−18 | 0.65 | 1.52 × 10−17 |
| ZWINT | SUC2C PCF Dream Team Cell 2015 | 0.62 | 1.24 × 10−13 | 0.66 | 3.83 × 10−16 |
| ZWINT | SUC2C PCF Dream Team PNAS 2019 | 0.54 | 1.80 × 10−17 | 0.53 | 2.87 × 10−16 |
| ZWINT | TCGA, Cell 2015 | 0.39 | 1.08 × 10−11 | 0.50 | 1.90 × 10−19 |
| ZWINT | TCGA, Firehose Legacy | 0.35 | 4.99 × 10−16 | 0.42 | 5.98 × 10−23 |
| ZWINT | TCGA, PanCancer Atlas | 0.34 | 2.55 × 10−14 | 0.41 | 1.01 × 10−20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salamini-Montemurri, M.; Vizoso-Vázquez, Á.; Barreiro-Alonso, A.; Lorenzo-Catoira, L.; Rodríguez-Belmonte, E.; Cerdán, M.-E.; Lamas-Maceiras, M. The Effect of HMGB1 and HMGB2 on Transcriptional Regulation Differs in Neuroendocrine and Adenocarcinoma Models of Prostate Cancer. Int. J. Mol. Sci. 2024, 25, 3106. https://doi.org/10.3390/ijms25063106
Salamini-Montemurri M, Vizoso-Vázquez Á, Barreiro-Alonso A, Lorenzo-Catoira L, Rodríguez-Belmonte E, Cerdán M-E, Lamas-Maceiras M. The Effect of HMGB1 and HMGB2 on Transcriptional Regulation Differs in Neuroendocrine and Adenocarcinoma Models of Prostate Cancer. International Journal of Molecular Sciences. 2024; 25(6):3106. https://doi.org/10.3390/ijms25063106
Chicago/Turabian StyleSalamini-Montemurri, Martín, Ángel Vizoso-Vázquez, Aida Barreiro-Alonso, Lidia Lorenzo-Catoira, Esther Rodríguez-Belmonte, María-Esperanza Cerdán, and Mónica Lamas-Maceiras. 2024. "The Effect of HMGB1 and HMGB2 on Transcriptional Regulation Differs in Neuroendocrine and Adenocarcinoma Models of Prostate Cancer" International Journal of Molecular Sciences 25, no. 6: 3106. https://doi.org/10.3390/ijms25063106
APA StyleSalamini-Montemurri, M., Vizoso-Vázquez, Á., Barreiro-Alonso, A., Lorenzo-Catoira, L., Rodríguez-Belmonte, E., Cerdán, M.-E., & Lamas-Maceiras, M. (2024). The Effect of HMGB1 and HMGB2 on Transcriptional Regulation Differs in Neuroendocrine and Adenocarcinoma Models of Prostate Cancer. International Journal of Molecular Sciences, 25(6), 3106. https://doi.org/10.3390/ijms25063106









