Gold and Silver Nanoparticles as Biosensors: Characterization of Surface and Changes in the Adsorption of Leucine Dipeptide under the Influence of Substituent Changes
Abstract
1. Introduction
2. Results
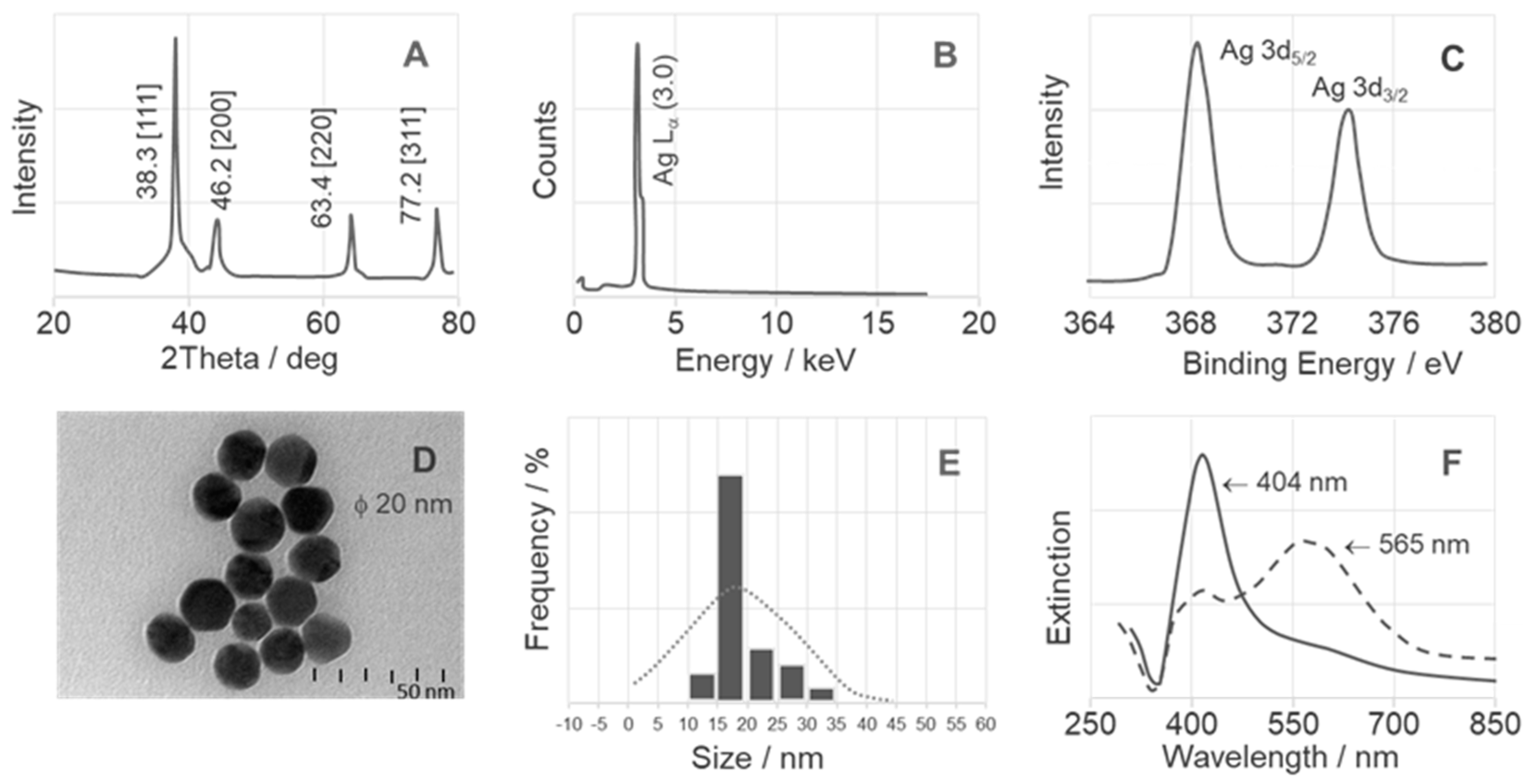
2.1. Surface Analysis of NPs
2.2. SERS Studies
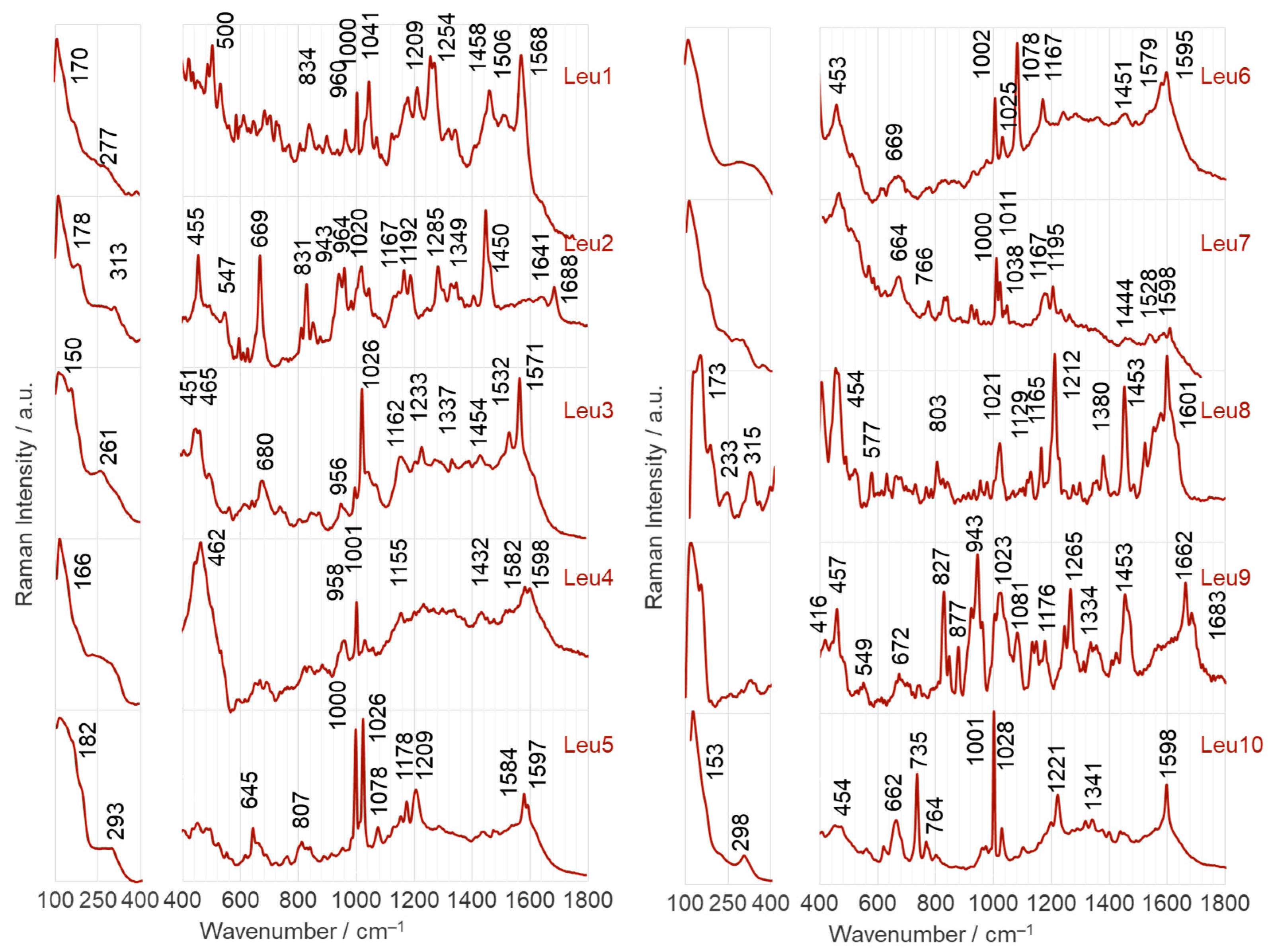
2.2.1. Adsorption on the Surface of AgNPs
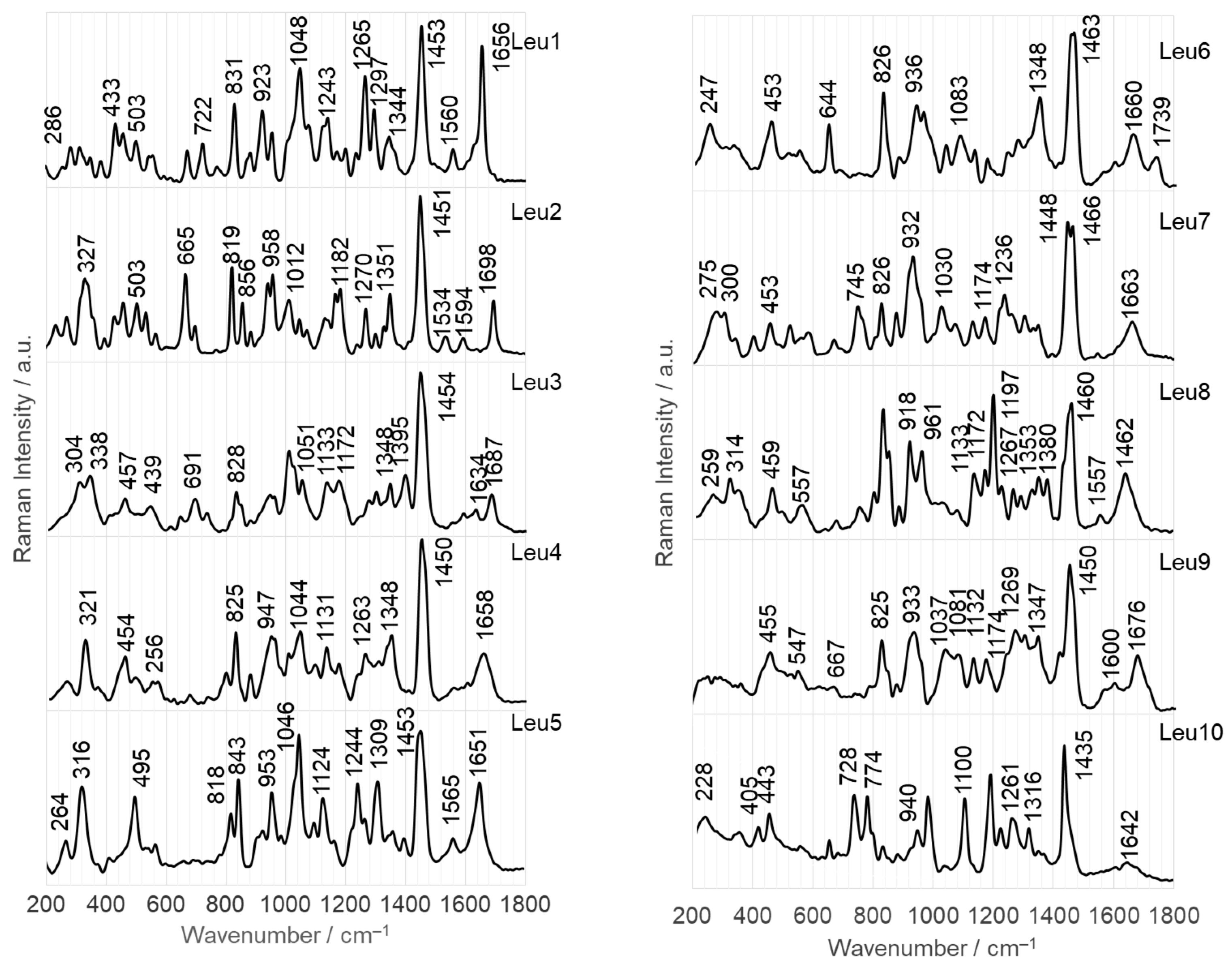
2.2.2. Adsorption at the Surface on AuNPs
3. Materials and Methods
3.1. Peptide Synthesis
3.2. Preparation of Silver Sol
3.3. Preparation of Gold Sol
3.4. Ultraviolet-Visible Spectroscopy (UV-Vis)
3.5. Dynamic Light Scattering (DLS)
3.6. X-ray Powder Diffraction (XRD)
3.7. X-ray Photoelectron (XPS)
3.8. Transmission Electron Microscopy with Energy-Dispersive X-ray Analysis (TEM-EDS)
3.9. Raman and Surface-Enhanced Raman (SERS)
3.10. Spectral Analysis and Graphics
4. Discussion
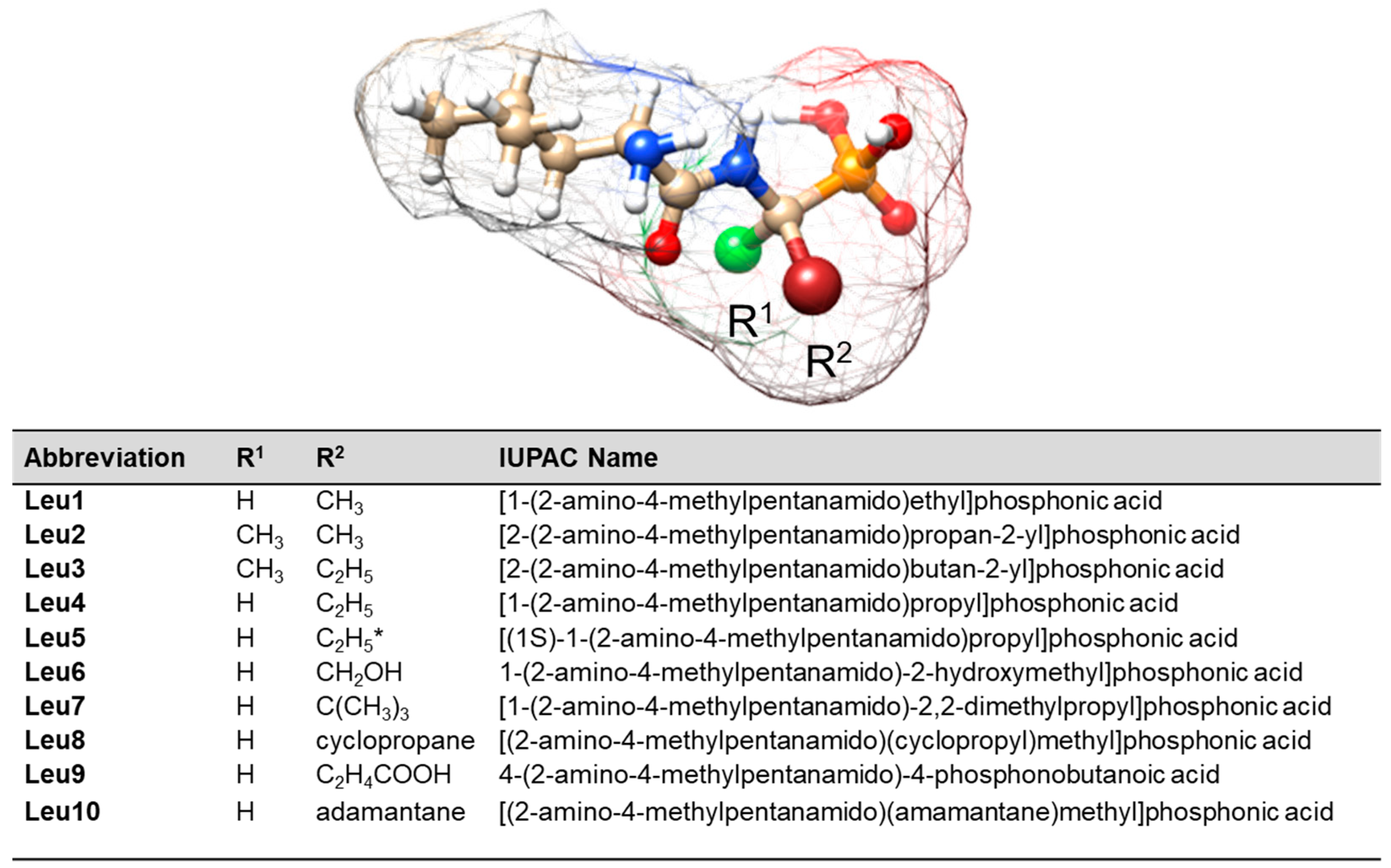
- Leu1 interacts with the AgNP surface through an amide bond, which is arranged to allow direct contact between the lone pair on the nitrogen and the surface. Phosphonate and amine groups are located in close proximity to the surface. The dipeptide is adsorbed onto the AuNP surface via a phosphonate group accompanied by the N(H)–Ca fragment of the peptide bond.
- Leu2 is adsorbed on AgNPs via N(H)–Ca(=O), while the PO3H− and C–NH2 groups are in close proximity. On the AuNP surface, the phosphonate group of Leu2 forms an O–H bond that is almost perpendicular to the nearby AuNP surface, and there is also a Ca=O bond in close proximity.
- The Cα(NH2)Ca(=O)Na(H) fragment of Leu3 is positioned on the AgNP surface, allowing for a weak interaction with the P=O group. In contrast, on the AuNP surface, the lone pair of electrons on the oxygen in the POH group interacts directly with AuNPs. This interaction occurs in a peptide orientation that allows for both an almost-vertical arrangement of the O–H bond and a Ca–N(H) interaction with AgNPs.
- The lone pairs of electrons on the =O atoms of the amide bond and the P=O bond of the Leu4 peptide make direct contact with the AgNPs. In addition, the amide oxygen is in contact with the surface of AuNPs, but is located at a certain distance from the surface, similar to the vertically arranged O–H bond of the phosphonate group.
- Similar to the case of Leu4 on AgNPs, both free electron pairs of electrons on the O= atoms of Leu5 are in contact with the AgNP surface. However, the Ca=O and P=O bonds are similarly tilted towards this surface. A lone pair of electrons on the oxygen of the P–O bond is also involved in the interactions between Leu5 and AgNPs. Leu5 is in contact with AuNPs via Ca–N and POH fragments. Similarly, the P–O and O–H bonds are tilted towards the AuNP surface.
- Leu6 interacts with the AgNP surface via the POH fragment and amide bond. The bands of C–OH and NH2 groups indicate their proximity to AgNPs. In contrast, the interaction between AuNPs and Leu6 is caused by the vertical P–O bond at a certain distance from the AuNP surface, as well as the Ca(=O)Na(H) fragment.
- The Leu7 fragment Cα(N)Ca(=O)NaCP(=O) interacts with the surface of AgNPs, while the vertical P–O bond mainly adsorbs onto AuNPs.
- The lone electron pair of the amide nitrogen is in contact with the AgNP surface, allowing the Na–H and Ca=O bonds to be similarly tilted to this surface and the amino and phosphonate groups to be located near it. The CaNH fragment of Leu8 is also responsible for the interaction of this dipeptide with AuNPs. Regarding the surface, the interaction of Leu8 with the vertically arranged P–O bond is weak.
- The contact between Leu9 and the AgNP surface is mainly due to the R2 fragment, with support from the amide, P=O, and POH groups. In contrast, Leu9 adsorbs onto AuNPs mainly with P=O, POH (with a more or less vertical arrangement of the O-H bond), and Ca=O.
- The Leu10 fragment, specifically Cα(N)Ca(=O)Na, with weak involvement of the POH moiety, is accountable for the adsorption of Leu10 onto AgNPs. Additionally, the free electron pair on the oxygen of the vertically arranged P–O bond is primarily responsible for the peptide’s interactions with the surface of AuNP. The CaNaH fragment aids in this interaction.
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviation
References
- Definition of Nanomaterial—European Observatory for Nanomaterials (europa.eu). European Chemicals Agency: Helsinki, Finland. Available online: https://euon.echa.europa.eu/definition-of-nanomaterial (accessed on 24 March 2024).
- Joudeh, N.; Linke, D. Review: Nanoparticle classification, physicochemical properties, characterization, and applications: A comprehensive review for biologists. J. Nanobiotechnol. 2022, 20, 262. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Saeed, K.; Khan, I. Nanoparticles: Properties, applications and Toxicities. Arab. J. Chem. 2019, 12, 908–931. [Google Scholar] [CrossRef]
- Talebian, S.; Rodrigues, T.; das Neves, J.; Sarmento, B.; Langer, R.; Conde, J. Facts and Figures on Materials Science and Nanotechnology Progress and Investment. ACS Nano 2021, 15, 15940–15952. [Google Scholar] [CrossRef] [PubMed]
- Malik, S.; Muhmmad, K.; Waheed, Y. Review: Nanotechnology: A Revolution in Modern Industry. Molecules 2023, 28, 661. [Google Scholar] [CrossRef] [PubMed]
- Jeevanandam, J.; Barhoum, A.; Chan, Y.S.; Dufresne, A.; Danquah, M.K. Review on nanoparticles and nanostruc-tured materials: History, sources, toxicity and regulations. Beilstein J. Nanotechnol. 2018, 9, 1050–1074. [Google Scholar] [CrossRef] [PubMed]
- Hossain, N.; Mobarak, H.; Mimona, M.A.; Islam, A.; Hossain, A.; Zohura, F.T.; Chowdhury, M.A. Advances and significances of nanoparticles in semiconductor applications—A review. Results Eng. 2023, 19, 10. [Google Scholar] [CrossRef]
- Haleem, A.; Javaid, M.; Singh, R.P.; Rab, S.; Suman, R. Applications of nanotechnology in medical field: A brief review. Glob. Health J. 2023, 7, 70–77. [Google Scholar] [CrossRef]
- Beni, A.A.; Jabbari, H. Nanomaterials for Environmental Applications. Results Eng. 2022, 15, 100467. [Google Scholar] [CrossRef]
- Altammar, K.A. A review on nanoparticles: Characteristics, synthesis, applications, and challenges. Front. Microbiol. 2023, 14, 100467. [Google Scholar] [CrossRef]
- Alexander, J.W. History of medicinal use of silver. Surg. Infect. 2009, 10, 289–292. [Google Scholar] [CrossRef]
- Paladini, F.; Pollini, M. Antimicrobial silver nanoparticles for wound healing application: Progress and future trends. Materials 2019, 12, 2540. [Google Scholar] [CrossRef] [PubMed]
- Biagioni, B.; Cavicchioli, M.; Massabni, A. Silver complexes for tuberculosis treatment: A short review. Quimica Nova 2022, 45, 83–88. [Google Scholar] [CrossRef]
- Jose, P.P.; Vivekanandan, V.; Sobhanakumari, K. Gonorrhea: Historical outlook. History 2020, 2, 110–114. [Google Scholar] [CrossRef]
- Chu, C.-S.; McManus, A.T.; Pruit, B.A.; Mason, A.D. Therapeutic Effects of Silver Nylon Dressings with Weak Direct Current on Pseudomonas aeruginosa-lnfected Burn Wounds. J. Trauma 1988, 28, 1488–1492. [Google Scholar] [CrossRef] [PubMed]
- Medici, S.; Peana, M.F.; Nurchi, V.M.; Zoroddu, M.A. Medical uses of silver: History, myths, and scientific evidence. J. Med. Chem. 2019, 62, 5923–5943. [Google Scholar] [CrossRef]
- Taufikurohmah, T.; Soepardjo, D.; Armadianto, H.; Rusmini, R. Synthesis and Characterization of Nanogold and Nanosilver as Leprosy Drug Candidates and Their Activity Tests in Leprosy Patients; Case Study. In Proceedings of the Mathematics, Informatics, Science, and Education International Conference, Surabaya, Indonesia, 28 September 2019. [Google Scholar] [CrossRef]
- Bruna, T.; Maldonado-Bravo, F.; Jara, P.; Caro, N. Silver Nanoparticles and Their Antibacterial Applications. Int. J. Mol. Sci. 2021, 22, 7202. [Google Scholar] [CrossRef] [PubMed]
- Matras, E.; Gorczyca, A.; Przemieniecki, S.W.; Oćwieja, M. Surface properties-dependent antifungal activity of silver nanoparticles. Sci. Rep. 2022, 12, 18046. [Google Scholar] [CrossRef]
- Luceri, A.; Francese, R.; Lembo, D.; Ferraris, M.; Balagna, C. Review: Silver Nanoparticles: Review of Antiviral Properties, Mechanism of Action and Applications. Microorganisms 2023, 11, 629. [Google Scholar] [CrossRef] [PubMed]
- Anik, M.I.; Mahmud, N.; Al Masud, A.; Hasan, M. Gold nanoparticles (GNPs) in biomedical and clinical applications: A review. Nano Sel. 2021, 3, 792–828. [Google Scholar] [CrossRef]
- Zhan, X.; Yan, J.; Tang, H.; Xia, D.; Lin, H. Antibacterial Properties of Gold Nanoparticles in the Modification of Medical Implants: A Systematic Review. Pharmaceuticals 2022, 14, 2654. [Google Scholar] [CrossRef]
- Madkhali, O.A. Review: A comprehensive review on potential applications of metallic nanoparticles as antifungal therapies to combat human fungal diseases. Saudi Pharm. J. 2023, 31, 101733. [Google Scholar] [CrossRef] [PubMed]
- Basova, T.V.; Vikulova, E.S.; Dorovskikh, S.I.; Hassan, A.; Morozova, N.B. The use of noble metal coatings and nanoparticles for the modification of medical implant materials. Mater. Des. 2021, 204, 109672. [Google Scholar] [CrossRef]
- Alamer, F.A.; Beyari, R.F. Overview of the Influence of Silver, Gold, and Titanium Nanoparticles on the Physical Properties of PEDOT:PSS-Coated Cotton Fabrics. Nanomaterials 2022, 12, 1609. [Google Scholar] [CrossRef] [PubMed]
- Neethu, S.; Midhun, S.J.; Radhakrishnan, E.; Jyothis, M. Surface functionalization of central venous catheter with mycofabricated silver nanoparticles and its antibiofilm activity on multidrug resistant Acinetobacter baumannii. Microb. Pathog. 2020, 138, 103832. [Google Scholar] [CrossRef] [PubMed]
- Couto, C.; Almeida, A. Metallic Nanoparticles in the Food Sector: A Mini-Review. Foods 2022, 11, 402. [Google Scholar] [CrossRef] [PubMed]
- Pulit-Prociak, J.; Grabowska, A.; Chwastowski, J.; Majka, T.M.; Banach, M. Safety of the application of nanosilver and nanogold in topical cosmetic preparations. Colloids Surf. B 2019, 183, 110416. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Castañón, G.A.; Niño-Martínez, N.; Martínez-Gutierrez, F.; Martínez-Mendoza, J.R.; Ruiz, F. Synthesis and antibacterial activity of silver nanoparticles with different sizes. J. Nanoparticle Res. 2008, 10, 1343–1348. [Google Scholar] [CrossRef]
- Xiu, Z.-M.; Zhang, Q.-B.; Puppala, H.L.; Colvin, V.L.; Alvarez, P.J.J. Negligible particle-specific antibacterial activity of silver nanoparticles. Nano Lett. 2012, 12, 4271–4275. [Google Scholar] [CrossRef]
- Budama-Kilinc, Y.; Cakir-Koc, R.; Zorlu, T.; Ozdemir, B.; Karavelioglu, Z.; Egil, A.C.; Kecel-Gunduz, S. Assessment of Nano-toxicity and Safety Profiles of Silver Nanoparticles. In Silver Nanoparticles—Fabrication, Characterization and Applications; Maaz, K., Ed.; IntechOpen: London, UK, 2018; Volume 10, pp. 185–208. [Google Scholar] [CrossRef]
- Gratton, S.E.; Ropp, P.A.; Pohlhaus, P.D.; Luft, J.C.; Madden, V.J.; Napier, M.E.; DeSimone, J.M. The effect of particle design on cellular internalization pathways. Proc. Natl. Acad. Sci. USA 2008, 105, 11613–11618. [Google Scholar] [CrossRef]
- Alkilany, A.M.; Murphy, C.J. Toxicity and cellular uptake of gold nanoparticles: What we have learned so far? J. Nanoparticle Res. 2010, 12, 2313–2333. [Google Scholar] [CrossRef]
- Chithrani, B.D.; Ghazani, A.A.; Chan, W.C.W. Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells. Nano Lett. 2006, 6, 662–668. [Google Scholar] [CrossRef] [PubMed]
- El Badawy, A.M.; Silva, R.G.; Morris, B.; Scheckel, K.G.; Suidan, M.T.; Tolaymat, T.M. Surface charge-dependent toxicity of silver nanoparticles. Environ. Sci. Technol. 2011, 45, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Van Phu, D.; Quoc, L.A.; Duy, N.N.; Lan, N.T.K.; Du, B.D.; Luan, L.Q.; Hien, N.Q. Study on antibacterial activity of silver nanoparticles synthesized by gamma irradiation method using different stabilizers. Nanoscale Res. Lett. 2014, 9, 162. [Google Scholar] [CrossRef] [PubMed]
- Suresh, A.K.; Pelletier, D.A.; Wang, W.; Morrell-Falvey, J.L.; Gu, B.; Doktycz, M.J. Cytotoxicity induced by engi-neered silver nanocrystallites is dependent on surface coatings and cell types. Langmuir 2012, 28, 2727–2735. [Google Scholar] [CrossRef] [PubMed]
- Akter, M.; Sikder, M.T.; Rahman, M.M.; Ullah, A.K.M.A.; Hossain, K.F.B.; Banik, S.; Hosokawa, T.; Saito, T.; Kurasaki, M. A systematic review on silver nanoparticles-induced cytotoxicity: Physicochemical properties and perspectives. J. Adv. Res. 2018, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Bojic, D.; Liu, M. Review: Applications and safety of gold nanoparticles as therapeutic devices in clinical trials. J. Pharmaceut. Anal. 2023, 13, 960–967. [Google Scholar] [CrossRef]
- Pem, B.; Pongrac, I.M.; Ulm, L.; Pavičić, I.; Vrček, V.; Jurašin, D.D.; Ljubojević, M.; Krivohlavek, A.; Vrček, I.V. Toxicity and safety study of silver and gold nanoparticles functionalized with cysteine and glutathione. Beilstein J. Nanotechnol. 2019, 10, 1802–1817. [Google Scholar] [CrossRef] [PubMed]
- Shooshtari, M.; Vollebregt, S.; Vaseghi, Y.; Rajati, M.; Pahlavan, S. The sensitivity enhancement of TiO2-based VOCs sensor decorated by gold at room temperature. Nanotechnology 2023, 34, 255501. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.C.; Kuo, C.C. Enhanced dual function of Ag nanoparticles decorated one-dimensional polymorphic TiO2 composites for sustainable environmental applications. J. Sci.-Adv. Mater. Dev. 2024, 9, 100696. [Google Scholar] [CrossRef]
- Li, J.-F.; Zhang, Y.-J.; Ding, S.-Y.; Panneerselvam, R.; Tian, Z.-Q. Core−Shell Nanoparticle-Enhanced Raman Spectroscopy. Chem. Rev. 2017, 117, 5002–5069. [Google Scholar] [CrossRef]
- Singh, R.; Bhateria, R. Core-shell nanostructures: A simplest two-component system with enhanced properties and multiple applications. Environ. Geochem. Health 2021, 43, 2459–2482. [Google Scholar] [CrossRef] [PubMed]
- Woźniak-Budych, M.J.; Staszek, K.; Staszek, M. Copper and Copper-Based Nanoparticles in Medicine—Perspectives and Challenges. Molecules 2023, 28, 6687. [Google Scholar] [CrossRef]
- Jiang, J.; Pi, J.; Cai, J. Review: The Advancing of Zinc Oxide Nanoparticles for Biomedical Applications. Bioinorg. Chem. Appl. 2018, 2018, 1062562. [Google Scholar] [CrossRef] [PubMed]
- Ziental, D.; Czarczynska-Goslinska, B.; Mlynarczyk, D.T.; Glowacka-Sobotta, A.; Stanisz, B.; Goslinski, T.; Sobotta, L. Titanium Dioxide Nanoparticles: Prospects and Applications in Medicine. Nanomaterials 2020, 10, 387. [Google Scholar] [CrossRef] [PubMed]
- Fahmy, H.M.; El-Hakim, M.H.; Nady, D.S.; Elkaramany, Y.; Mohamed, F.A.; Yasien, A.M.; Moustafa, M.A.; Elmsery, B.E.; Yousef, H.A. Review on MgO nanoparticles multifunctional role in the biomedical field: Properties and applications. Nanomed. J. 2022, 9, 1–14. [Google Scholar] [CrossRef]
- Schneider, M.G.M.; Martín, M.J.; Otarola, J.; Vakarelska, E.; Simeonov, V.; Lassalle, V.; Nedyalkova, M. Biomedical Applications of Iron Oxide Nanoparticles: Current Insights Progress and Perspectives. Pharmaceutics 2022, 14, 204. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.R.; Al Mamun, M.S.; Ara, M.H. Review on platinum nanoparticles: Synthesis, characterization, and applications. Microchem. J. 2021, 171, 106840. [Google Scholar] [CrossRef]
- Nagchowdhury, P.; Patra, C.R. Tin-based nanomaterials for biomedical applications. Nanomedicine 2023, 18, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Shahbazi, N.; Zare-Dorabei, R.; Naghib, S.M. Multifunctional nanoparticles as optical biosensing probe for breast cancer detection: A review. Mater. Sci. Eng. C 2021, 127, 112249. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, N.; Jamaluddin, N.D.; Tan, L.L.; Yusof, N.Y.M. A Review on the Development of Gold and Silver Nano-particles-Based Biosensor as a Detection Strategy of Emerging and Pathogenic RNA Virus. Sensors 2021, 21, 5114. [Google Scholar] [CrossRef]
- Yuan, K.; Jurado-Sánchez, B.; Escarpa, A. Nanomaterials meet surface-enhanced Raman scattering towards en-hanced clinical diagnosis: A review. J. Nanobiotechnology 2022, 20, 537. [Google Scholar] [CrossRef] [PubMed]
- Puntmann, V.O. How-to guide on biomarkers: Biomarker definitions, validation and applications with examples from cardiovascular disease. Postgrad. Med. J. 2009, 85, 538–545. [Google Scholar] [CrossRef] [PubMed]
- Tilea, I.; Varga, A.; Serban, R.C. Review: Past, Present, and Future of Blood Biomarkers for the Diagnosis of Acute Myocardial Infarction—Promises and Challenges. Diagnostics 2021, 11, 881. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yang, Y.; Zhang, B.; Lin, X.; Fu, X.; An, Y.; Zou, Y.; Wang, J.-X.; Wang, Z.; Yu, T. Review: Lactate metabolism in human health and disease. Signal Transduct. Target Ther. 2022, 7, 305. [Google Scholar] [CrossRef] [PubMed]
- Alba-Patiño, A.; Vaquer, A.; Barón, E.; Russell, S.M.; Borges, M.; de la Rica, R. Micro- and nanosensors for detecting blood pathogens and biomarkers at different points of sepsis care. Microchim. Acta 2022, 189, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Loonen, A.J.M.; de Jager, C.P.C.; Tosserams, J.; Kusters, R.; Hilbink, M.; Wever, P.C.; van den Brule, A.J.C. Bi-omarkers and Molecular Analysis to Improve Bloodstream Infection Diagnostics in an Emergency Care Unit. PLoS ONE 2014, 9, e87315. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Cai, Y.; Yao, H.; Lin Ch Xie, Y.; Tang, S.; Zhang, A. Review: Small molecule metabolites: Discovery of biomarkers and therapeutic targets. Signal Transduct. Target Ther. 2023, 8, 132. [Google Scholar] [CrossRef] [PubMed]
- Chiu, F.-Y.; Yen, Y. Review: Imaging biomarkers for clinical applications in neuro-oncology: Current status and future Perspectives. Biomarker Res. 2023, 11, 35. [Google Scholar] [CrossRef] [PubMed]
- Korczowska-Łącka, I.; Hurła, M.; Banaszek, N.; Kobylarek, D.; Szymanowicz, O.; Kozubski, W.; Dorszewska, J. Selected Biomarkers of Oxidative Stress and Energy Metabolism Disorders in Neurological Diseases. Mol. Neurobiol. 2023, 60, 4132–4149. [Google Scholar] [CrossRef] [PubMed]
- Ealdman, S.A.; Terzic, A. Molecular Diagnostics. Clin. Transl. Sci. 2009, 2, 6–8. [Google Scholar] [CrossRef]
- Wieszczycka, K.; Staszak, K.; Wozniak-Budych, M.J.; Litowczenko, J.; Maciejewska, B.M.; Jurga, S. Review: Surface functionalization—The way for advanced applications of smart materials. Co-ord. Chem. Rev. 2021, 436, 213846. [Google Scholar] [CrossRef]
- Rezić, I.; Meštrović, E. Review: Characterization of Nanoparticles in Antimicrobial Coatings for Medical Applications—A Review. Coatings 2023, 13, 1830. [Google Scholar] [CrossRef]
- Konoeda, H.; Yang, H.; Yang Ch Gower, A.; Xu Ch Zhang, W.; Liu, M. Protein Kinase C-delta Inhibitor Peptide Formulation using Gold Nanoparticles. J. Vis. Exp. 2019, 145, e58741. [Google Scholar] [CrossRef]
- Jia, C.; Yang, K.-W.; Liu, C.-C.; Feng, L.; Xiao, J.-M.; Zhou, L.-S.; Zhang, Y.-L. Synthesis, characterization and activity of new phosphonate dipeptides as potential inhibitors of VanX. Bioorganic Med. Chem. Lett. 2012, 22, 482–484. [Google Scholar] [CrossRef]
- Lejczak, B.; Kafarski, P.; Sztajer, H.; Mastalerz, P. Antibacterial activity of phosphono dipeptides related to alafosfalin. J. Med. Chem. 1986, 29, 2212–2217. [Google Scholar] [CrossRef] [PubMed]
- Galezowska, J.; Gumienna-Kontecka, E. Phosphonates, their complexes and bio-applications: A spectrum of surprising diversity. Co-ord. Chem. Rev. 2012, 256, 105–124. [Google Scholar] [CrossRef]
- Leucine: Everything You Need to Know. Available online: https://www.verywellhealth.com/leucine-uses-benefits-side-effects-dosage-7567617 (accessed on 3 February 2024).
- Van Loon, L.J. Review: Leucine as a pharmaconutrient in health and disease. Curr. Opin. Clin. Nutr. Metab. Care 2012, 15, 71–77. [Google Scholar] [CrossRef]
- Rehman, S.U.; Ali, R.; Zhang, H.; Zafar, M.H.; Wang, M. Research progress in the role and mechanism of Leucine in regulating animal growth and development. Front. Physiol. 2023, 14, 1252089. [Google Scholar] [CrossRef] [PubMed]
- Kalogeropoulou, D.; LaFave, L.; Schweim, K.; Gannon, M.C.; Nuttall, F.Q. Leucine, when ingested with glucose, synergistically stimulates insulin secretion and lowers blood glucose. Metab. Clin. Exper. 2008, 57, 1747–1752. [Google Scholar] [CrossRef]
- Chen, J.; Ou, Y.; Luo, R.; Wang, J.; Wang, D.; Guan, J.; Li, Y.; Xia, P.; Chen, P.R.; Liu, Y. SAR1B senses leucine levels to regulate mTORC1 signalling. Nature 2021, 596, 281–303. [Google Scholar] [CrossRef]
- Wolfson, R.L.; Chantranupong, L.; Saxton, R.A.; Shen, K.; Scaria, S.M.; Cantor, J.R.; Sabatini, D.M. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 2016, 351, 6268. [Google Scholar] [CrossRef]
- Wang, B.; Liu, Y.; Ai, C.; Chu, R.; Chen, M.; Ye, H.; Wang, H.; Zhou, F. Highly sensitive SERS detection in a non-volatile liquid-phase system with nanocluster-patterned optical fiber SERS probes. Opt. Express 2022, 30, 15846. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Panwar, N.; Yap, S.H.K.; Wu, Q.; Zeng, S.; Xu, J.; Tjin, S.C.; Song, J.; Qu, J.; Yong, K.-T. Review: SERS-based ultrasensitive sensing platform: An insight into design and practical applications. Conduct. Chem. Rev. 2017, 337, 1–33. [Google Scholar] [CrossRef]
- Surface Analysis. Available online: https://www.sciencedirect.com/topics/chemistry/surface-analysis (accessed on 15 January 2024).
- Kosa, S.A.; Zaheer, Z. Biogenic fabrication of silver nanoparticles, oxidative dissolution and antimicrobial activities J. Saudi Chem. Soc. 2022, 26, 101414. [Google Scholar] [CrossRef]
- Syed, B.; Prasad, N.; Dhananjaya, B.L.; Yallappa, S.; Satish, S. Synthesis of silver nanoparticles by endosymbiont Pseudomonas fluorescens CA 417 and their bactericidal activity. Enzym. Microb. Technol. 2016, 95, 128–136. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, X.; Xin, Z.; Deng, M.; Wen, Y.; Song, Y. Synthesis of monodisperse silver nanoparticles for ink-jet printed flexible electronics. Nanotechnology 2011, 22, 425601. [Google Scholar] [CrossRef] [PubMed]
- Agnihotri, S.; Mukherji, S.; Mukherji, S. Size-controlled silver nanoparticles synthesized over the range 5–100 nm using the same protocol and their antibacterial efficacy. RSC Adv. 2014, 4, 3974. [Google Scholar] [CrossRef]
- Ramalingam, K.; Devasena, T.; Senthil, B.; Kalpana, R.; Jayavel, R. Silver nanoparticles for melamine detection in milk based on transmitted light intensity. IET Sci. Meas. Technol. 2017, 11, 171–178. [Google Scholar] [CrossRef]
- Unser, S.; Bruzas, I.; He, J.; Sagle, L. Localized Surface Plasmon Resonance Biosensing: Current Challenges and Approaches. Sensors 2015, 15, 15684–15716. [Google Scholar] [CrossRef]
- He, Y.Q.; Liu, S.P.; Kong, L.; Liu, Z.F. A study on the sizes and concentrations of gold nanoparticles by spectra of absorption, resonance Rayleigh scattering and resonance non-linear scattering. Spectrochim. Acta A 2005, 61, 2861–2866. [Google Scholar] [CrossRef]
- Debnath, D.; Kim, S.H.; Geckeler, K.E. The first solid-phase route to fabricate and size-tune gold nanoparticles at room temperature. J. Mater. Chem. 2009, 19, 8810–8816. [Google Scholar] [CrossRef]
- Thanh, N.T.K.; Rosenzweig, Z. Development of an Aggregation-Based Immunoassay for Anti-Protein A Using Gold Nanoparticles. Anal. Chem. 2002, 74, 1624–1628. [Google Scholar] [CrossRef] [PubMed]
- Long, N.V.; Ohtaki, M.; Yuasa, M.; Yoshida, S.; Kuragaki, T.; Thi, C.M.; Nogami, M. Synthesis and Self-Assembly of Gold Nanoparticles by Chemically Modified Polyol Methods under Experimental Control. J. Nanomater. 2013, 2013, 793125. [Google Scholar] [CrossRef]
- Krishnamurthy, S.; Esterle, A.; Sharma, N.C.; Sahi, S.V. Yucca-derived synthesis of gold nanomaterial and their catalytic potential. Nanoscale Res. Lett. 2014, 9, 627. [Google Scholar] [CrossRef] [PubMed]
- Yakimchuk, D.V.; Bundyukova, V.D.; Ustarroz, J.; Terryn, H.; Baert, K.; Kozlovskiy, A.L.; Zdorovets, M.V.; Khubezhov, S.A.; Trukhanov, A.V.; Trukhanov, S.V.; et al. Morphology and Microstructure Evolution of Gold Nanostructures in the Limited Volume Porous Matrices. Sensors 2020, 20, 4397. [Google Scholar] [CrossRef] [PubMed]
- Podstawka, E.; Kafarski, P.; Proniewicz, L.M. Effect of an aliphatic spacer group on the adsorption mechanism of phosphonodipeptides containing N-terminal glycine on the colloidal silver surface. J. Raman Spectrosc. 2008, 39, 1396–1407. [Google Scholar] [CrossRef]
- Podstawka, E.; Andrzejak, M.; Kafarski, P.; Proniewicz, L.M. Comparison of adsorption mechanism on colloidal silver surface of alafosfalin and its analogues. J. Raman Spectrosc. 2008, 39, 1238–1249. [Google Scholar] [CrossRef]
- Podstawka-Proniewicz, E.; Andrzejak, M.; Kafarski, P.; Kim, Y.; Proniewicz, L.M. Vibrational Characterization of L-Valine Phosphonate Dipeptides: FT-IR, FT-RS, and SERS Spectroscopy Studies and DFT Calculations. J. Raman Spectrosc. 2011, 42, 958–979. [Google Scholar] [CrossRef]
- Podstawka, E.; Ozaki, Y.; Proniewicz, L.M. Part II: Surface enhanced Raman spectroscopy investigation of hetero-dipeptides containing methionine adsorbed on colloidal silver. Appl. Spectrosc. 2004, 58, 581–590. [Google Scholar] [CrossRef]
- Podstawka, E.; Ozaki, Y.; Proniewicz, L.M. Part I: Surface enhanced Raman spectroscopy investigation of amino acids and their homodipeptides adsorbed on colloidal silver. Appl. Spectrosc. 2004, 58, 570–580. [Google Scholar] [CrossRef]
- Podstawka, E.; Ozaki, Y.; Proniewicz, L.M. Part III: Surface-enhanced Raman scattering of amino acids and their homodipeptides monolayers deposited onto colloidal gold surface. Appl. Spectrosc. 2005, 59, 1516–1526. [Google Scholar] [CrossRef] [PubMed]
- Khoury, Y.E.; Hielscher, R.; Voicescu, M.; Gross, J.; Hellwig, P. On the specificity of the amide VI band for the secondary structure of proteins. Vib. Spectrosc. 2011, 55, 258–266. [Google Scholar] [CrossRef]
- Sexton, B.A.; Madix, R.J. Vibrational spectra of molecular and atomic oxygen on Ag(100). Chem. Phys. Lett. 1980, 76, 294–297. [Google Scholar] [CrossRef]
- Kudzin, Z.; Depczyński, R.; Andrijewski, G.; Drabowicz, J.; Łuczak, J. 1-(N-Acylamino)alkanephosphonates. Part IV. N-Acylation of 1-Aminoalkanephosphonic Acids. Polish J. Chem. 2005, 79, 499–513. [Google Scholar]


| Assignment | Leu1 | Leu2 | Leu3 | Leu4 | Leu5 | Leu6 | Leu7 | Leu8 | Leu9 | Leu10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Ag–molecule | 216 | 210 | 223 | |||||||
| Ag–O | 338 | 350 | 326 | 369 | 363 | |||||
| Ag–N | 496 | |||||||||
| Ca(=O)NaCP | 544 | |||||||||
| Amide VI, ρoopb(Ca=O) | 572 | 570 | 562 | 587 | 584 | 578 | 584 | 560 | 571 | 573 |
| δ(NaC(P)C), δoop(CaNa(H)C), ν(CP) | 641 | |||||||||
| AmideIV, ρipb(Ca=O) | 686 | 692 | ||||||||
| AmideV, ρoopb(NH) | 740 | 741 | 745 | 747 | 738 | 738 | ||||
| ν(PO), ρr/ρt(CC(H,Y)C) | 771 | 780 | ||||||||
| ν(PO) | 815 | 804 | 797 | 810 | ||||||
| ρr(CαNH2), ρt(C–P=O), ν(CN) | 851 | 861 | 841 | |||||||
| δoop(CαNH2), ρt(CPO3H−), ρt(CαCa(=O)Na | 894 | 896 | 899 | 898 | 874 | 900 | ||||
| ν(PO) + ρb(POH)//ν(C–COOH) | 928 | 928 | 930 | 929 | 930 | 914 | 910 | 938 | ||
| δoop(CαNH2) | 952 | 963 | 962 | |||||||
| δ(CPO3H−), δoop(CαNH2), ν(CαN) | 979 | 973 | 983 | 983 | 982 | 971 | 980 | |||
| ν(PO), ρb(POH) | 1000 | 1001 | ||||||||
| ρb(POH), ν(CαN), ν(CCα) | 1026 | 1054 | 1029 | 1053 | ||||||
| δoop(CαNH2), ρr(CCH3)L, ν(CC)L, ρt/ρr(CH2Cα(H,N)Ca | 1082 | 1075 | 1070 | 1069 | 1082 | |||||
| ν(NaC), ρr(CC(Y)C), δ(NaC(P)C), ν(CC), ρr(CCH3) | 1105 | 1108 | 1120 | |||||||
| ρr(CαNH2), ν(CCα), ν(CN), ρr (Cα(H,N)Ca) | 1141 | 1147 | ||||||||
| ν(P=O) | 1168 | 1172 | 1161 | 1160 | 1191 | |||||
| AmideIII, ρipb(NaH) + ν(CaNa) | 1221 | 1224 | 1226 | 1218 | ||||||
| ν(CaNa), ν(CNa) | 1244 | |||||||||
| ν(P=O) | 1260 | 1269 | 1253 | 1251 | 1262 | 1256 | 1270 | 1263 | ||
| ν(CaNa)//ν(CO), ρb(COH) | 1285 | 1289 | 1286 | 1284 | 1290 | 1287 | 1302 | |||
| ρr(CC(H,C)Cβ(H2)Cα(H,N)Ca) | 1336 | 1338 | 1345 | 1347 | ||||||
| δ/ρr(CCH3)//ρw/ρt(CCH2O), ρb(COH) | 1432 | 1456 | 1447 | |||||||
| δ/ρr(CH3) | 1481 | 1482 | 1484 | 1480 | 1481 | 1474 | 1475 | |||
| AmideII, ρipb(NaH) + ν(CaNa) | 1580 | 1566 | 1580 | 1580 | 1550 | 1574 | ||||
| δ(NH2), ρb(C-NH2) | 1642 | 1632 | 1606 | 1604 | 1610 | 1634 | 1632 | 1601 | ||
| AmideI, ν(Ca=O) | 1671 | 1675 | 1675 | 1670 | 1666 | 1674 | ||||
| ν(COOH) | 1725 |
| Assignment | Leu1 | Leu2 | Leu3 | Leu4 | Leu5 | Leu6 | Leu7 | Leu8 | Leu9 | Leu10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Ag–molecule | 170 | 178 | 150 | 166 | 182 | 173 | 153 | |||
| Ag–O | 277 | 313 | 261 | 293 | 233,315 | 320 | 298 | |||
| Ag–N, Ca(=O)NaCP | 500 | 455 | 458 | 462 | 453 | 451 | 405,454 | 457 | 454 | |
| Amide VI, ρoopb(Ca=O) | 547 | 549 | ||||||||
| δ(NaC(P)C), δoop(CaNa(H)C), ν(CP) | 645 | |||||||||
| AmideIV, ρipb(Ca=O) | 669 | 680 | 669 | 664 | 672 | 662 | ||||
| AmideV, ρoopb(NH) | 766 | 735,764 | ||||||||
| ρr(CαNH2), ρt(C–P=O), ν(CN) | 834 | 831 | 827 | |||||||
| δoop(CαNH2), ρt(CPO3H−), ρt(CαCa(=O)Na | 877 | |||||||||
| δoop(CαNH2)//ν(C–COOH) | 943 | 956 | 958 | 943 | ||||||
| δ(CPO3H−), δoop(CαNH2), ν(CαN) | 960 | 964 | ||||||||
| ν(PO), ρb(POH) | 1000 | 1001 | 1000 | 1001 | 1000 | 1002 | 1000 | 1001 | 1002 | 1001 |
| ρb(POH) | 1041 | 1020 | 1026 | 1030 | 1026 | 1025 | 1011 | 1021 | 1023 | 1028 |
| δoop(CαNH2), ρr(CCH3)L, ν(CC)L, ρt/ρr(CH2Cα(H,N)Ca | 1078 | 1078 | 1081 | |||||||
| ρr(CαNH2), ν(CCα), ν(CN), ρr(Cα(H,N)Ca), ν(P=O | 1162 | 1167 | 1162 | 1155 | 1167 | 1167 | 1165 | 1176 | ||
| AmideIII, ρipb(NaH) + ν(CaNa) | 1209 | 1192 | 1233 | 1209 | 1195 | 1212 | 1221 | |||
| ν(P=O) | 1254 | 1265 | ||||||||
| ν(CaNa)//ν(CO), ρb(COH) | 1285 | |||||||||
| ρr(CC(H,C)Cβ(H2)Cα(H,N)Ca) | 1349 | 1337 | 1334 | 1341 | ||||||
| δ/ρr(CCH3)//ρw/ρt(CCH2O), ρb(COH) | 1458 | 1450 | 1454 | 1432 | 1451 | 1444 | 1453 | 1453 | ||
| δ/ρr(CH3)L, δ(CCH3)L, δ(CC(H2)Cα), ρr(Cα(H,N)Ca) | 1506 | 1532 | 1528 | 1528 | ||||||
| AmideII, ρipb(NaH) + ν(CaNa) | 1568 | 1588 | 1571 | 1582 | 1584 | 1579 | 1581 | |||
| δ(NH2), ρb(C-NH2) | 1598 | 1597 | 1595 | 1598 | 1601 | 1598 | ||||
| AmideI, ν(Ca=O) | 1641 | 1662 | ||||||||
| ν(COOH) | 1683 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Proniewicz, E. Gold and Silver Nanoparticles as Biosensors: Characterization of Surface and Changes in the Adsorption of Leucine Dipeptide under the Influence of Substituent Changes. Int. J. Mol. Sci. 2024, 25, 3720. https://doi.org/10.3390/ijms25073720
Proniewicz E. Gold and Silver Nanoparticles as Biosensors: Characterization of Surface and Changes in the Adsorption of Leucine Dipeptide under the Influence of Substituent Changes. International Journal of Molecular Sciences. 2024; 25(7):3720. https://doi.org/10.3390/ijms25073720
Chicago/Turabian StyleProniewicz, Edyta. 2024. "Gold and Silver Nanoparticles as Biosensors: Characterization of Surface and Changes in the Adsorption of Leucine Dipeptide under the Influence of Substituent Changes" International Journal of Molecular Sciences 25, no. 7: 3720. https://doi.org/10.3390/ijms25073720
APA StyleProniewicz, E. (2024). Gold and Silver Nanoparticles as Biosensors: Characterization of Surface and Changes in the Adsorption of Leucine Dipeptide under the Influence of Substituent Changes. International Journal of Molecular Sciences, 25(7), 3720. https://doi.org/10.3390/ijms25073720





