Optimization of Enzymatic and Chemical Decellularization of Native Porcine Heart Valves for the Generation of Decellularized Xenografts
Abstract
1. Introduction
2. Results
2.1. Visual Appearance
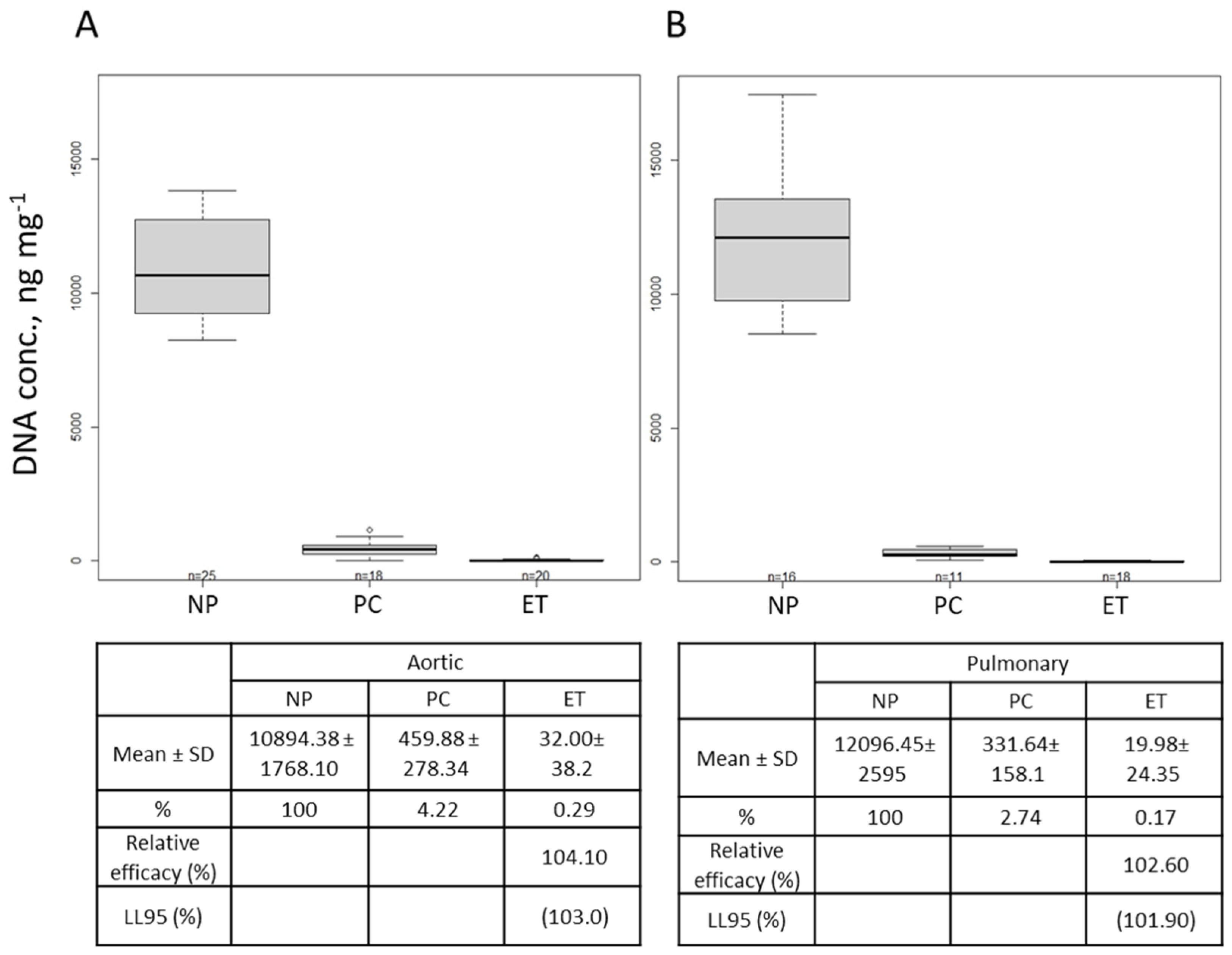
2.2. DNA Quantification
2.3. Biomechanical Test
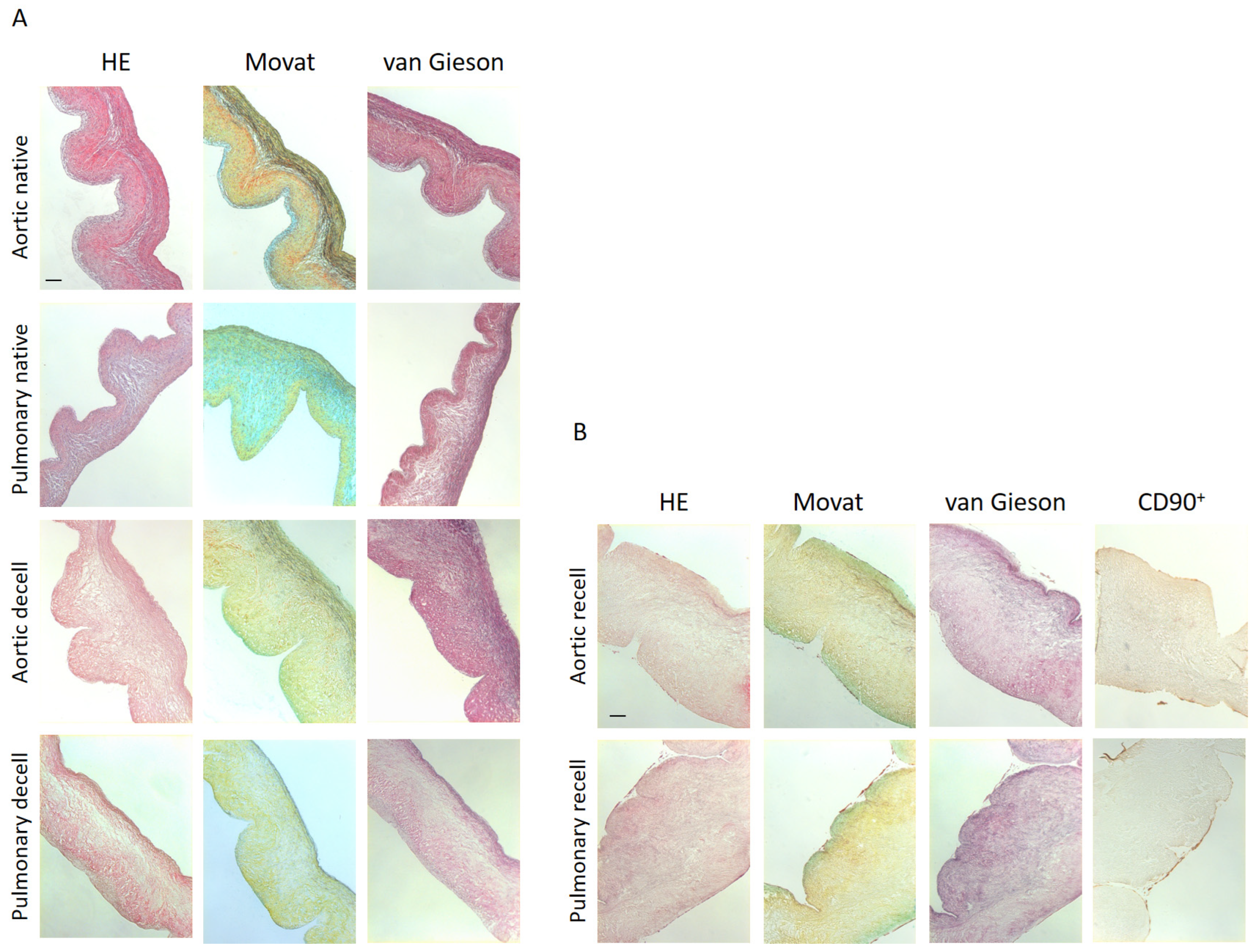
2.4. Histology and Immunohistochemistry Analysis
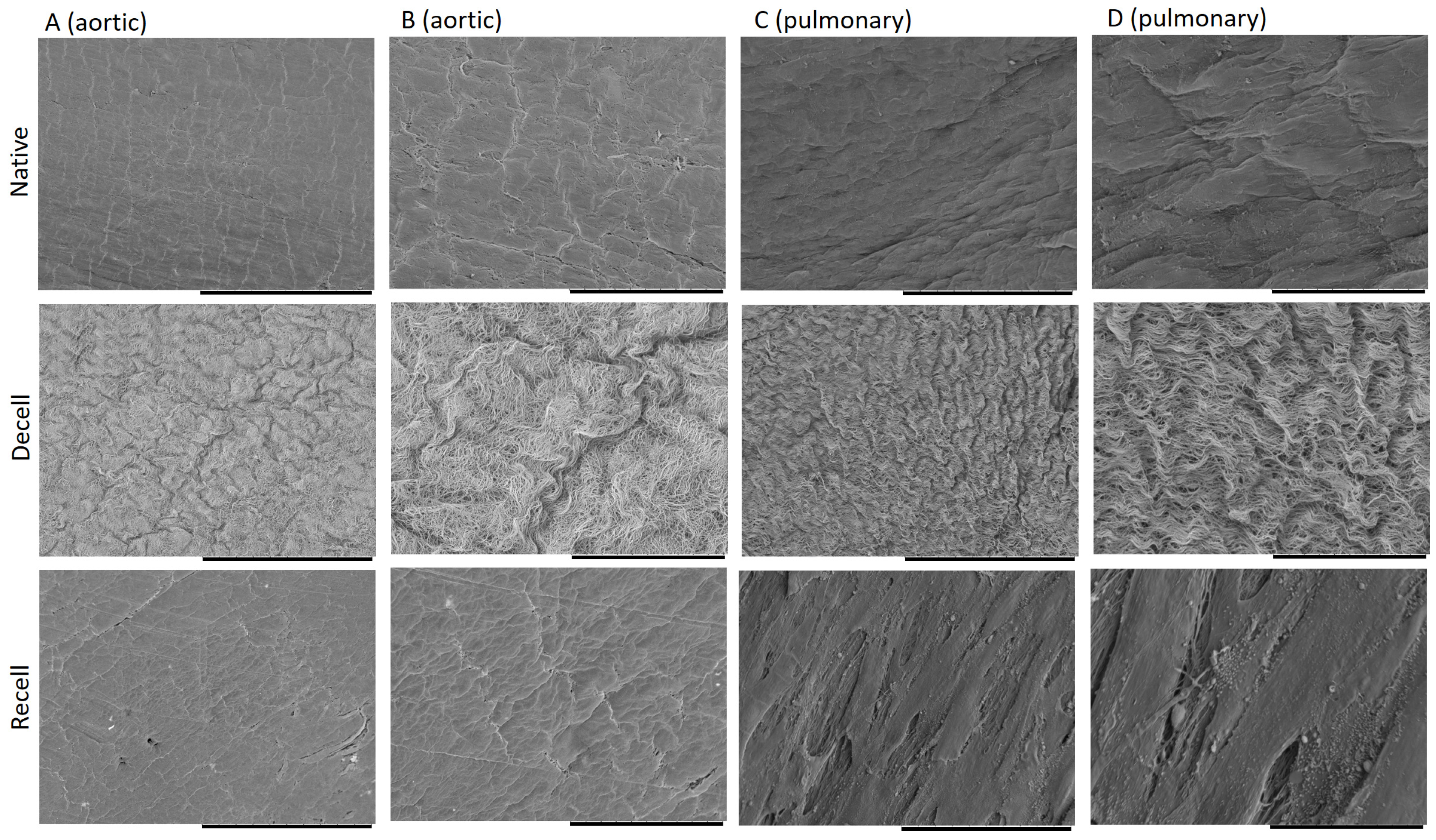
2.5. Scanning Electron Microscopy and ECM Topography
2.6. Morphological Results of the Valved Stent
3. Discussion
4. Materials and Methods
4.1. Valve Stent Preparation and Methods for Decellularization Process
4.2. Home Decellularization Method
4.3. Modified Decellularization Method
4.4. DNA Isolation and Quantification
4.5. Histology and Immunohistochemistry
4.5.1. HE Staining
4.5.2. Movat Pentachrome
4.5.3. Elastica Van Gieson
4.5.4. Immunohistochemistry: CD90+ Antibody Staining
4.6. Scanning Electron Microscopy (SEM)
4.7. Recellularization
4.8. Biomechanical Test
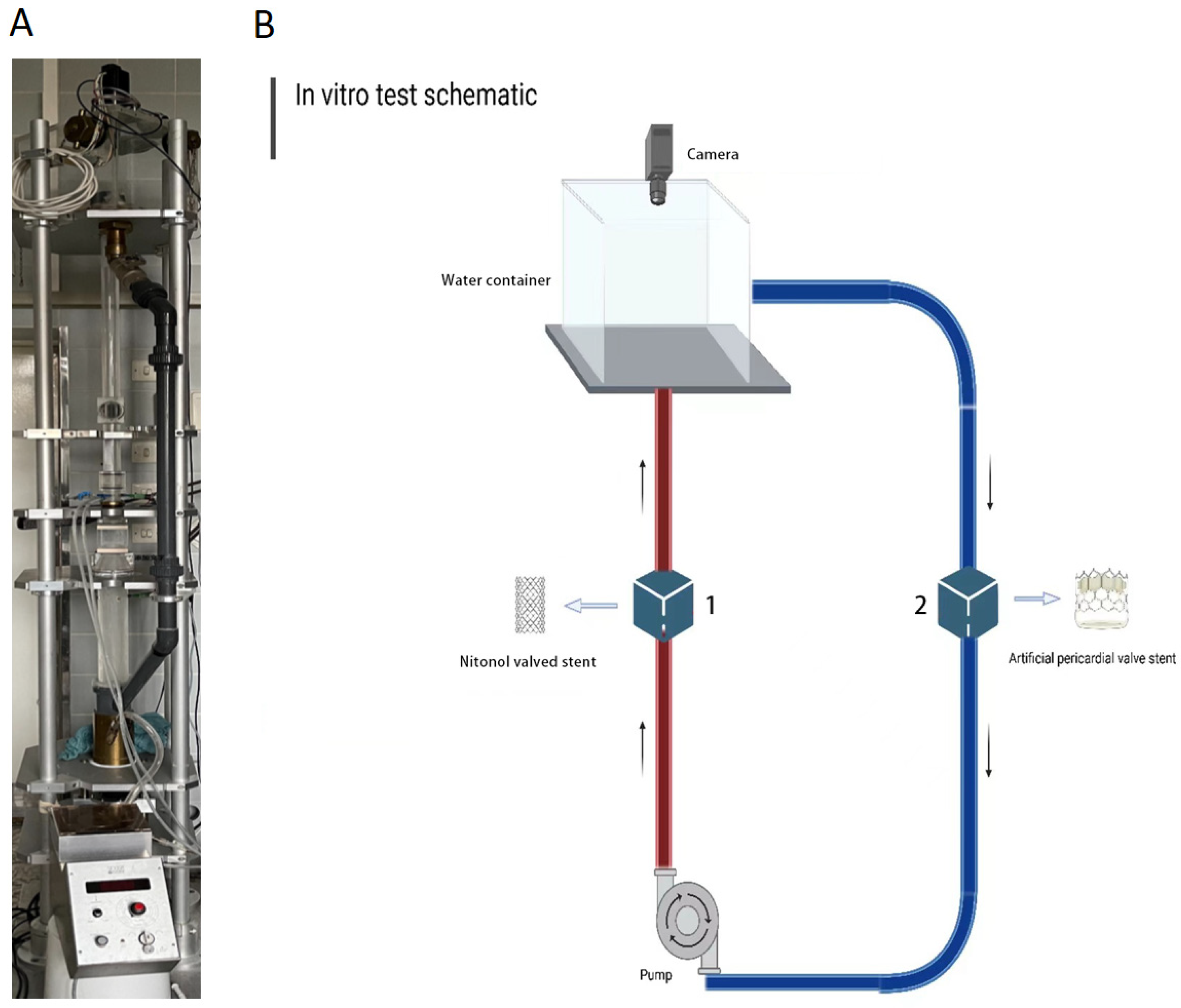
4.9. Heart Valve Test
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roudaut, R.; Serri, K.; Lafitte, S. Thrombosis of prosthetic heart valves: Diagnosis and therapeutic considerations. Heart 2007, 93, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Dangas, G.D.; Weitz, J.I.; Giustino, G.; Makkar, R.; Mehran, R. Prosthetic Heart Valve Thrombosis. J. Am. Coll. Cardiol. 2016, 68, 2670–2689. [Google Scholar] [CrossRef] [PubMed]
- Faggioli, M.; Moro, A.; Butt, S.; Todesco, M.; Sandrin, D.; Borile, G.; Bagno, A.; Fabozzo, A.; Romanato, F.; Marchesan, M.; et al. A new decellularization protocol of porcine aortic valves using tergitol to characterize the scaffold with the biocompatibility profile using human bone marrow mesenchymal stem cells. Polymers 2022, 14, 1226. [Google Scholar] [CrossRef]
- Haupt, J.; Lutter, G.; Gorb, S.N.; Simionescu, D.T.; Frank, D.; Seiler, J.; Paur, A.; Haben, I. Detergent-based decellularization strategy preserves macro- and microstructure of heart valves. Interact. Cardiovasc. Thorac. Surg. 2018, 26, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Neishabouri, A.; Soltani Khaboushan, A.; Daghigh, F.; Kajbafzadeh, A.-M.; Majidi Zolbin, M. Decellularization in tissue engineering and regenerative medicine: Evaluation, modification, and application methods. Front. Bioeng. Biotechnol. 2022, 10, 805299. [Google Scholar] [CrossRef] [PubMed]
- Soltani Khaboushan, A.; Shakibaei, M.; Kajbafzadeh, A.M.; Majidi Zolbin, M. Prenatal neural tube anomalies: A decade of intrauterine stem cell transplantation using advanced tissue engineering methods. Stem Cell Rev. Rep. 2022, 18, 752–767. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, T.W.; Sellaro, T.L.; Badylak, S.F. Decellularization of tissues and organs. Biomaterials 2006, 27, 3675–3683. [Google Scholar] [CrossRef] [PubMed]
- Crapo, P.M.; Gilbert, T.W.; Badylak, S.F. An overview of tissue and whole organ decellularization processes. Biomaterials 2011, 32, 3233–3243. [Google Scholar] [CrossRef] [PubMed]
- Jana, S.; Tefft, B.J.; Spoon, D.B.; Simari, R.D. Scaffolds for tissue engineering of cardiac valves. Acta Biomater. 2014, 10, 2877–2893. [Google Scholar] [CrossRef]
- Simionescu, D.T.; Chen, J.; Jaeggli, M.; Wang, B.; Liao, J. Form follows function: Advances in trilayered structure replication for aortic heart valve tissue engineering. J. Healthc. Eng. 2012, 3, 179–202. [Google Scholar] [CrossRef]
- van Steenberghe, M.; Schubert, T.; Gerelli, S.; Bouzin, C.; Guiot, Y.; Xhema, D.; Bollen, X.; Abdelhamid, K.; Gianello, P. Porcine pulmonary valve decellularization with NaOH-based vs detergent process: Preliminary in vitro and in vivo assessments. J. Cardiothorac. Surg. 2018, 13, 34. [Google Scholar] [CrossRef] [PubMed]
- Bozso, S.J.; Kang, J.J.H.; EL-Andari, R.; Fialka, N.; Zhu, L.F.; Meyer, S.R.; Freed, D.H.; Nagendran, Jayan, Nagendran, Jeevan. Recellularization of xenograft heart valves reduces the xenoreactive immune response in an in vivo rat model. Eur. J. Cardiothorac. Surg. 2022, 61, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Guruswamy Damodaran, R.; Vermette, P. Tissue and organ decellularization in regenerative medicine. Biotechnol. Prog. 2018, 34, 1494–1505. [Google Scholar] [CrossRef] [PubMed]
- Dijkman, P.E.; Driessen-Mol, A.; Frese, L.; Hoerstrup, S.P.; Baaijens, F.P.T. Decellularized homologous tissue-engineered heart valves as off-the-shelf alternatives to xeno- and homografts. Biomaterials 2012, 33, 4545–4554. [Google Scholar] [CrossRef] [PubMed]
- Rieder, E.; Kasimir, M.T.; Silberhumer, G.; Seebacher, G.; Wolner, E.; Simon, P.; Weigel, G. Decellularization protocols of porcine heart valves differ importantly in efficiency of cell removal and susceptibility of the matrix to recellularization with human vascular cells. J. Thorac. Cardiovasc. Surg. 2004, 127, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Deborde, C.; Simionescu, D.T.; Wright, C.; Liao, J.; Sierad, L.N.; Simionescu, A. Stabilized collagen and elastin-based scaffolds for mitral valve tissue engineering. Tissue Eng. Part A 2016, 22, 1241–1251. [Google Scholar] [CrossRef]
- Luo, Y.; Lou, D.; Ma, L.; Gao, C. Optimizing detergent concentration and processing time to balance the decellularization efficiency and properties of bioprosthetic heart valves. J. Biomed. Mater. Res. Part A 2019, 107, 2235–2243. [Google Scholar] [CrossRef]
- Wu, L.C.; Kuo, Y.J.; Sun, F.W.; Chen, C.H.; Chiang, C.J.; Weng, P.W.; Tsuang, Y.H.; Huang, Y.Y. Optimized decellularization protocol including α-Gal epitope reduction for fabrication of an acellular porcine annulus fibrosus scaffold. Cell Tissue Bank 2017, 18, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Lutter, G.; Puehler, T.; Cyganek, L.; Seiler, J.; Rogler, A.; Herberth, T.; Knueppel, P.; Gorb, S.N.; Sathananthan, J.; Sellers, S.; et al. Biodegradable poly-ε-caprolactone scaffolds with ECFCs and iMSCs for tissue-engineered heart valves. Int. J. Mol. Sci. 2022, 23, 527. [Google Scholar] [CrossRef]
- Chen, J.-H.; Simmons, C.A. Cell-matrix interactions in the pathobiology of calcific aortic valve disease: Critical roles for matricellular, matricrine, and matrix mechanics cues. Circ. Res. 2011, 108, 1510–1524. [Google Scholar] [CrossRef]
- Gumpangseth, T.; Lekawanvijit, S.; Mahakkanukrauh, P. Histological assessment of the human heart valves and its relationship with age. Anat. Cell Biol. 2020, 53, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Joyce, E.M.; Sacks, M.S. Effects of decellularization on the mechanical and structural properties of the porcine aortic valve leaflet. Biomaterials 2008, 29, 1065–1074. [Google Scholar] [CrossRef] [PubMed]
- Blum, K.M.; Drews, J.D.; Breuer, C.K. Tissue-engineered heart valves: A call for mechanistic studies. Tissue Eng. Part B Rev. 2018, 24, 240–253. [Google Scholar] [CrossRef] [PubMed]
- Whelan, A.; Williams, E.; Fitzpatrick, E.; Murphy, B.P.; Gunning, P.S.; O’Reilly, D.; Lally, C. Collagen fibre-mediated mechanical damage increases calcification of bovine pericardium for use in bioprosthetic heart valves. Acta Biomater. 2021, 128, 384–392. [Google Scholar] [CrossRef]
- Yang, M.; Chen, C.-Z.; Wang, X.-N.; Zhu, Y.-B.; Gu, Y.J. Favorable effects of the detergent and enzyme extraction method for preparing decellularized bovine pericardium scaffold for tissue engineered heart valves. J. Biomed. Mater. Res. B Appl. Biomater. 2009, 91, 354–361. [Google Scholar] [CrossRef]
- Schenke-Layland, K.; Vasilevski, O.; Opitz, F.; König, K.; Riemann, I.; Halbhuber, K.J.; Wahlers, T.; Stock, U.A. Impact of decellularization of xenogeneic tissue on extracellular matrix integrity for tissue engineering of heart valves. J. Struct. Biol. 2003, 143, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Sierad, L.N.; Shaw, E.L.; Bina, A.; Brazile, B.; Rierson, N.; Patnaik, S.S.; Kennamer, A.; Odum, R.; Cotoi, O.; Terezia, P.; et al. Functional heart valve scaffolds obtained by complete decellularization of porcine aortic roots in a novel differential pressure gradient perfusion system. Tissue Eng. Part C Methods 2015, 21, 1284–1296. [Google Scholar] [CrossRef]
- He, M.; Callanan, A.; Lagaras, K.; Steele, J.A.M.; Stevens, M.M. Optimization of SDS exposure on preservation of ECM characteristics in whole organ decellularization of rat kidneys. J. Biomed. Mater. Res. B Appl. Biomater. 2017, 105, 1352–1360. [Google Scholar] [CrossRef] [PubMed]
- Poornejad, N.; Momtahan, N.; Salehi, A.S.M.; Scott, D.R.; Fronk, C.A.; Roeder, B.L.; Reynolds, P.R.; Bundy, B.C.; Cook, A.D. Efficient decellularization of whole porcine kidneys improves reseeded cell behavior. Biomed. Mater. 2016, 11, 025003. [Google Scholar] [CrossRef]
- Xu, K.; Kuntz, L.A.; Foehr, P.; Kuempel, K.; Wagner, A.; Tuebel, J.; Deimling, C.V.; Burgkart, R.H. Efficient decellularization for tissue engineering of the tendon-bone interface with preservation of biomechanics. PLoS ONE 2017, 12, e0171577. [Google Scholar] [CrossRef]
- Gilbert, T.W. Strategies for tissue and organ decellularization. J. Cell. Biochem. 2012, 113, 2217–2222. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, X.; Hong, H.; Hu, R.; Liu, J.; Liu, C. Decellularized extracellular matrix scaffolds: Recent trends and emerging strategies in tissue engineering. Bioact. Mater. 2022, 10, 15–31. [Google Scholar] [CrossRef] [PubMed]
- Willemse, J.; Verstegen, M.M.A.; Vermeulen, A.; Schurink, I.J.; Roest, H.P.; van der Laan, L.J.W.; de Jonge, J. Fast, robust and effective decellularization of whole human livers using mild detergents and pressure-controlled perfusion. Mater. Sci. Eng. C Mater. Biol. Appl. 2020, 108, 110200. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Pérez, J.; Ahearne, M. The impact of decellularization methods on extracellular matrix derived hydrogels. Sci Rep. 2019, 9, 14933. [Google Scholar] [CrossRef] [PubMed]
- Gilpin, A.; Yang, Y. Decellularization strategies for regenerative medicine: From processing techniques to applications. Biomed. Res. Int. 2017, 2017, 9831534. [Google Scholar] [CrossRef] [PubMed]
- Moffat, D.; Ye, K.; Jin, S. Decellularization for the retention of tissue niches. J. Tissue. Eng. 2022, 13, 20417314221101150. [Google Scholar] [CrossRef] [PubMed]
- Tondato, S.; Moro, A.; Butt, S.; Todesco, M.; Sandrin, D.; Borile, G.; Marchesan, M.; Fabozzo, A.; Bagno, A.; Romanato, F.; et al. Tergitol based decellularization protocol improves the prerequisites for pulmonary xenografts: Characterization and biocompatibility assessment. Polymers 2023, 15, 819. [Google Scholar] [CrossRef] [PubMed]
- VeDepo, M.C.; Detamore, M.S.; Hopkins, R.A.; Converse, G.L. Recellularization of decellularized heart valves: Progress toward the tissue-engineered heart valve. J. Tissue Eng. 2017, 8, 2041731417726327. [Google Scholar] [CrossRef] [PubMed]
- Grauss, R.W.; Hazekamp, M.G.; van Vliet, S.; Gittenberger-de Groot, A.C.; DeRuiter, M.C. Decellularization of rat aortic valve allografts reduces leaflet destruction and extracellular matrix remodeling. J. Thorac. Cardiovasc. Surg. 2003, 126, 2003–2010. [Google Scholar] [CrossRef]
- Olsen, B.R. Electron microscope studies on collagen. Z. Zellforsch. 1964, 61, 913–919. [Google Scholar] [CrossRef]
- Irvin, R.T.; MacAlister, T.J.; Costerton, J.W. Tris(hydroxymethyl)aminomethane buffer modification of Escherichia coli outer membrane permeability. J. Bacteriol. 1981, 145, 1397–1403. [Google Scholar] [CrossRef]
- Scherge, M.; Gorb, S.S. Biological Micro- and Nanotribology, Nanoscience and Technology; Springer: Berlin/Heidelberg, Germany, 2001. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023; ISBN 3-900051-07-0. Available online: http://www.R-project.org/ (accessed on 28 February 2024).
- Carroll, R.J.; Ruppert, D. Transformation and Weighting in Regression, Chapman and Hall; CRC: New York, NY, USA, 1988. [Google Scholar] [CrossRef]
- Kozak, M.; Piepho, H.P. What’s normal anyway? Residual plots are more telling than significance tests when checking ANOVA assumptions. J. Agro. Crop Sci. 2018, 204, 86–98. [Google Scholar] [CrossRef]
- Hasler, M.; Vonk, R.; Hothorn, L.A. Assessing non-inferiority of a new treatment in a three-arm trial in the presence of heteroscedasticity. Stat. Med. 2008, 27, 490–503. [Google Scholar] [CrossRef]
- Rogers, J.L.; Howard, K.I.; Vessey, J.T. Using significance tests to evaluate equivalence between two experimental groups. Psychol. Bull. 1993, 113, 553–565. [Google Scholar] [CrossRef]
- Snapinn, S.M. Noninferiority trials. Curr. Control Trials Cardiovasc. Med. 2000, 1, 19. [Google Scholar] [CrossRef]

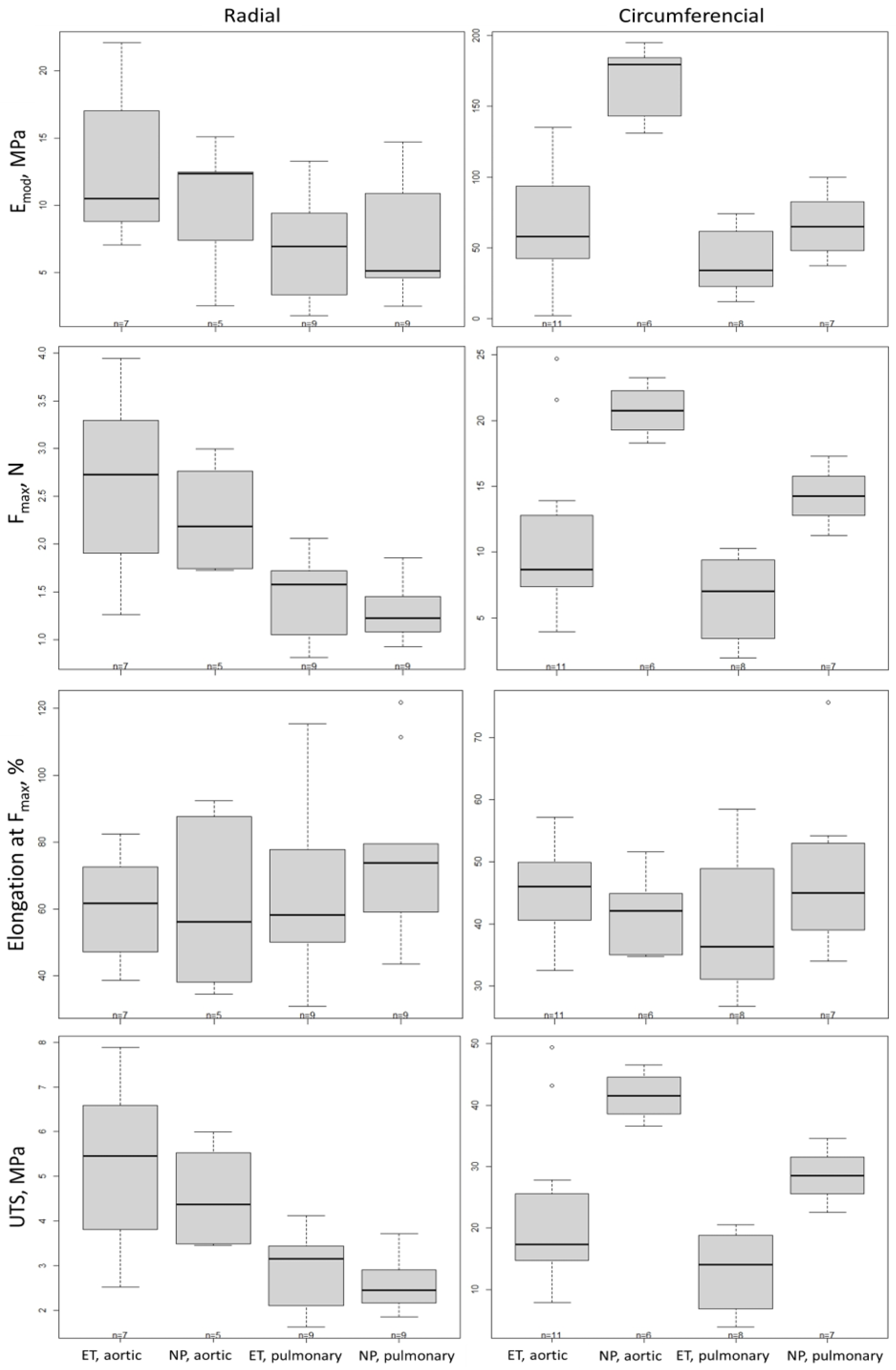

| Emod (MPa) | Radial | Circumferential | ||||||
| Aortic | Pulmonary | Aortic | Pulmonary | |||||
| ET | NP | ET | NP | ET | NP | ET | NP | |
| Mean ± SD | 13.05 ± 5.71 | 9.98 ± 5.01 | 6.64 ± 3.95 | 7.02 ± 4.33 | 64.22 ± 43.44 | 168.65 ± 25.43 | 40.46 ± 22.98 | 66.26 ± 23.63 |
| Difference | 3.06 | −0.38 | −104.43 | −25.81 | ||||
| CI 90 | (−2.53; 8.66) | (−4.88; 4.12) | (−137.68; −71.18) | (−59.72; 8.10) | ||||
| Fmax (N) | Radial | Circumferential | ||||||
| Aortic | Pulmonary | Aortic | Pulmonary | |||||
| ET | NP | ET | NP | ET | NP | ET | NP | |
| Mean ± SD | 2.62 ± 0.99 | 2.28 ± 0.58 | 1.44 ± 0.45 | 1.30 ± 0.29 | 11.09 ± 6.59 | 20.78 ± 1.87 | 6.50 ± 3.28 | 14.28 ± 2.16 |
| Difference | 0.33 | 0.13 | −9.69 | −7.78 | ||||
| CI 90 | (−0.39; 1.06) | (−0.45; 0.72) | (−14.28; −5.10) | (−12.46; −3.10) | ||||
| Elongation at Fmax (%) | Radial | Circumferential | ||||||
| Aortic | Pulmonary | Aortic | Pulmonary | |||||
| ET | NP | ET | NP | ET | NP | ET | NP | |
| Mean ± SD | 60.30 ± 17.61 | 61.74 ± 27.06 | 65.81 ± 28.21 | 77.23 ± 25.29 | 45.48 ± 7.91 | 41.79 ± 6.52 | 39.75 ± 11.68 | 48.38 ± 14.09 |
| Difference | −1.44 | −11.42 | 3.69 | − 8.64 | ||||
| CI 90 | (−31.28; 28.40) | (−35.45; 12.60) | (−6.95; 14.33) | (−19.49; 2.22) | ||||
| UTS (MPa) | Radial | Circumferential | ||||||
| Aortic | Pulmonary | Aortic | Pulmonary | |||||
| ET | NP | ET | NP | ET | NP | ET | NP | |
| Mean ± SD | 5.24 ± 1.99 | 4.57 ± 1.16 | 2.88 ± 0.89 | 2.61 ± 0.58 | 22.19 ± 13.18 | 41.57 ± 3.74 | 13.00 ± 6.56 | 28.57 ± 4.32 |
| Difference | 0.67 | 0.27 | −19.38 | −15.96 | ||||
| CI 90 | (−0.78; 2.11) | (−0.89; 1.43) | (−28.56; −10.20) | (−24.92; −6.20) | ||||
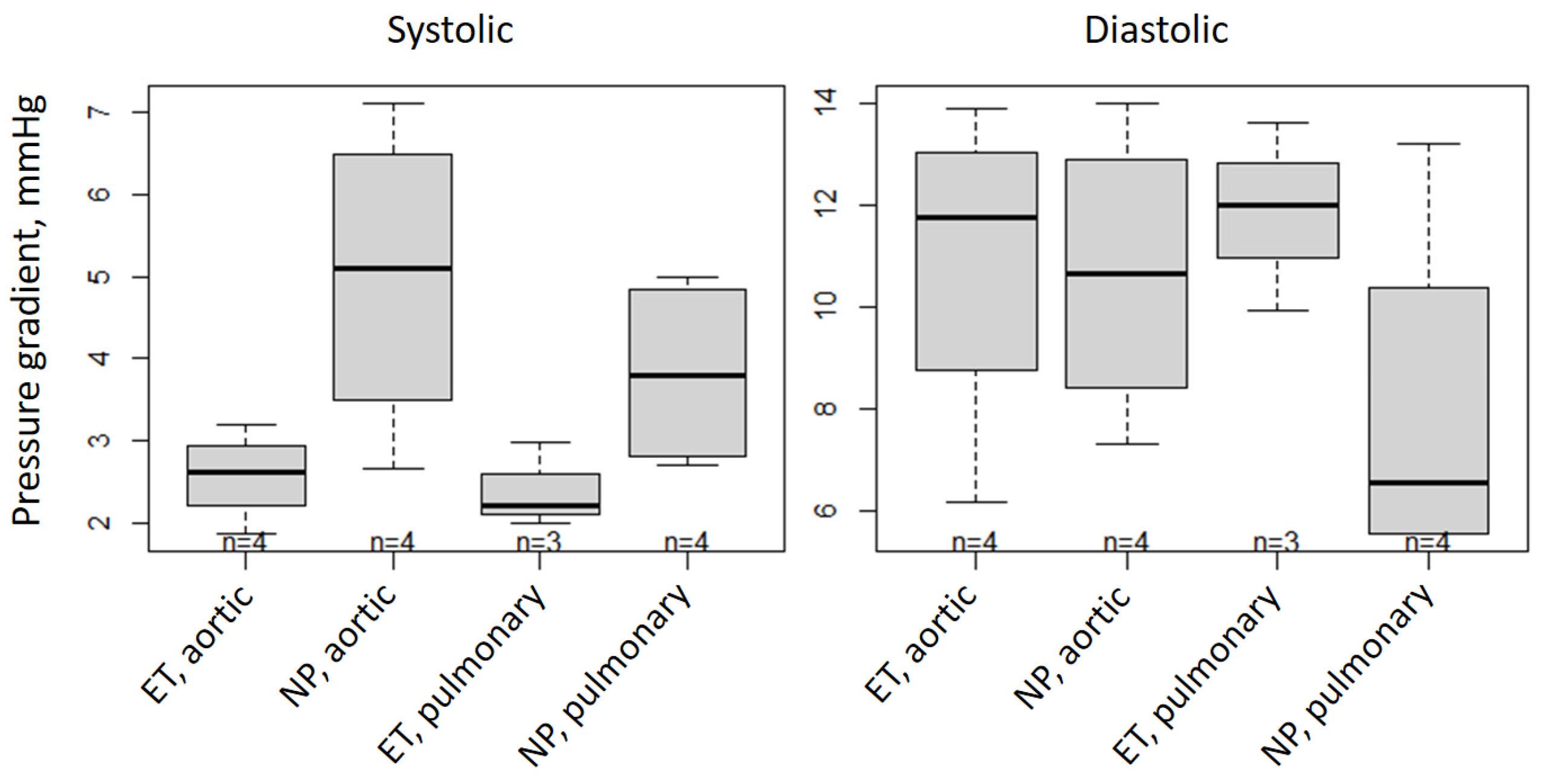
| Pressure Gradient (mmHg) | Systolic | Diastolic | ||||||
|---|---|---|---|---|---|---|---|---|
| Aortic | Pulmonary | Aortic | Pulmonary | |||||
| ET | NP | ET | NP | ET | NP | ET | NP | |
| Mean ± SD | 2.58 ± 0.55 | 4.99 ±1.92 | 2.40 ± 0.51 | 3.83 ± 1.18 | 10.89 ± 3.31 | 10.65 ± 2.88 | 11.85 ± 1.85 | 7.96 ± 3.62 |
| Difference | −2.41 | −1.42 | 0.24 | 3.89 | ||||
| CI 90 | (−4.3; −0.53) | (−3.46; 0.61) | (−4.48; 4.96) | (−1.21; 8.99) | ||||
| Opening Area (mm2) | At 60 bpm | |||
|---|---|---|---|---|
| Aortic | Pulmonary | |||
| ET | NP | ET | NP | |
| Mean ± SD | 115.24 ± 37.13 | 62.84 ± 18.48 | 104.61 ± 18.1 | 92.40 ± 17.92 |
| Difference | 52.4 | 12.21 | ||
| CI 90 | (15.41; 89.38) | (−24.77; 49.20) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saeid Nia, M.; Floder, L.M.; Seiler, J.A.; Puehler, T.; Pommert, N.S.; Berndt, R.; Meier, D.; Sellers, S.L.; Sathananthan, J.; Zhang, X.; et al. Optimization of Enzymatic and Chemical Decellularization of Native Porcine Heart Valves for the Generation of Decellularized Xenografts. Int. J. Mol. Sci. 2024, 25, 4026. https://doi.org/10.3390/ijms25074026
Saeid Nia M, Floder LM, Seiler JA, Puehler T, Pommert NS, Berndt R, Meier D, Sellers SL, Sathananthan J, Zhang X, et al. Optimization of Enzymatic and Chemical Decellularization of Native Porcine Heart Valves for the Generation of Decellularized Xenografts. International Journal of Molecular Sciences. 2024; 25(7):4026. https://doi.org/10.3390/ijms25074026
Chicago/Turabian StyleSaeid Nia, Monireh, Lena Maria Floder, Jette Anika Seiler, Thomas Puehler, Nina Sophie Pommert, Rouven Berndt, David Meier, Stephanie L. Sellers, Janarthanan Sathananthan, Xiling Zhang, and et al. 2024. "Optimization of Enzymatic and Chemical Decellularization of Native Porcine Heart Valves for the Generation of Decellularized Xenografts" International Journal of Molecular Sciences 25, no. 7: 4026. https://doi.org/10.3390/ijms25074026
APA StyleSaeid Nia, M., Floder, L. M., Seiler, J. A., Puehler, T., Pommert, N. S., Berndt, R., Meier, D., Sellers, S. L., Sathananthan, J., Zhang, X., Hasler, M., Gorb, S. N., Warnecke, G., & Lutter, G. (2024). Optimization of Enzymatic and Chemical Decellularization of Native Porcine Heart Valves for the Generation of Decellularized Xenografts. International Journal of Molecular Sciences, 25(7), 4026. https://doi.org/10.3390/ijms25074026







