Landscape of FLT3 Variations Associated with Structural and Functional Impact on Acute Myeloid Leukemia: A Computational Study
Abstract
1. Introduction
2. Results
2.1. Association of FLT3 with AML
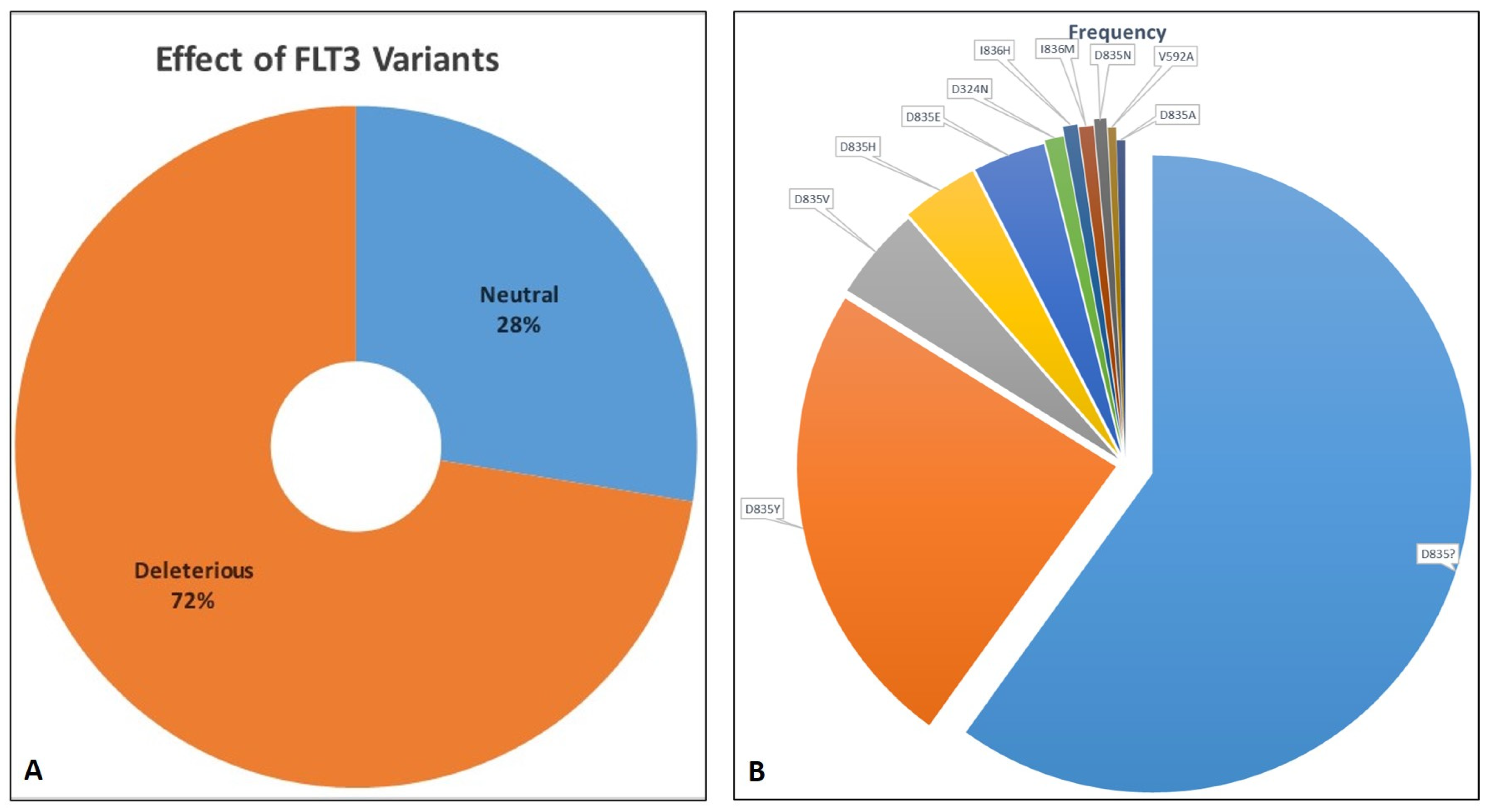
2.2. Retrieval of SNVs and Deleterious Variants
2.3. Deleterious Variants and Their Effect on FLT3 Function
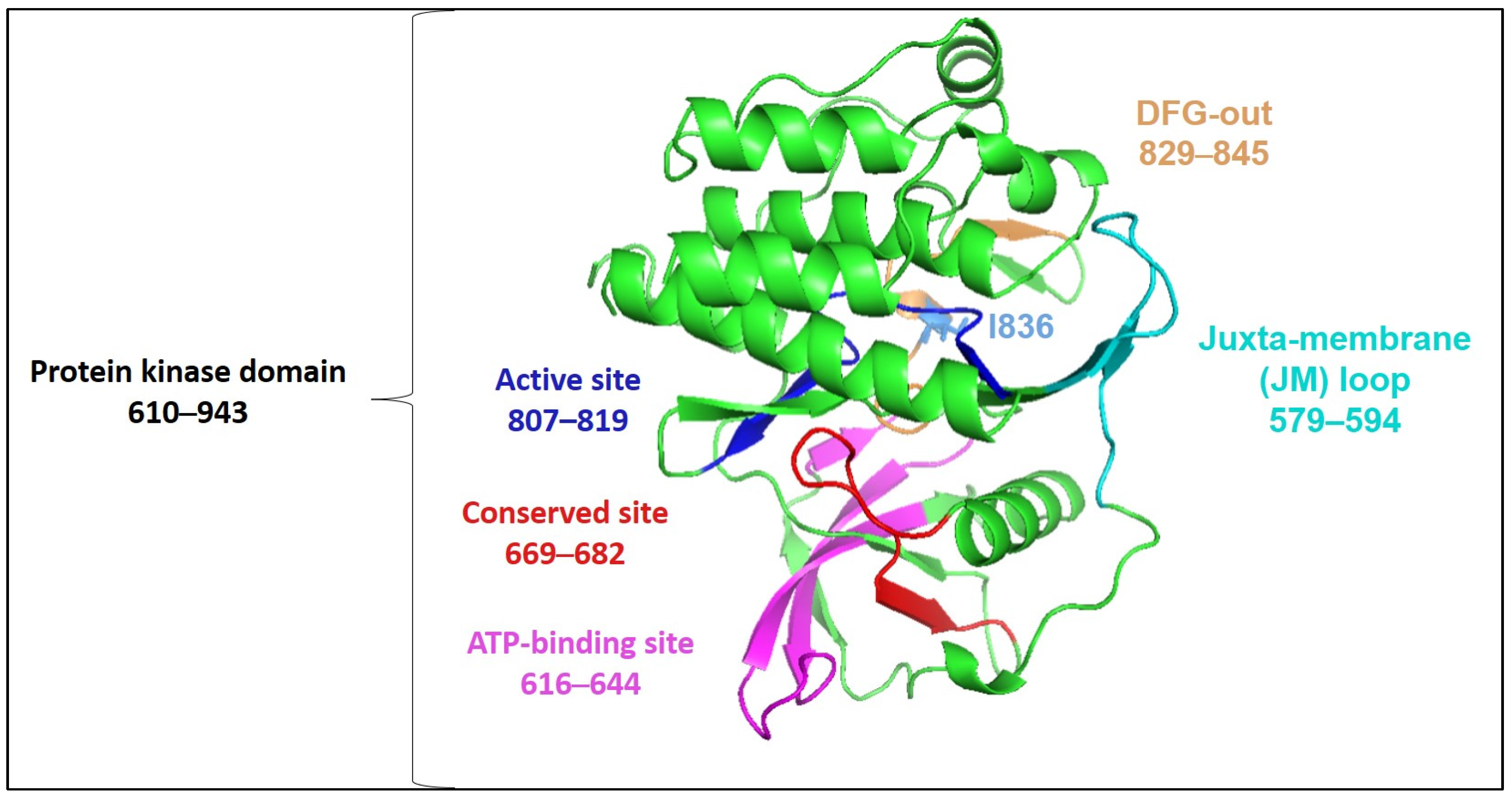
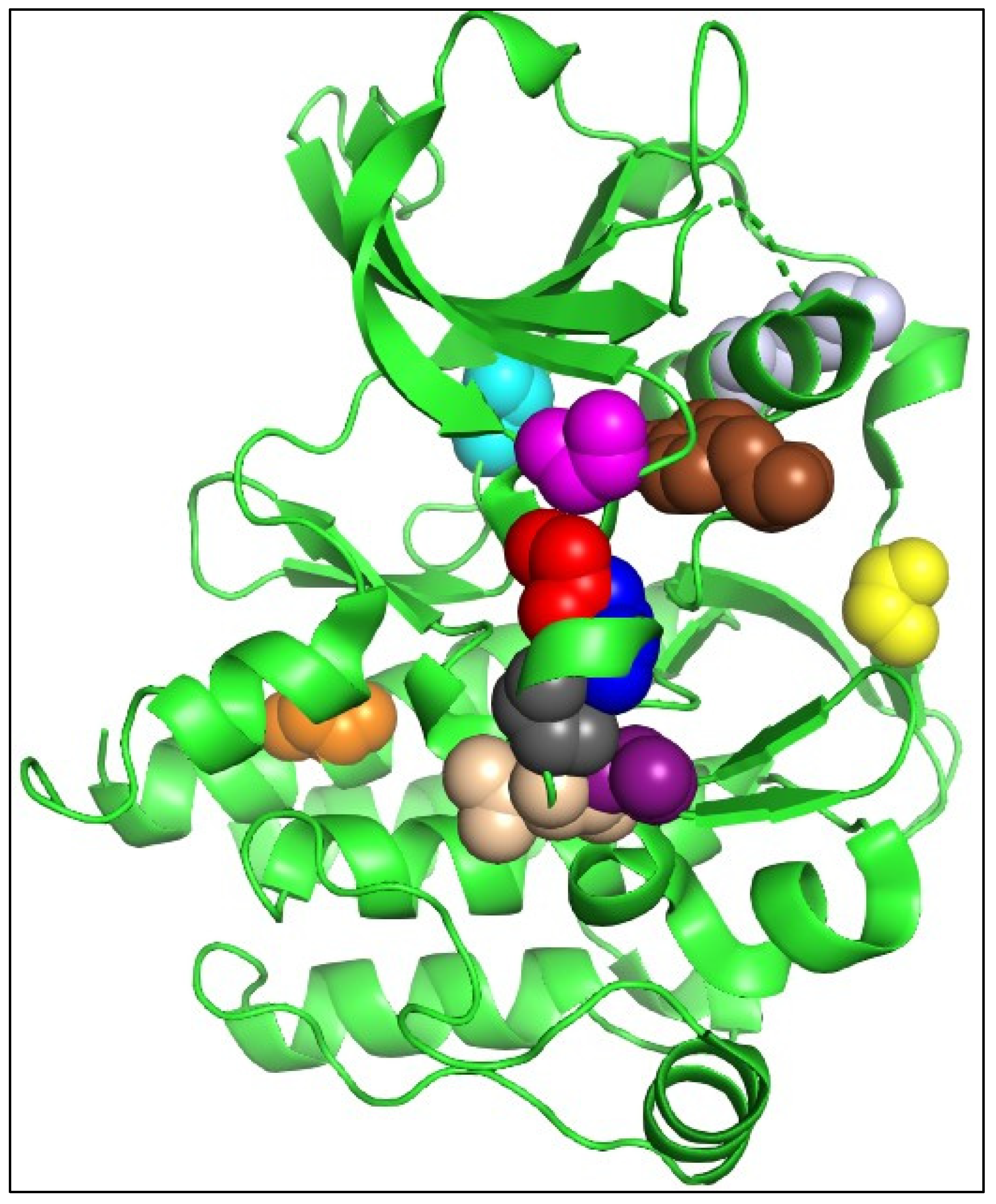
2.4. Deleterious Variants and Their Effects on FLT3 Structure
2.5. Molecular Docking and Interaction Analysis
3. Discussion
4. Materials and Methods
4.1. Association of FLT3 with AML
4.2. Variant Annotation
4.3. Variant Deleterious Effects
4.4. FLT3 Protein and Its Re-Modeling
4.5. Protein Stability Analysis for the Mutation Hotspot
4.6. Molecular Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ehsan, H.; Iqbal, Q.; Masood, A.; Grunwald, M.R. Durable remission of acute myeloid leukemia in an elderly patient following a limited course of azacitidine and venetoclax. Leuk. Res. Rep. 2022, 18, 100345. [Google Scholar] [CrossRef]
- de Rooij, J.D.; Zwaan, C.M.; van den Heuvel-Eibrink, M. Pediatric AML: From Biology to Clinical Management. J. Clin. Med. 2015, 4, 127–149. [Google Scholar] [CrossRef]
- Leukemia—Acute Myeloid—AML: Statistics. Available online: https://www.cancer.net/cancer-types/leukemia-acute-myeloid-aml/statistics#:~:text=The%205%2Dyear%20relative%20survival%20rate%20for%20people%2020%20and,well%20the%20treatment%20plan%20works (accessed on 24 February 2024).
- Tawfik, B.; Pardee, T.S.; Isom, S.; Sliesoraitis, S.; Winter, A.; Lawrence, J.; Powell, B.L.; Klepin, H.D. Comorbidity, age, and mortality among adults treated intensively for acute myeloid leukemia (AML). J. Geriatr. Oncol. 2016, 7, 24–31. [Google Scholar] [CrossRef]
- Rasche, M.; Zimmermann, M.; Borschel, L.; Bourquin, J.P.; Dworzak, M.; Klingebiel, T.; Lehrnbecher, T.; Creutzig, U.; Klusmann, J.H.; Reinhardt, D. Successes and challenges in the treatment of pediatric acute myeloid leukemia: A retrospective analysis of the AML-BFM trials from 1987 to 2012. Leukemia 2018, 32, 2167–2177. [Google Scholar] [CrossRef]
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719. [Google Scholar] [CrossRef]
- DiNardo, C.D.; Cortes, J.E. Mutations in AML: Prognostic and therapeutic implications. Hematol. Am. Soc. Hematol. Educ. Program 2016, 2016, 348–355. [Google Scholar] [CrossRef]
- Makkar, H.; Majhi, R.K.; Goel, H.; Gupta, A.K.; Chopra, A.; Tanwar, P.; Seth, R. Acute myeloid leukemia: Novel mutations and their clinical implications. Am. J. Blood Res. 2023, 13, 12–27. [Google Scholar]
- Baldus, C.D.; Mrózek, K.; Marcucci, G.; Bloomfield, C.D. Clinical outcome of de novo acute myeloid leukaemia patients with normal cytogenetics is affected by molecular genetic alterations: A concise review. Br. J. Haematol. 2007, 137, 387–400. [Google Scholar] [CrossRef]
- Kennedy, V.E.; Smith, C.C. FLT3 Mutations in Acute Myeloid Leukemia: Key Concepts and Emerging Controversies. Front. Oncol. 2020, 10, 612880. [Google Scholar] [CrossRef]
- Li, S.; Li, N.; Chen, Y.; Zheng, Z.; Guo, Y. FLT3-TKD in the prognosis of patients with acute myeloid leukemia: A meta-analysis. Front. Oncol. 2023, 13, 1086846. [Google Scholar] [CrossRef]
- Aung, N.E.E.; Yamsri, S.; Teawtrakul, N.; Kamsaen, P.; Fucharoen, S. FLT3 Gene Mutations in Acute Myeloid Leukemia Patients in Northeast Thailand. Med. Sci. Monit. Basic Res. 2022, 28, e937446. [Google Scholar] [CrossRef]
- Sheikhha, M.H.; Awan, A.; Tobal, K.; Liu Yin, J.A. Prognostic significance of FLT3 ITD and D835 mutations in AML patients. Hematol. J. Off. J. Eur. Haematol. Assoc. 2003, 4, 41–46. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Kiyoi, H.; Nakano, Y.; Suzuki, R.; Kodera, Y.; Miyawaki, S.; Asou, N.; Kuriyama, K.; Yagasaki, F.; Shimazaki, C.; et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood 2001, 97, 2434–2439. [Google Scholar] [CrossRef] [PubMed]
- Gilliland, D.G.; Griffin, J.D. The roles of FLT3 in hematopoiesis and leukemia. Blood 2002, 100, 1532–1542. [Google Scholar] [CrossRef]
- Griffith, J.; Black, J.; Faerman, C.; Swenson, L.; Wynn, M.; Lu, F.; Lippke, J.; Saxena, K. The structural basis for autoinhibition of FLT3 by the juxtamembrane domain. Mol. Cell 2004, 13, 169–178. [Google Scholar] [CrossRef]
- Zorn, J.A.; Wang, Q.; Fujimura, E.; Barros, T.; Kuriyan, J. Crystal structure of the FLT3 kinase domain bound to the inhibitor Quizartinib (AC220). PLoS ONE 2015, 10, e0121177. [Google Scholar] [CrossRef]
- Stone, R.M.; DeAngelo, D.J.; Klimek, V.; Galinsky, I.; Estey, E.; Nimer, S.D.; Grandin, W.; Lebwohl, D.; Wang, Y.; Cohen, P.; et al. Patients with acute myeloid leukemia and an activating mutation in FLT3 respond to a small-molecule FLT3 tyrosine kinase inhibitor, PKC412. Blood 2005, 105, 54–60. [Google Scholar] [CrossRef]
- Ravandi, F.; Kantarjian, H.; Faderl, S.; Garcia-Manero, G.; O’Brien, S.; Koller, C.; Pierce, S.; Brandt, M.; Kennedy, D.; Cortes, J.; et al. Outcome of patients with FLT3-mutated acute myeloid leukemia in first relapse. Leuk. Res. 2010, 34, 752–756. [Google Scholar] [CrossRef]
- Nitika; Wei, J.; Hui, A.-M. Role of Biomarkers in FLT3 AML. Cancers 2022, 14, 1164. [Google Scholar] [CrossRef]
- Spiekermann, K.; Bagrintseva, K.; Schoch, C.; Haferlach, T.; Hiddemann, W.; Schnittger, S. A new and recurrent activating length mutation in exon 20 of the FLT3 gene in acute myeloid leukemia. Blood 2002, 100, 3423–3425. [Google Scholar] [CrossRef]
- Döhner, H.; Estey, E.H.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Burnett, A.K.; Dombret, H.; Fenaux, P.; Grimwade, D.; Larson, R.A.; et al. Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood 2010, 115, 453–474. [Google Scholar] [CrossRef]
- Friedman, R. The molecular mechanisms behind activation of FLT3 in acute myeloid leukemia and resistance to therapy by selective inhibitors. Biochim. Biophys. Acta BBA Rev. Cancer 2022, 1877, 188666. [Google Scholar] [CrossRef]
- Georgoulia, P.S.; Bjelic, S.; Friedman, R. Deciphering the molecular mechanism of FLT3 resistance mutations. FEBS J. 2020, 287, 3200–3220. [Google Scholar] [CrossRef]
- Scholl, S.; Fleischmann, M.; Schnetzke, U.; Heidel, F.H. Molecular Mechanisms of Resistance to FLT3 Inhibitors in Acute Myeloid Leukemia: Ongoing Challenges and Future Treatments. Cells 2020, 9, 2493. [Google Scholar] [CrossRef] [PubMed]
- Antar, A.I.; Otrock, Z.K.; Jabbour, E.; Mohty, M.; Bazarbachi, A. FLT3 inhibitors in acute myeloid leukemia: Ten frequently asked questions. Leukemia 2020, 34, 682–696. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cai, J.; Cheng, J.; Yang, W.; Zhu, Y.; Li, H.; Lu, T.; Chen, Y.; Lu, S. FLT3 Inhibitors in Acute Myeloid Leukemia: Challenges and Recent Developments in Overcoming Resistance. J. Med. Chem. 2021, 64, 2878–2900. [Google Scholar] [CrossRef] [PubMed]
- Daver, N.; Schlenk, R.F.; Russell, N.H.; Levis, M.J. Targeting FLT3 mutations in AML: Review of current knowledge and evidence. Leukemia 2019, 33, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Yalniz, F.; Abou Dalle, I.; Kantarjian, H.; Borthakur, G.; Kadia, T.; Patel, K.; Loghavi, S.; Garcia-Manero, G.; Sasaki, K.; Daver, N.; et al. Prognostic significance of baseline FLT3-ITD mutant allele level in acute myeloid leukemia treated with intensive chemotherapy with/without sorafenib. Am. J. Hematol. 2019, 94, 984–991. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Kim, H.W.; Lee, I.Y.; Lee, J.; Lee, J.; Jung, D.S.; Lee, S.Y.; Park, S.H.; Hwang, H.; Choi, J.-S.; et al. G-749, a novel FLT3 kinase inhibitor, can overcome drug resistance for the treatment of acute myeloid leukemia. Blood 2014, 123, 2209–2219. [Google Scholar] [CrossRef]
- Pon, J.R.; Marra, M.A. Driver and Passenger Mutations in Cancer. Annu. Rev. Pathol. Mech. Dis. 2015, 10, 25–50. [Google Scholar] [CrossRef]
- Bhattacharya, R.; Rose, P.W.; Burley, S.K.; Prlić, A. Impact of genetic variation on three dimensional structure and function of proteins. PLoS ONE 2017, 12, e0171355. [Google Scholar] [CrossRef]
- Dixit, A.; Yi, L.; Gowthaman, R.; Torkamani, A.; Schork, N.J.; Verkhivker, G.M. Sequence and structure signatures of cancer mutation hotspots in protein kinases. PLoS ONE 2009, 4, e7485. [Google Scholar] [CrossRef] [PubMed]
- Carow, C.E.; Levenstein, M.; Kaufmann, S.H.; Chen, J.; Amin, S.; Rockwell, P.; Witte, L.; Borowitz, M.J.; Civin, C.I.; Small, D. Expression of the hematopoietic growth factor receptor FLT3 (STK-1/Flk2) in human leukemias. Blood 1996, 87, 1089–1096. [Google Scholar] [CrossRef] [PubMed]
- Kuchenbauer, F.; Kern, W.; Schoch, C.; Kohlmann, A.; Hiddemann, W.; Haferlach, T.; Schnittger, S. Detailed analysis of FLT3 expression levels in acute myeloid leukemia. Haematologica 2005, 90, 1617–1625. [Google Scholar]
- Lilakos, K.; Viniou, N.A.; Mavrogianni, D.; Vassilakopoulos, T.P.; Dimopoulou, M.N.; Plata, E.; Angelopoulou, M.K.; Variami, E.; Stavrogianni, N.; Liapi, D.; et al. FLT3 overexpression in acute promyelocytic leukemia patients without detectable FLT3-ITD or codon 835-836 mutations: A pilot study. Anticancer Res. 2006, 26, 1201–1207. [Google Scholar] [PubMed]
- Ozeki, K.; Kiyoi, H.; Hirose, Y.; Iwai, M.; Ninomiya, M.; Kodera, Y.; Miyawaki, S.; Kuriyama, K.; Shimazaki, C.; Akiyama, H.; et al. Biologic and clinical significance of the FLT3 transcript level in acute myeloid leukemia. Blood 2004, 103, 1901–1908. [Google Scholar] [CrossRef]
- Armstrong, S.A.; Kung, A.L.; Mabon, M.E.; Silverman, L.B.; Stam, R.W.; Den Boer, M.L.; Pieters, R.; Kersey, J.H.; Sallan, S.E.; Fletcher, J.A.; et al. Inhibition of FLT3 in MLL. Validation of a therapeutic target identified by gene expression based classification. Cancer Cell 2003, 3, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Parcells, B.W.; Ikeda, A.K.; Simms-Waldrip, T.; Moore, T.B.; Sakamoto, K.M. FMS-like tyrosine kinase 3 in normal hematopoiesis and acute myeloid leukemia. Stem Cells 2006, 24, 1174–1184. [Google Scholar] [CrossRef] [PubMed]
- Ruvolo, P.P. GSK-3 as a novel prognostic indicator in leukemia. Adv. Biol. Regul. 2017, 65, 26–35. [Google Scholar] [CrossRef]
- Jiang, J.; Griffin, J.D. Wnt/β-catenin Pathway Modulates the Sensitivity of the Mutant FLT3 Receptor Kinase Inhibitors in a GSK-3β Dependent Manner. Genes Cancer 2010, 1, 164–176. [Google Scholar] [CrossRef]
- Xia, J.; Feng, S.; Zhou, J.; Zhang, L.; Shi, D.; Wang, M.; Zhu, Y.; Bu, C.; Xu, D.; Li, T. GSK3 inhibitor suppresses cell growth and metabolic process in FLT3-ITD leukemia cells. Med. Oncol. 2022, 40, 44. [Google Scholar] [CrossRef]
- Larrosa-Garcia, M.; Baer, M.R. FLT3 Inhibitors in Acute Myeloid Leukemia: Current Status and Future Directions. Mol. Cancer Ther. 2017, 16, 991–1001. [Google Scholar] [CrossRef]
- Smith, C.C.; Wang, Q.; Chin, C.S.; Salerno, S.; Damon, L.E.; Levis, M.J.; Perl, A.E.; Travers, K.J.; Wang, S.; Hunt, J.P.; et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature 2012, 485, 260–263. [Google Scholar] [CrossRef]
- Lee, C.-C.; Chuang, Y.-C.; Liu, Y.-L.; Yang, C.-N. A molecular dynamics simulation study for variant drug responses due to FMS-like tyrosine kinase 3 G697R mutation. RSC Adv. 2017, 7, 29871–29881. [Google Scholar] [CrossRef]
- Short, N.J.; Kantarjian, H.; Ravandi, F.; Daver, N. Emerging treatment paradigms with FLT3 inhibitors in acute myeloid leukemia. Ther. Adv. Hematol. 2019, 10, 2040620719827310. [Google Scholar] [CrossRef]
- Meshinchi, S.; Appelbaum, F.R. Structural and functional alterations of FLT3 in acute myeloid leukemia. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2009, 15, 4263–4269. [Google Scholar] [CrossRef] [PubMed]
- Carter, J.L.; Hege, K.; Yang, J.; Kalpage, H.A.; Su, Y.; Edwards, H.; Hüttemann, M.; Taub, J.W.; Ge, Y. Targeting multiple signaling pathways: The new approach to acute myeloid leukemia therapy. Signal Transduct. Target. Ther. 2020, 5, 288. [Google Scholar] [CrossRef] [PubMed]
- Manley, P.W.; Caravatti, G.; Furet, P.; Roesel, J.; Tran, P.; Wagner, T.; Wartmann, M. Comparison of the Kinase Profile of Midostaurin (Rydapt) with That of Its Predominant Metabolites and the Potential Relevance of Some Newly Identified Targets to Leukemia Therapy. Biochemistry 2018, 57, 5576–5590. [Google Scholar] [CrossRef] [PubMed]
- Stirewalt, D.L.; Meshinchi, S.; Kopecky, K.J.; Fan, W.; Pogosova-Agadjanyan, E.L.; Engel, J.H.; Cronk, M.R.; Dorcy, K.S.; McQuary, A.R.; Hockenbery, D.; et al. Identification of genes with abnormal expression changes in acute myeloid leukemia. Genes Chromosom. Cancer 2008, 47, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Gentles, A.J.; Plevritis, S.K.; Majeti, R.; Alizadeh, A.A. Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. JAMA 2010, 304, 2706–2715. [Google Scholar] [CrossRef]
- Haghi, A.; Mohammadi Kian, M.; Salemi, M.; Eghdami, M.R.; Nikbakht, M. The Question of Survival or Death: What Is the Role of Autophagy in Acute Myeloid Leukemia (AML)? Int. J. Hematol.-Oncol. Stem Cell Res. 2022, 16, 250–263. [Google Scholar] [CrossRef]
- Díaz-Beyá, M.; García-Fortes, M.; Valls, R.; Artigas, L.; Gómez-Casares, M.T.; Montesinos, P.; Sánchez-Guijo, F.; Coma, M.; Vendranes, M.; Martínez-López, J. A Systems Biology- and Machine Learning-Based Study to Unravel Potential Therapeutic Mechanisms of Midostaurin as a Multitarget Therapy on FLT3-Mutated AML. BioMedInformatics 2022, 2, 375–397. [Google Scholar] [CrossRef]
- Tang, G.; Cho, M.; Wang, X. OncoDB: An interactive online database for analysis of gene expression and viral infection in cancer. Nucleic Acids Res. 2022, 50, D1334–D1339. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Consortium, T.U. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2022, 51, D523–D531. [Google Scholar] [CrossRef] [PubMed]
- Sondka, Z.; Dhir, N.B.; Carvalho-Silva, D.; Jupe, S.; Madhumita; McLaren, K.; Starkey, M.; Ward, S.; Wilding, J.; Ahmed, M.; et al. COSMIC: A curated database of somatic variants and clinical data for cancer. Nucleic Acids Res. 2023, 52, D1210–D1217. [Google Scholar] [CrossRef] [PubMed]
- Richardson, C.J.; Gao, Q.; Mitsopoulous, C.; Zvelebil, M.; Pearl, L.H.; Pearl, F.M.G. MoKCa database--mutations of kinases in cancer. Nucleic Acids Res. 2009, 37, D824–D831. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sim, N.-L.; Kumar, P.; Hu, J.; Henikoff, S.; Schneider, G.; Ng, P.C. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012, 40, W452–W457. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 76, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Bendl, J.; Stourac, J.; Salanda, O.; Pavelka, A.; Wieben, E.D.; Zendulka, J.; Brezovsky, J.; Damborsky, J. PredictSNP: Robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Comput. Biol. 2014, 10, e1003440. [Google Scholar] [CrossRef]
- Calabrese, R.; Capriotti, E.; Fariselli, P.; Martelli, P.L.; Casadio, R. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum. Mutat. 2009, 30, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef] [PubMed]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Khanna, T.; Hanna, G.; Sternberg, M.J.E.; David, A. Missense3D-DB web catalogue: An atom-based analysis and repository of 4M human protein-coding genetic variants. Hum. Genet. 2021, 140, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Pennica, C.; Hanna, G.; Islam, S.A.; Sternberg, M.J.E.; David, A. Missense3D-PPI: A Web Resource to Predict the Impact of Missense Variants at Protein Interfaces Using 3D Structural Data. J. Mol. Biol. 2023, 435, 168060. [Google Scholar] [CrossRef]
- Rodrigues, C.H.; Pires, D.E.; Ascher, D.B. DynaMut: Predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018, 46, W350–W355. [Google Scholar] [CrossRef]
- Parthiban, V.; Gromiha, M.M.; Schomburg, D. CUPSAT: Prediction of protein stability upon point mutations. Nucleic Acids Res. 2006, 34, W239–W242. [Google Scholar] [CrossRef]




| Variants ID | Mutation | Mutation Type | SIFT | Polyphen-2 | PredictSNP | |
|---|---|---|---|---|---|---|
| Effect | Confidence | |||||
| RCV000444818 | Y572C | Substitution—Missense | 0 | 1.000 | Deleterious | 87% |
| RCV000445102 | V579A | Substitution—Missense | 0.01 | 0.551 | Neutral | 63% |
| RCV000420236 | Y591C | Substitution—Missense | 0 | 1.000 | Deleterious | 72% |
| RCV000441431 | Y591D | Substitution—Missense | 0 | 0.996 | Deleterious | 55% |
| RCV000435462 | V592A | Substitution—Missense | 0 | 0.742 | Neutral | 63% |
| RCV000432251 | F594L | Substitution—Missense | 0.48 | 0.999 | Neutral | 68% |
| RCV000437384 | G619C | Insertion—In frame | 0 | 1.000 | Deleterious | 87% |
| RCV000426662 | D651G | Substitution—Missense | 0.23 | 0.326 | Neutral | 83% |
| RCV000422333 | K663Q | Substitution—Missense | 0 | 0.981 | Deleterious | 51% |
| RCV000427705 | N676K | Substitution—Missense | 0 | 1.000 | Deleterious | 72% |
| RCV000443196 | I687F | Substitution—Missense | 0.05 | 0.010 | Neutral | 63% |
| RCV000420978 | F691I | Substitution—Missense | 0 | 0.881 | Deleterious | 61% |
| RCV000444069 | D835A | Substitution—Missense | 0 | 1.000 | Deleterious | 87% |
| RCV000424615 | D835E | Substitution—Missense | 0 | 0.959 | Deleterious | 87% |
| RCV000017663 | D835F | Substitution—Missense | 0 | 0.995 | Deleterious | 87% |
| RCV000017662 | D835H | Substitution—Missense | 0 | 1.000 | Deleterious | 72% |
| RCV000017663 | D835N | Substitution—Missense | 0 | 0.938 | Deleterious | 61% |
| RCV000017660 | D835V | Substitution—Missense | 0 | 0.999 | Deleterious | 87% |
| RCV000017665 | D835Y | Substitution—Missense | 0 | 1.000 | Deleterious | 87% |
| RCV000417837 | I836F | Substitution—Missense | 0 | 0.991 | Deleterious | 61% |
| RCV000432941 | I836L | Substitution—Missense | 0 | 0.204 | Neutral | 75% |
| RCV000422249 | I836M | Substitution—Missense | 0 | 1.000 | Deleterious | 61% |
| RCV000444162 | I836S | Substitution—Missense | 0 | 1.000 | Deleterious | 76% |
| RCV000428691 | I836V | Substitution—Missense | 0 | 0.230 | Neutral | 83% |
| RCV000429280 | D839G | Substitution—Missense | 0 | 0.880 | Neutral | 61% |
| RCV000440005 | N841H | Substitution—Missense | 0 | 0.022 | Deleterious | 51% |
| RCV000427616 | N841K | Substitution—Missense | 0.01 | 0.246 | Deleterious | 61% |
| RCV000421989 | Y842C | Substitution—Missense | 0 | 1.000 | Deleterious | 87% |
| RCV000431811 | Y842H | Substitution—Missense | 0 | 1.000 | Deleterious | 76% |
| Variants | MutPred2 | SNPs&GO | |||
|---|---|---|---|---|---|
| Score | Molecular Mechanisms | p-Values | Effect | Reliability Index | |
| Y572C | 0.66 | Altered transmembrane protein | 6.00 × 10−3 | Neutral | 2 |
| V579A | 0.37 | - | - | Neutral | 5 |
| Y591C | 0.69 | Altered ordered interface Loss of phosphorylation at Y591 Loss of sulfation at Y591 Altered transmembrane protein | 3.6 × 10−3 0.02 4.7 × 10−4 3.1 × 10−3 | Disease | 5 |
| Y591D | 0.85 | Altered ordered interface Gain of relative solvent accessibility Loss of phosphorylation at Y591 Loss of sulfation at Y591 Altered transmembrane protein | 9.0 × 10−3 0.01 0.02 4.7 × 10−4 3.9 × 10−3 | Disease | 5 |
| V592A | 0.27 | - | - | Neutral | 4 |
| F594L | 0.44 | - | - | Neutral | 1 |
| G619C | 0.91 | Loss of acetylation at K614 Gain of relative solvent accessibility Loss of methylation at K623 Altered transmembrane protein | 7.5 × 10−3 0.03 0.01 0.03 | Disease | 5 |
| D651G | 0.31 | - | - | Neutral | 3 |
| K663Q | 0.46 | - | - | Neutral | 7 |
| N676K | 0.89 | Gain of helix Altered transmembrane protein | 0.05 0.04 | Disease | 0 |
| I687F | 0.69 | Altered transmembrane protein | 0.02 | Neutral | 0 |
| F691I | 0.76 | - | - | Disease | 2 |
| D835A | 0.85 | Loss of relative solvent accessibility Altered ordered interface Loss of loop Loss of allosteric site at R834 Altered transmembrane protein Altered metal binding Altered DNA binding | 8.3 × 10−3 0.05 0.03 8.6 × 10−3 1.5 × 10−3 0.03 0.04 | Disease | 3 |
| D835E | 0.70 | Loss of allosteric site at R834 Loss of relative solvent accessibility Altered transmembrane protein Altered metal binding Altered DNA binding | 0.01 0.03 2.9 × 10−3 0.05 0.05 | Neutral | 0 |
| D835F | 0.9 | Loss of relative solvent accessibility Loss of allosteric site at R834 Altered ordered interface Altered transmembrane protein Altered metal binding Altered DNA binding | 4.1 × 10−3 5.3 × 10−3 0.04 1.1 × 10−3 0.03 0.04 | Disease | 7 |
| D835H | 0.87 | Altered metal binding Loss of relative solvent accessibility Altered ordered interface Loss of loop Loss of allosteric site at R834 Altered transmembrane protein Altered DNA binding | 0.01 0.01 0.04 0.04 9.9 × 10−3 1.3 × 10−3 0.04 | Disease | 4 |
| D835N | 0.75 | Altered ordered interface Loss of loop Loss of relative solvent accessibility Gain of allosteric site at R834 Altered transmembrane protein Altered metal binding Altered DNA binding | 0.02 0.03 0.03 8.5 × 10−3 2.6 × 10−3 0.05 0.04 | Disease | 0 |
| D835V | 0.87 | Loss of relative solvent accessibility Altered ordered interface Loss of loop Loss of allosteric site at R834 Altered transmembrane protein Altered metal binding Altered DNA binding | 4.3 × 10−3 0.03 0.02 8.5 × 10−3 7.7 × 10−4 0.03 0.04 | Disease | 6 |
| D835Y | 0.90 | Loss of relative solvent accessibility Loss of allosteric site at R834 Loss of loop Altered transmembrane protein Altered metal binding Altered DNA binding | 8.7 × 10−3 7.8 × 10−3 0.04 1.4 × 10−3 0.03 0.04 | Disease | 6 |
| I836F | 0.80 | Gain of allosteric site at R834 Gain of loop Altered transmembrane protein Gain of relative solvent accessibility Altered DNA binding Altered metal binding Gain of proteolytic cleavage at D835 | 3.3 × 10−4 0.02 1.5 × 10−3 0.04 0.04 0.02 0.04 | Disease | 2 |
| I836L | 0.54 | Gain of allosteric site at R834 Altered ordered interface Gain of relative solvent accessibility Altered transmembrane protein Altered metal binding Altered DNA binding Gain of proteolytic cleavage at D835 | 1.3 × 10−3 0.02 0.04 2.8 × 10−3 0.02 0.04 0.04 | Neutral | 4 |
| I836M | 0.63 | Gain of allosteric site at R834 Gain of relative solvent accessibility Altered ordered interface Altered transmembrane protein Altered metal binding Altered DNA binding | 4.0 × 10−3 0.03 0.05 2.7 × 10−3 0.05 0.04 | Neutral | 3 |
| I836S | 0.89 | Gain of relative solvent accessibility Altered ordered interface Gain of allosteric site at R834 Gain of loop Gain of B-factor Altered transmembrane protein Altered DNA binding Altered metal binding Gain of proteolytic cleavage at D835 Altered stability | 6.3 × 10−3 0.02 2.5 × 10−3 0.03 0.02 2.2 × 10−3 0.02 0.05 9.6 × 10−3 0.03 | Disease | 6 |
| I836V | 0.30 | - | - | Neutral | 8 |
| D839G | 0.871 | Altered ordered interface Loss of relative solvent accessibility Loss of allosteric site at R834 Altered transmembrane protein Altered metal binding Altered DNA binding | 0.03 0.02 6.3 × 10−3 1.5 × 10−3 0.04 0.05 | Disease | 3 |
| N841H | 0.453 | - | - | Neutral | 3 |
| N841K | 0.622 | Gain of acetylation at N841 Altered ordered interface Loss of relative solvent accessibility Altered transmembrane protein Altered metal binding Gain of ubiquitylation at N841 Altered stability | 9.8 × 10−4 0.04 0.04 2.0 × 10−3 0.05 0.02 0.03 | Neutral | 2 |
| Y842C | 0.859 | Altered ordered interface Altered transmembrane protein Loss of relative solvent accessibility Loss of strand Altered metal binding Gain of disulfide linkage at Y842 | 6.3 × 10−4 7.1 × 10−4 0.03 0.04 0.04 0.04 | Disease | 2 |
| Y842H | 0.823 | Altered ordered interface Gain of relative solvent accessibility Altered transmembrane protein Altered DNA binding Altered metal binding Altered stability | 1.2 × 10−3 0.01 1.2 × 10−3 0.03 0.03 0.03 | Disease | 7 |
| Variants | Missense3D | DynaMut-Predicted ΔΔG (kcal/mol) | CUPSAT-predicted ΔΔG (kcal/mol) |
|---|---|---|---|
| Y572C | Structural damage detected Cavity altered | −0.936 kcal/mol | 6.96 |
| V579A | Structural damage detected Buried H-bond breakage | −1.173 kcal/mol | 2.06 |
| Y591C | No structural damage detected | −0.984 kcal/mol | -1.0 |
| Y591D | No structural damage detected | −1.325 kcal/mol | 2.98 |
| V592A | No structural damage detected | −1.41 kcal/mol | 3.67 |
| F594L | Structural damage detected Buried H-bond breakage | −1.432 kcal/mol | 1.73 |
| G619C | Structural damage detected Buried Gly replaced Buried/exposed switch | 0.076 kcal/mol | -2.78 |
| D651G | No structural damage detected | −0.071 kcal/mol | - |
| K663Q | Structural damage detected Buried charge replaced | −0.287 kcal/mol | 0.25 |
| N676K | Structural damage detected Buried charge introduced | 0.736 kcal/mol | 3.28 |
| I687F | Structural damage detected Buried/exposed switch | 1.402 kcal/mol | 1.35 |
| F691I | No structural damage detected | −0.048 kcal/mol | 4.43 |
| D835A | No structural damage detected | −0.531 kcal/mol | 0.76 |
| D835E | No structural damage detected | −0.164 kcal/mol | 1.1 |
| D835F | No structural damage detected | 1.538 kcal/mol | −0.44 |
| D835H | No structural damage detected | 0.029 kcal/mol | 0.0 |
| D835N | No structural damage detected | −0.0 kcal/mol | 0.01 |
| D835V | No structural damage detected | 0.77 kcal/mol | −0.04 |
| D835Y | No structural damage detected | 0.041 kcal/mol | 0.75 |
| I836F | Structural damage detected Cavity altered | 1.105 kcal/mol | −0.29 |
| I836L | No structural damage detected | 0.137 kcal/mol | 0.6 |
| I836M | No structural damage detected | 0.429 kcal/mol | 3.17 |
| I836S | No structural damage detected | −1.449 kcal/mol | 2.44 |
| I836V | No structural damage detected | −0.183 kcal/mol | 1.55 |
| D839G | No structural damage detected | −0.922 kcal/mol | 4.06 |
| N841H | No structural damage detected | 1.904 kcal/mol | 1.45 |
| N841K | No structural damage detected | 1.669 kcal/mol | 1.79 |
| Y842C | No structural damage detected | −1.037 kcal/mol | 2.37 |
| Y842H | No structural damage detected | −1.224 kcal/mol | 4.99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mirza, Z.; Al-Saedi, D.A.; Alganmi, N.; Karim, S. Landscape of FLT3 Variations Associated with Structural and Functional Impact on Acute Myeloid Leukemia: A Computational Study. Int. J. Mol. Sci. 2024, 25, 3419. https://doi.org/10.3390/ijms25063419
Mirza Z, Al-Saedi DA, Alganmi N, Karim S. Landscape of FLT3 Variations Associated with Structural and Functional Impact on Acute Myeloid Leukemia: A Computational Study. International Journal of Molecular Sciences. 2024; 25(6):3419. https://doi.org/10.3390/ijms25063419
Chicago/Turabian StyleMirza, Zeenat, Dalal A. Al-Saedi, Nofe Alganmi, and Sajjad Karim. 2024. "Landscape of FLT3 Variations Associated with Structural and Functional Impact on Acute Myeloid Leukemia: A Computational Study" International Journal of Molecular Sciences 25, no. 6: 3419. https://doi.org/10.3390/ijms25063419
APA StyleMirza, Z., Al-Saedi, D. A., Alganmi, N., & Karim, S. (2024). Landscape of FLT3 Variations Associated with Structural and Functional Impact on Acute Myeloid Leukemia: A Computational Study. International Journal of Molecular Sciences, 25(6), 3419. https://doi.org/10.3390/ijms25063419





