Identification of PTPN12 Phosphatase as a Novel Negative Regulator of Hippo Pathway Effectors YAP/TAZ in Breast Cancer
Abstract
:1. Introduction
2. Results
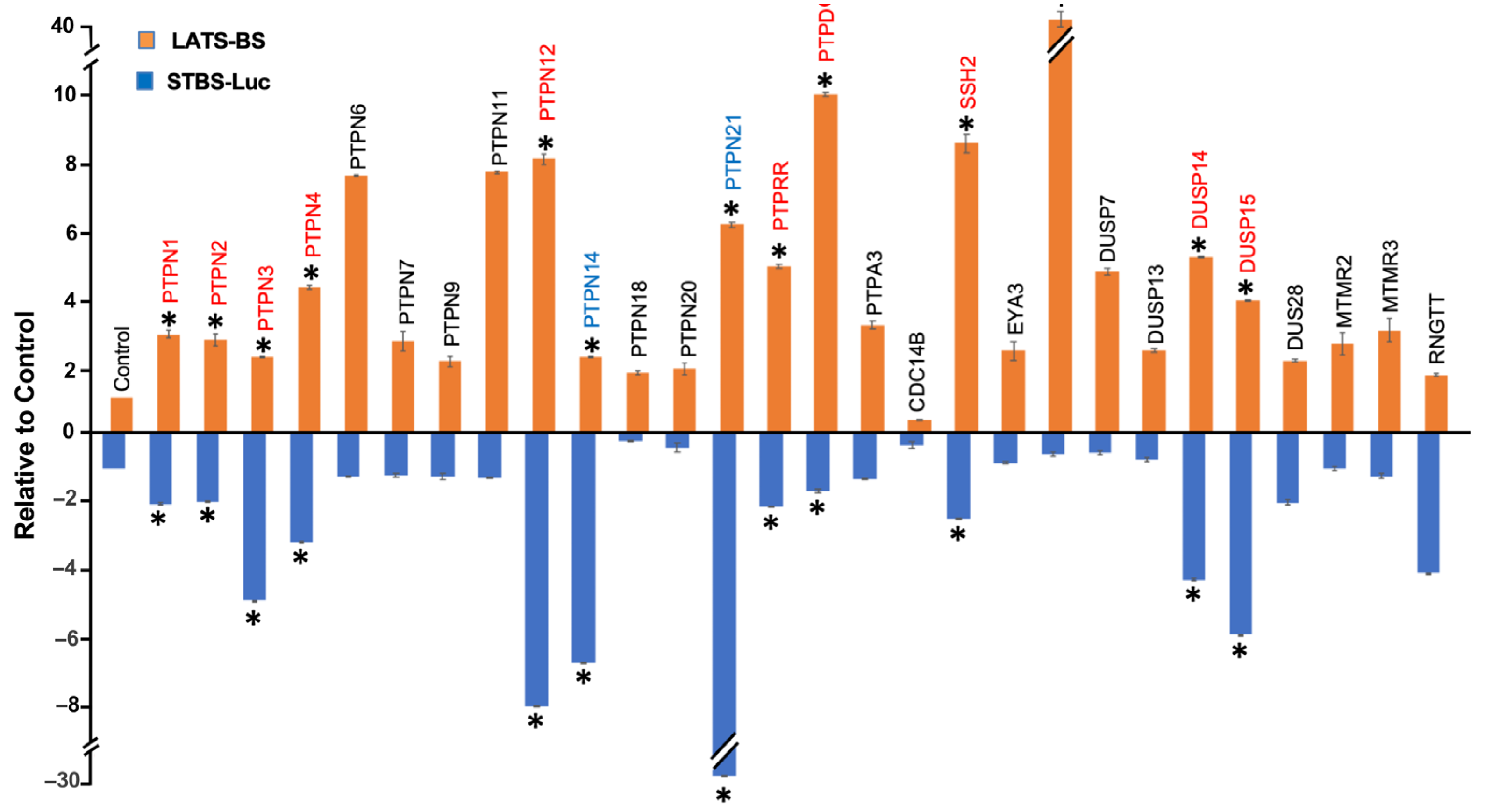
2.1. Gain-of-Functional Screening for PTPs Regulating the Hippo Pathway
2.2. Validation of Candidate PTPs Regulating the Hippo Pathway
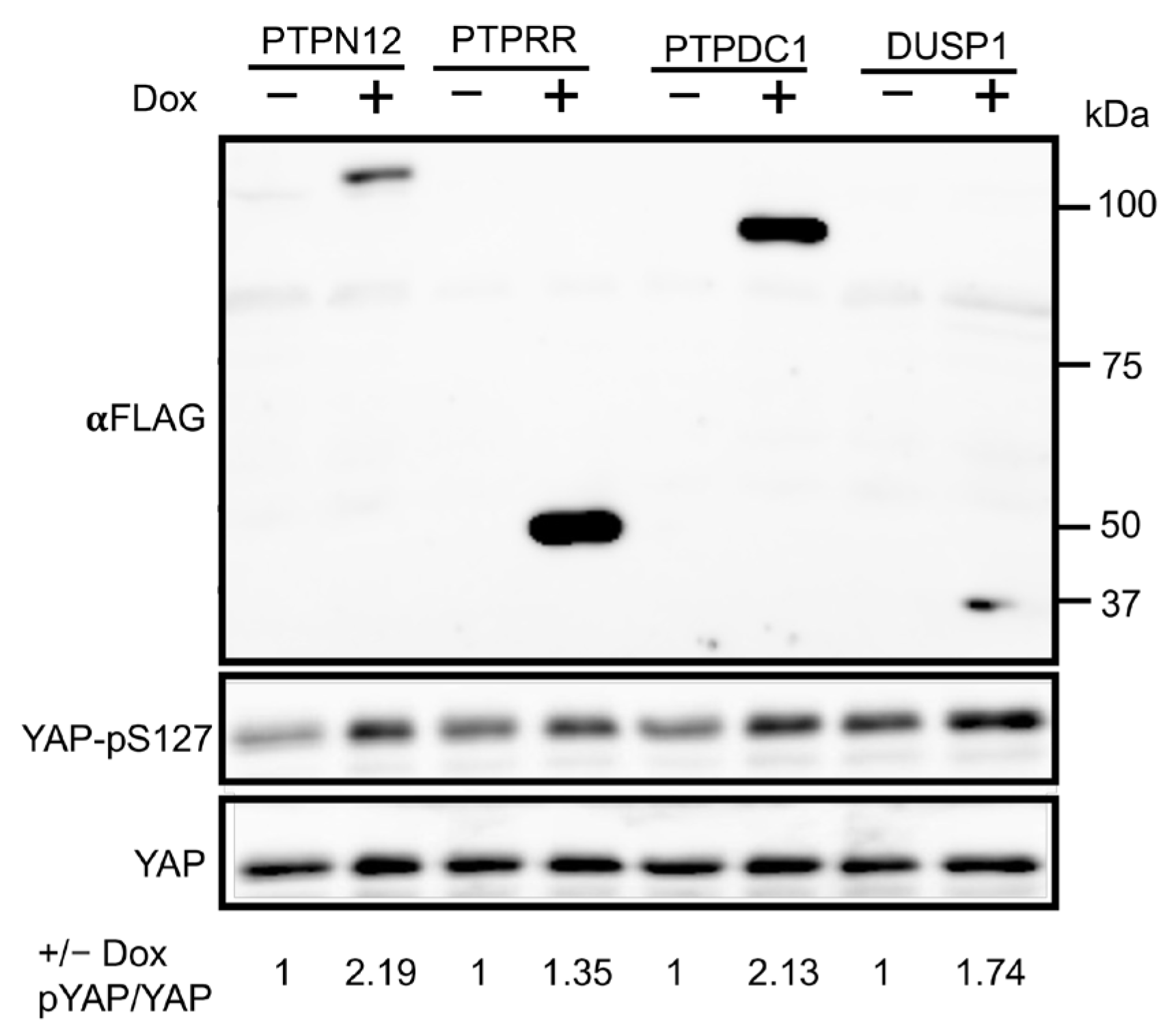
2.3. Validation of PTPN12 as a Novel Regulator of the Hippo Pathway
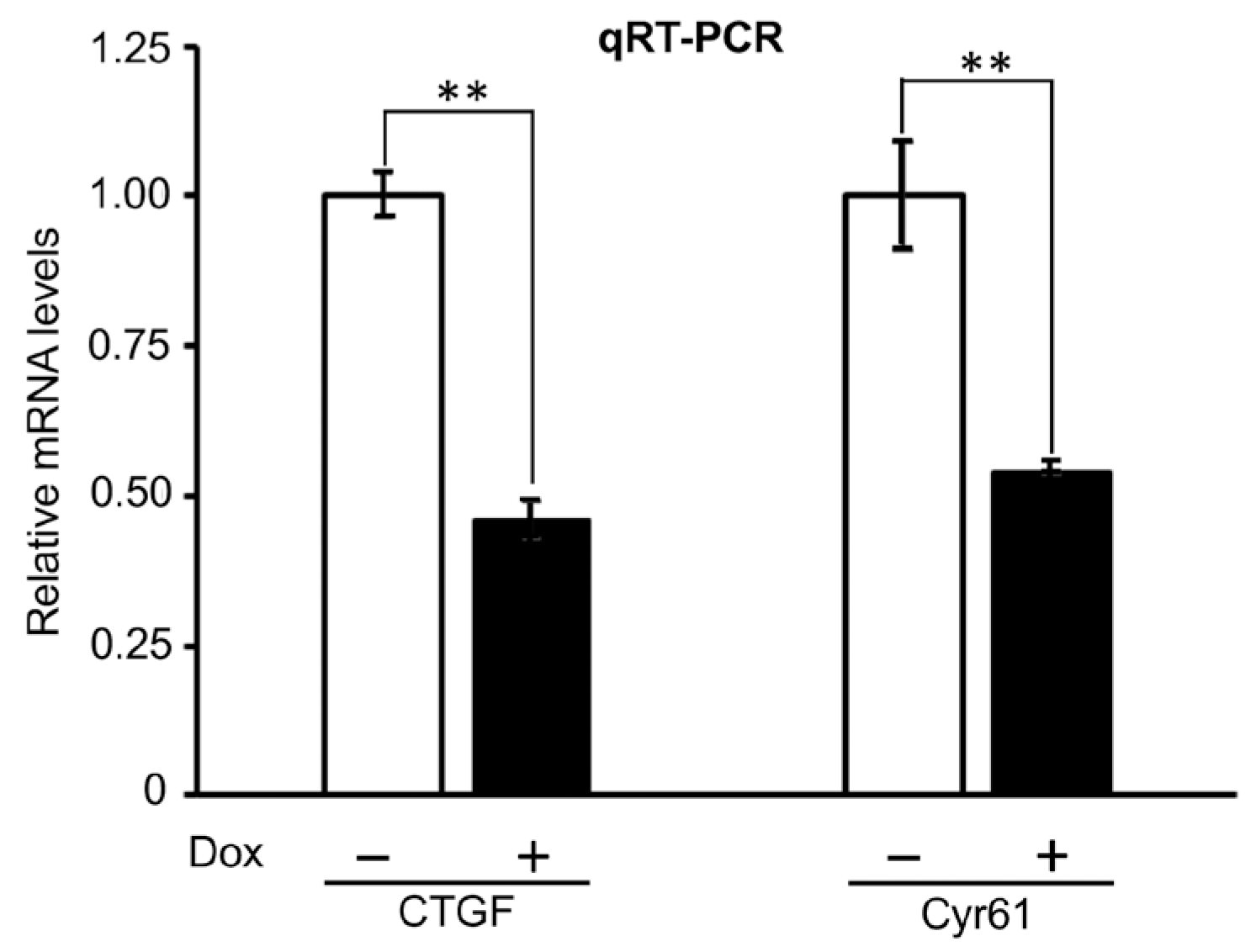
2.3.1. Effects of PTPN12 Overexpression on the mRNA Expression Level of Hippo-Targeted Genes in HEK293T-PTPN12
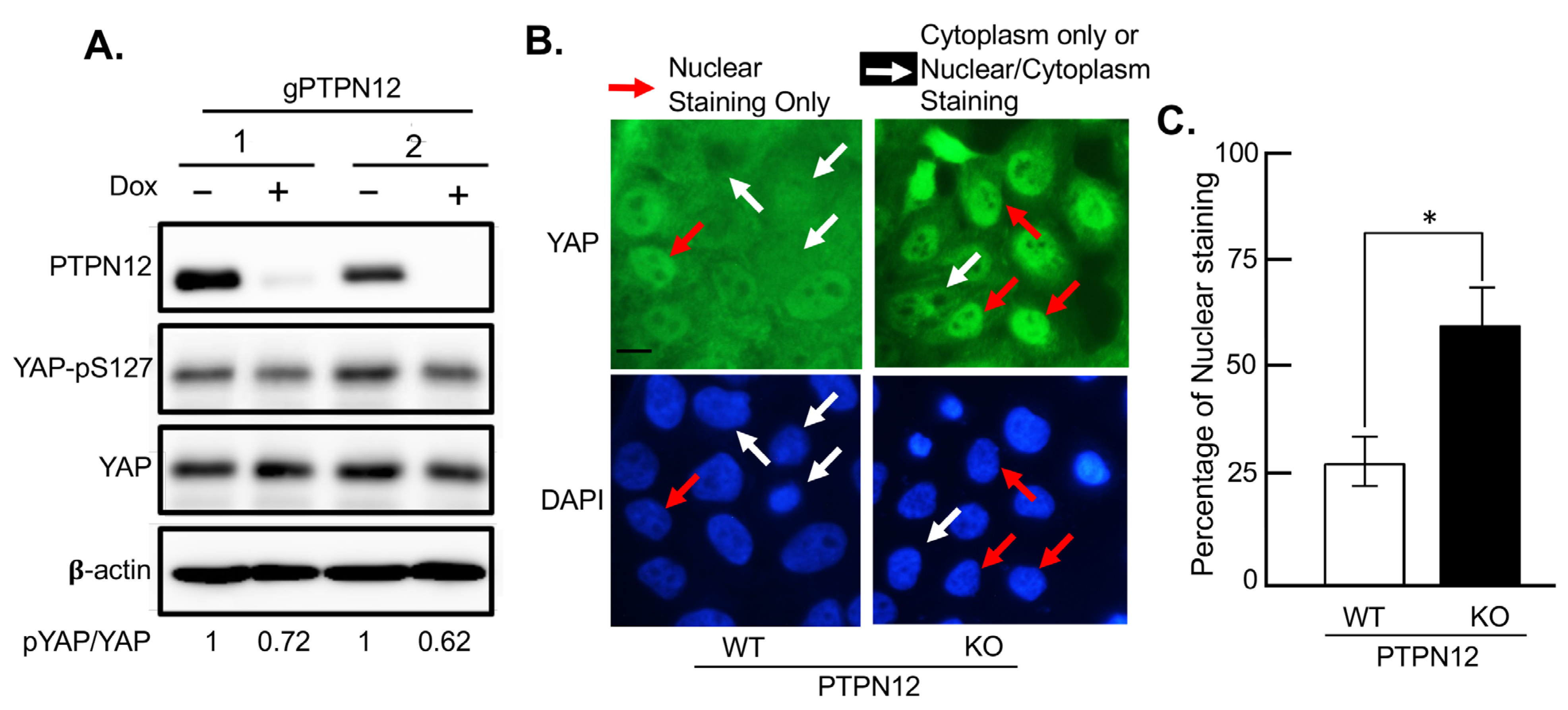
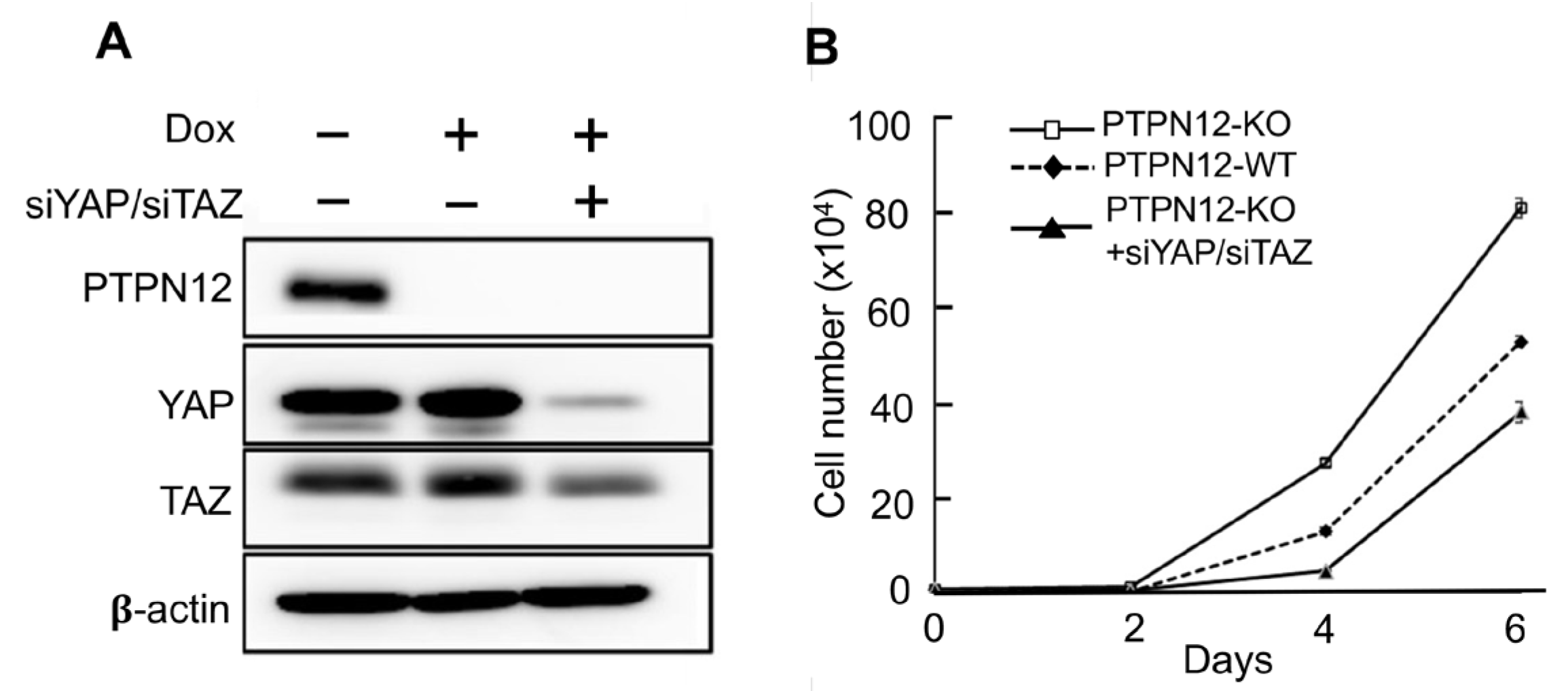
2.3.2. Effect of Loss-of-PTPN12 on Hippo Signaling and Cell Proliferation in Mammary Cells
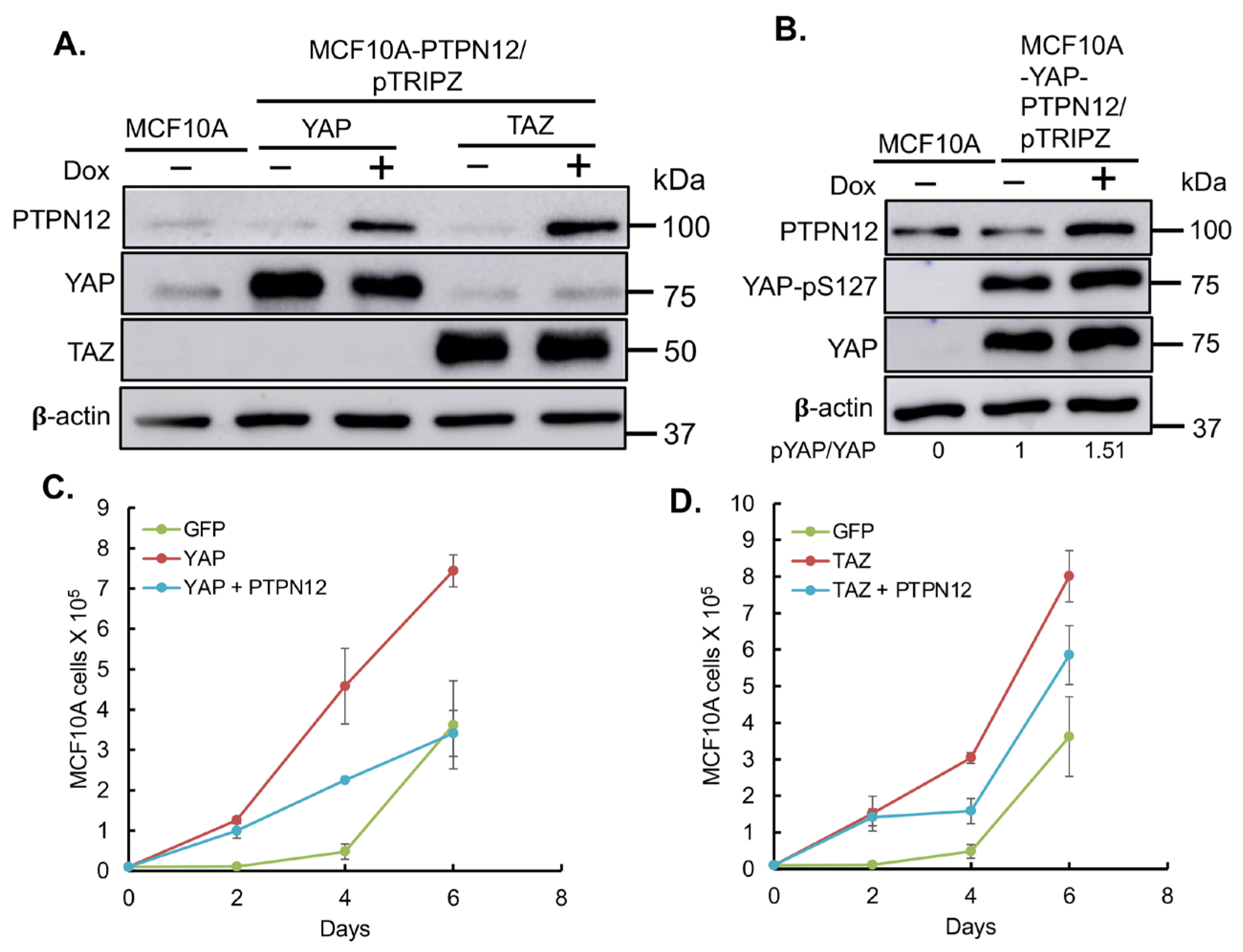
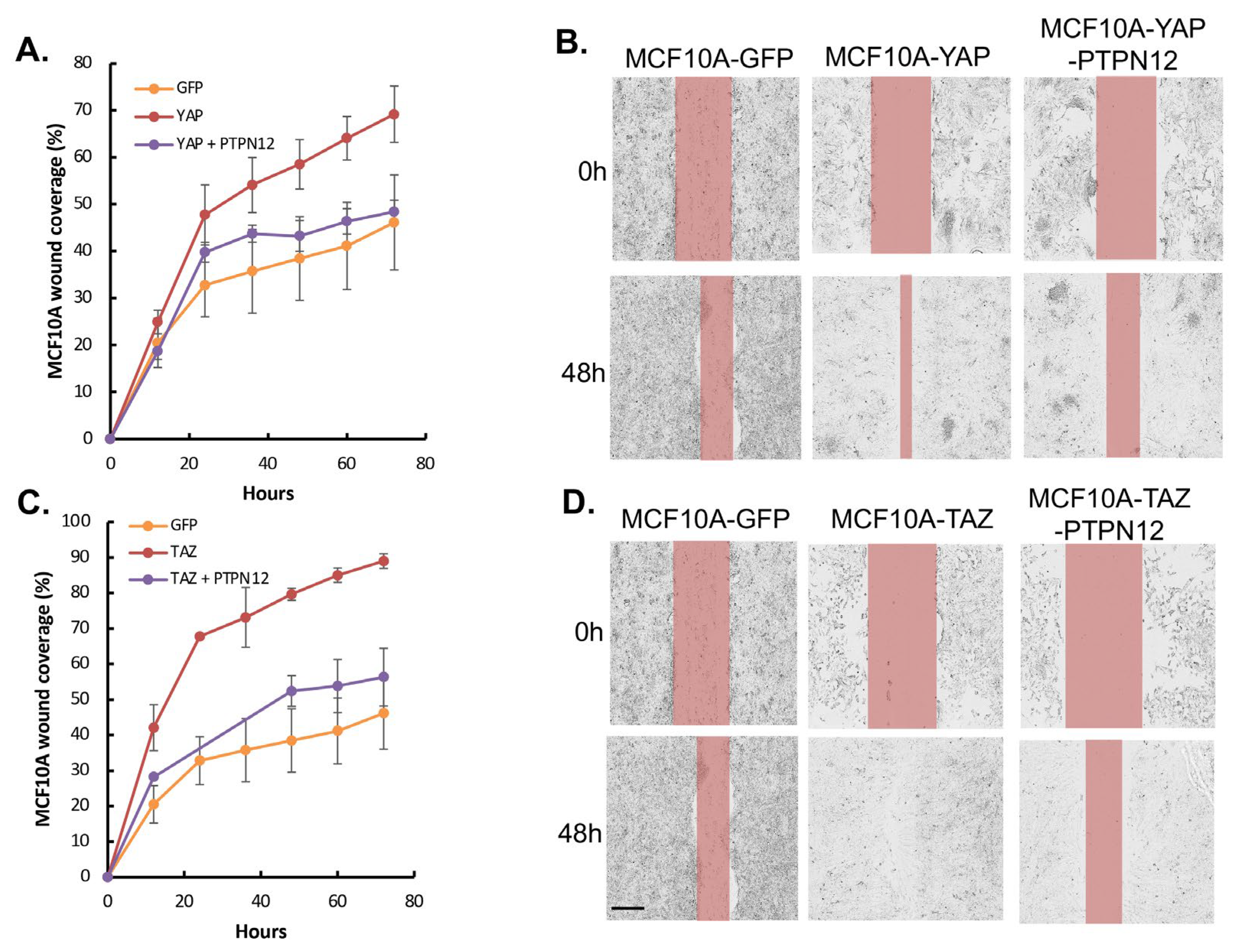
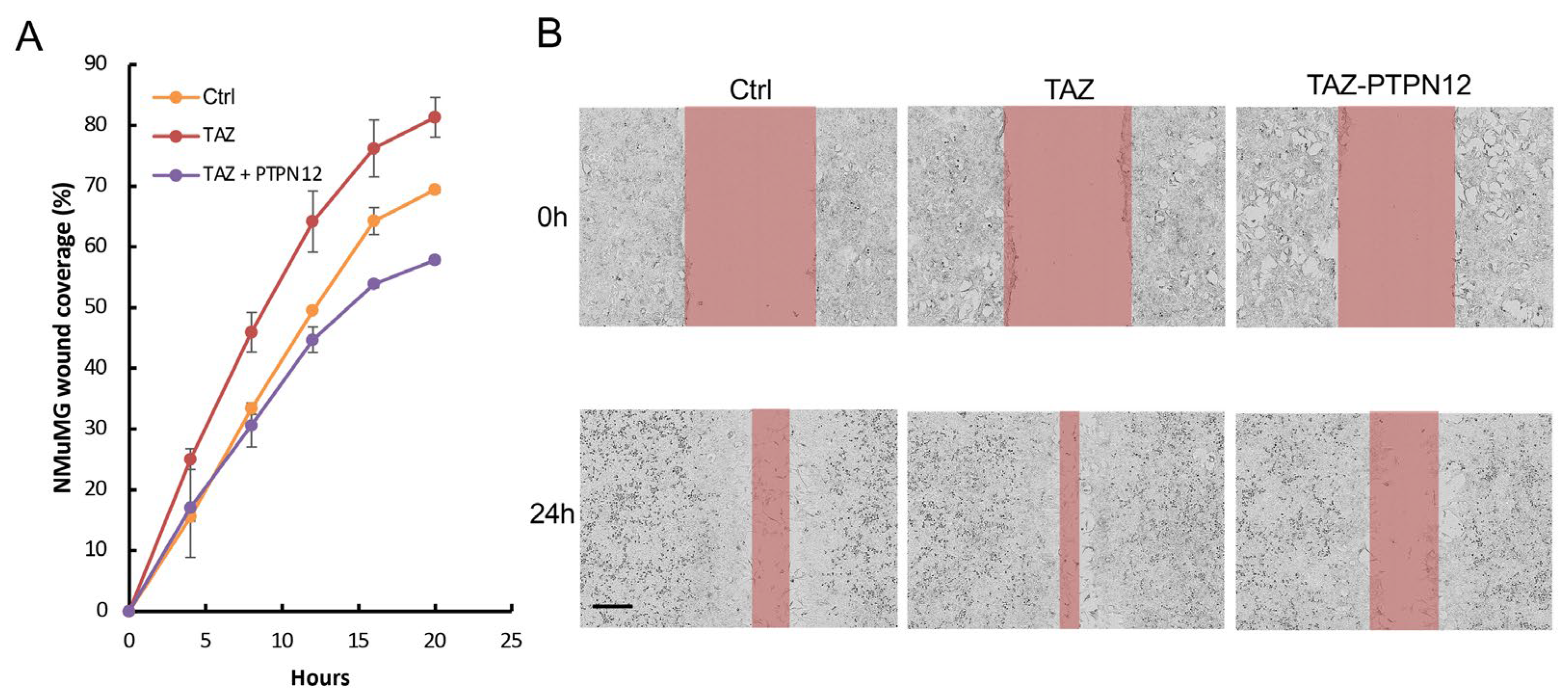
2.3.3. Effect of PTPN12 Overexpression on Hippo Signaling and Cell Proliferation and Cell Migration in Mammary Cells
3. Discussion
3.1. Identification of Novel PTP Regulating the Hippo Pathway
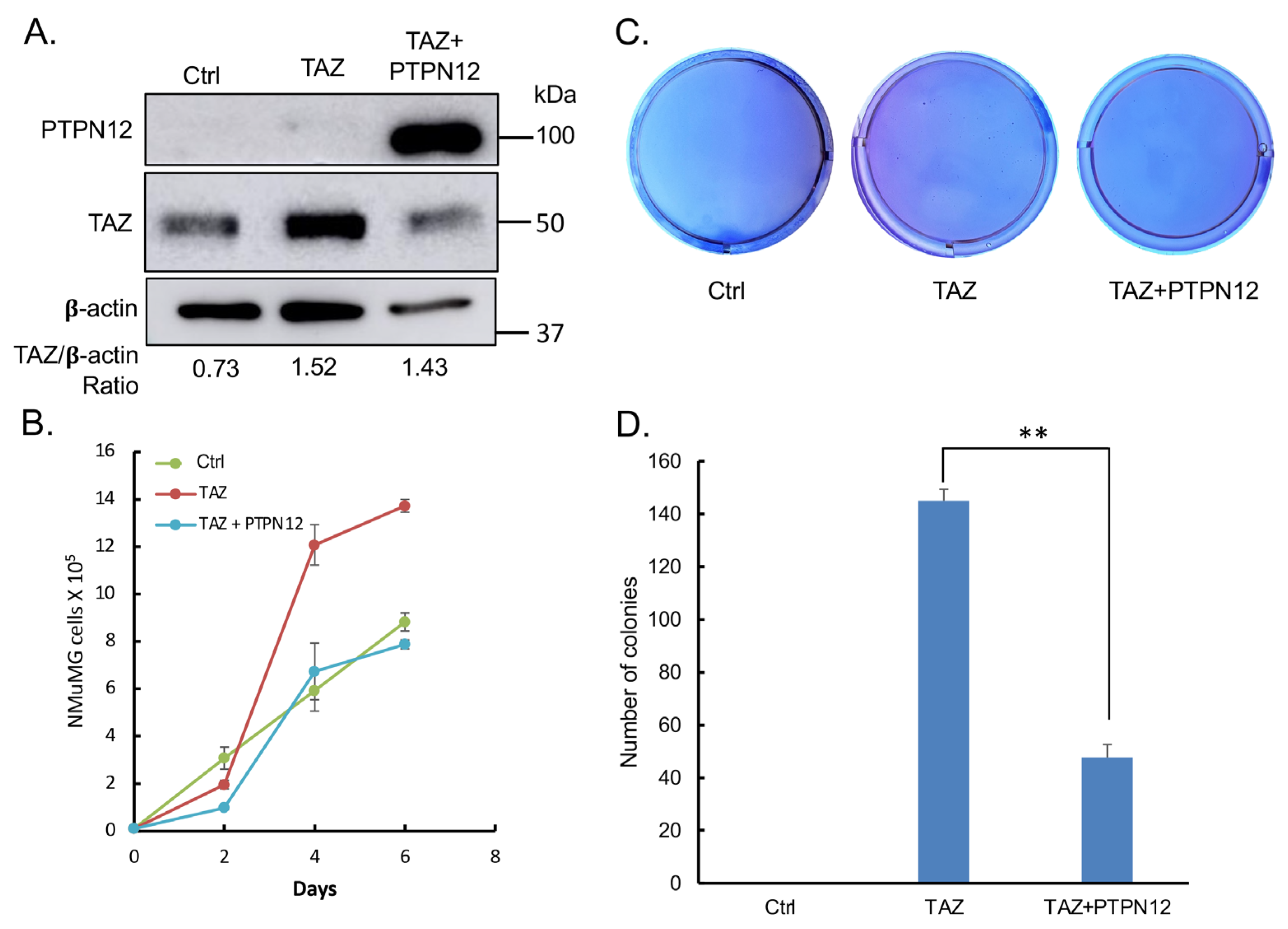
3.2. PTPN12 Tumor Suppressor Function in Cancer Is Mediated by the Hippo Pathway
4. Materials and Methods
4.1. Plasmid Construction and Purification
4.2. Cell Culture
4.3. Lentivirus Production
4.4. Establishment of Stable Cell Lines
4.5. Protein Extraction and Western Blot Analysis and Antibodies
4.6. RNA Extraction and qRT-PCR Analysis
4.7. Luciferase Assays and Gain-of-Functional Screening
4.8. Immunostaining
4.9. Transient Knockdown of YAP and TAZ in MCF10A-PTPN12-KO
4.10. Cell Proliferation Assay
4.11. Soft Agar Assay
4.12. Cell Migration Assay
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chan, E.H.; Nousiainen, M.; Chalamalasetty, R.B.; Schafer, A.; Nigg, E.A.; Sillje, H.H. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 2005, 24, 2076–2086. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wu, S.; Barrera, J.; Matthews, K.; Pan, D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 2005, 122, 421–434. [Google Scholar] [CrossRef] [PubMed]
- Harvey, K.F.; Pfleger, C.M.; Hariharan, I.K. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 2003, 114, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Udan, R.S.; Kango-Singh, M.; Nolo, R.; Tao, C.; Halder, G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 2003, 5, 914–920. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Huang, J.; Dong, J.; Pan, D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 2003, 114, 445–456. [Google Scholar] [CrossRef]
- Dong, J.; Feldmann, G.; Huang, J.; Wu, S.; Zhang, N.; Comerford, S.A.; Gayyed, M.F.; Anders, R.A.; Maitra, A.; Pan, D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 2007, 130, 1120–1133. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.C.; Wei, X.; Shimizu, T.; Ramos, E.; Rohrbaugh, M.; Nikolaidis, N.; Ho, L.L.; Li, Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell 2005, 120, 675–685. [Google Scholar] [CrossRef]
- Tapon, N.; Harvey, K.F.; Bell, D.W.; Wahrer, D.C.; Schiripo, T.A.; Haber, D.A.; Hariharan, I.K. Salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 2002, 110, 467–478. [Google Scholar] [CrossRef]
- Xu, T.; Wang, W.; Zhang, S.; Stewart, R.A.; Yu, W. Identifying tumor suppressors in genetic mosaics: The Drosophila lats gene encodes a putative protein kinase. Development 1995, 121, 1053–1063. [Google Scholar] [CrossRef]
- Kulkarni, A.; Chang, M.T.; Vissers, J.H.A.; Dey, A.; Harvey, K.F. The Hippo Pathway as a Driver of Select Human Cancers. Trends Cancer 2020, 6, 781–796. [Google Scholar] [CrossRef]
- Visser, S.; Yang, X. LATS tumor suppressor: A new governor of cellular homeostasis. Cell Cycle 2010, 9, 3892–3903. [Google Scholar] [CrossRef] [PubMed]
- Taha, Z.; J Janse van Rensburg, H.; Yang, X. The Hippo pathway: Immunity and cancer. Cancers 2018, 10, 94. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, F.; Gao, Z.; Zhang, P.; Gao, J.; Wu, X. Regulation of Hippo Signaling by Mechanical Signals and the Cytoskeleton. DNA Cell. Biol. 2020, 39, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yang, X. The Hippo pathway in chemotherapeutic drug resistance. Int. J. Cancer 2015, 137, 2767–2773. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Chun, A.; Cheung, K.; Rashidi, B.; Yang, X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 2008, 283, 5496–5509. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Wei, X.; Li, W.; Udan, R.S.; Yang, Q.; Kim, J.; Xie, J.; Ikenoue, T.; Yu, J.; Li, L.; et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007, 21, 2747–2761. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Ye, X.; Yu, J.; Li, L.; Li, W.; Li, S.; Yu, J.; Lin, J.D.; Wang, C.Y.; Chinnaiyan, A.M.; et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008, 22, 1962–1971. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.W.; Lim, C.J.; Loo, L.S.; Chong, Y.F.; Huang, C.; Hong, W. TEADs mediate nuclear retention of TAZ to promote oncogenic transformation. J. Biol. Chem. 2009, 284, 14347–14358. [Google Scholar] [CrossRef] [PubMed]
- Lei, Q.Y.; Zhang, H.; Zhao, B.; Zha, Z.Y.; Bai, F.; Pei, X.H.; Zhao, S.; Xiong, Y.; Guan, K.L. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol. Cell. Biol. 2008, 28, 2426–2436. [Google Scholar] [CrossRef]
- Dey, A.; Varelas, X.; Guan, K.L. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat. Rev. Drug Discov. 2020, 19, 480–494. [Google Scholar] [CrossRef]
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Lo Muzio, L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy. Int. J. Mol. Med. 2017, 40, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Hunter, T. Tyrosine phosphorylation: Thirty years and counting. Curr. Opin. Cell Biol. 2009, 21, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, M.A.; Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef]
- Day, E.K.; Sosale, N.G.; Lazzara, M.J. Cell signaling regulation by protein phosphorylation: A multivariate, heterogeneous, and context-dependent process. Curr. Opin. Biotechnol. 2016, 40, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Sarmasti Emami, S.; Zhang, D.; Yang, X. Interaction of the Hippo Pathway and Phosphatases in Tumorigenesis. Cancers 2020, 12, 2438. [Google Scholar] [CrossRef] [PubMed]
- Azad, T.N.K.; Janse van Rensburg, H.J.; Maritan, S.M.; Wu, L.; Hao, Y.; Montminy, T.; Yu, J.; Khanal, K.; Mulligan, L.M.; Yang, X. A gain-of-functional screen identifies the Hippo pathway as a central mediator of receptor tyrosine kinases during tumorigenesis. Oncogene 2020, 39, 334–355. [Google Scholar] [CrossRef]
- Reuven, N.; Shanzer, M.; Shaul, Y. Hippo Pathway Regulation by Tyrosine Kinases. Methods Mol. Biol. 2019, 1893, 215–236. [Google Scholar] [PubMed]
- Lamar, J.M.; Xiao, Y.; Norton, E.; Jiang, Z.G.; Gerhard, G.M.; Kooner, S.; Warren, J.S.A.; Hynes, R.O. SRC tyrosine kinase activates the YAP/TAZ axis and thereby drives tumor growth and metastasis. J. Biol. Chem. 2019, 294, 2302–2317. [Google Scholar] [CrossRef]
- Azad, T.; Janse van Rensburg, H.J.; Lightbody, E.D.; Neveu, B.; Champagne, A.; Ghaffari, A.; Kay, V.R.; Hao, Y.; Shen, H.; Yeung, B.; et al. A LATS biosensor functional screen identifies VEGFR as a novel regulator of the Hippo pathway in angiogenesis. Nat.Commun. 2018, 9, 1061. [Google Scholar] [CrossRef]
- Si, Y.; Ji, X.; Cao, X.; Dai, X.; Xu, L.; Zhao, H.; Guo, X.; Yan, H.; Zhang, H.; Zhu, C.; et al. Src Inhibits the Hippo Tumor Suppressor Pathway through Tyrosine Phosphorylation of Lats1. Cancer Res. 2017, 77, 4868–4880. [Google Scholar] [CrossRef]
- Wilson, K.E.; Yang, N.; Mussell, A.L.; Zhang, J. The Regulatory Role of KIBRA and PTPN14 in Hippo Signaling and Beyond. Genes 2016, 7, 23. [Google Scholar] [CrossRef]
- Liu, X.; Yang, N.; Figel, S.A.; Wilson, K.E.; Morrison, C.D.; Gelman, I.H.; Zhang, J. PTPN14 interacts with and negatively regulates the oncogenic function of YAP. Oncogene 2013, 32, 1266–1273. [Google Scholar] [CrossRef]
- Michaloglou, C.; Lehmann, W.; Martin, T.; Delaunay, C.; Hueber, A.; Barys, L.; Niu, H.; Billy, E.; Wartmann, M.; Ito, M.; et al. The tyrosine phosphatase PTPN14 is a negative regulator of YAP activity. PLoS ONE 2013, 8, e61916. [Google Scholar] [CrossRef]
- Li, X.; Tran, K.M.; Aziz, K.E.; Sorokin, A.V.; Chen, J.; Wang, W. Defining the Protein-Protein Interaction Network of the Human Protein Tyrosine Phosphatase Family. Mol. Cell. Proteom. 2016, 15, 3030–3044. [Google Scholar] [CrossRef]
- Wilson, K.E.; Li, Y.W.; Yang, N.; Shen, H.; Orillion, A.R.; Zhang, J. PTPN14 forms a complex with Kibra and LATS1 prot3eins and negatively regulates the YAP oncogenic function. J. Biol. Chem. 2014, 289, 23693–23700. [Google Scholar] [CrossRef]
- Xiao, L.; Chen, Y.; Ji, M.; Dong, J. KIBRA regulates Hippo signaling activity via interactions with large tumor suppressor kinases. J. Biol. Chem. 2011, 286, 7788–7796. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Chen, Y.; Ji, M.; Volle, D.J.; Lewis, R.E.; Tsai, M.-Y.; Dong, J. KIBRA protein phosphorylation is regulated by mitotic kinase aurora and protein phosphatase 1. J. Biol. Chem. 2011, 286, 36304–36315. [Google Scholar] [CrossRef]
- Tang, C.; Takahashi-Kanemitsu, A.; Kikuchi, I.; Ben, C.; Hatakeyama, M. Transcriptional co-activator functions of YAP and TAZ are inversely regulated by tyrosine phosphorylation status of Parafibromin. iScience 2018, 1, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Tsutsumi, R.; Masoudi, M.; Takahashi, A.; Fujii, Y.; Hayashi, T.; Kikuchi, I.; Satou, Y.; Taira, M.; Hatakeyama, M. YAP and TAZ, Hippo signaling targets, act as a rheostat for nuclear SHP2 function. Dev. Cell 2013, 26, 658–665. [Google Scholar] [CrossRef]
- Sun, T.; Aceto, N.; Meerbrey, K.L.; Kessler, J.D.; Zhou, C.; Migliaccio, I.; Nguyen, D.X.; Pavlova, N.N.; Botero, M.; Huang, J.; et al. Activation of multiple proto-oncogenic tyrosine kinases in breast cancer via loss of the PTPN12 phosphatase. Cell 2011, 144, 703–718. [Google Scholar] [CrossRef]
- Li, J.; Davidson, D.; Martins Souza, C.; Zhong, M.C.; Wu, N.; Park, M.; Muller, W.J.; Veillette, A. Loss of PTPN12 Stimulates Progression of ErbB2-Dependent Breast Cancer by Enhancing Cell Survival, Migration, and Epithelial-to-Mesenchymal Transition. Mol. Cell. Biol. 2015, 35, 4069–4082. [Google Scholar] [CrossRef]
- Shen, N.; Li, L.; Xu, W.; Tian, J.; Yang, Y.; Zhu, Y.; Gong, Y.; Ke, J.; Gong, J.; Chang, J.; et al. A missense variant in PTPN12 associated with the risk of colorectal cancer by modifying Ras/MEK/ERK signaling. Cancer Epidemiol. 2019, 59, 109–114. [Google Scholar] [CrossRef]
- Huo, Y.H.; Wang, Y.N.; Meng, L.B.; Zhang, A.L.; Liu, B. Progress in the correlation between PTPN12 gene expression and human tumors. Medicine 2020, 99, e20445. [Google Scholar] [CrossRef]
- Cao, J.; Wu, L.; Zhang, S.M.; Lu, M.; Cheung, W.K.; Cai, W.; Gale, M.; Xu, Q.; Yan, Q. An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Res. 2016, 44, e149. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Wang, M.; Cai, M.; Zhang, C.; Qiu, Y.; Wang, X.; Zhang, T.; Zhou, H.; Wang, J.; Zhao, W.; et al. Transcriptional co-activators YAP/TAZ: Potential therapeutic targets for metastatic breast cancer. Biomed. Pharmacother. 2021, 133, 110956. [Google Scholar] [CrossRef]
- Thompson, B.J. YAP/TAZ: Drivers of tumor growth, metastasis, and resistance to therapy. Bioessays 2020, 42, e1900162. [Google Scholar] [CrossRef]
- Chan, S.W.; Lim, C.J.; Guo, K.; Ng, C.P.; Lee, I.; Hunziker, W.; Zeng, Q.; Hong, W. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 2008, 68, 2592–2598. [Google Scholar] [CrossRef] [PubMed]
- Menigatti, M.; Cattaneo, E.; Sabates-Bellver, J.; Ilinsky, V.V.; Went, P.; Buffoli, F.; Marquez, V.E.; Jiricny, J.; Marra, G. The protein tyrosine phosphatase receptor type R gene is an early and frequent target of silencing in human colorectal tumorigenesis. Mol. Cancer 2009, 8, 124. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Shen, D.; Parsons, D.W.; Bardelli, A.; Sager, J.; Szabo, S.; Ptak, J.; Silliman, N.; Peters, B.A.; van der Heijden, M.S.; et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science 2004, 304, 1164–1166. [Google Scholar] [CrossRef]
- Munkley, J.; Lafferty, N.P.; Kalna, G.; Robson, C.N.; Leung, H.Y.; Rajan, P.; Elliott, D.J. Androgen-regulation of the protein tyrosine phosphatase PTPRR activates ERK1/2 signalling in prostate cancer cells. BMC Cancer 2015, 15, 9. [Google Scholar] [CrossRef]
- Chirivi, R.G.; Noordman, Y.E.; Van der Zee, C.E.; Hendriks, W.J. Altered MAP kinase phosphorylation and impaired motor coordination in PTPRR deficient mice. J. Neurochem. 2007, 101, 829–840. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cao, J.; Liu, W.; Zhang, J.; Wang, Z.; Zhang, Y.; Hou, L.; Chen, S.; Hao, P.; Zhang, L.; et al. Protein tyrosine phosphatase receptor type R (PTPRR) antagonizes the Wnt signaling pathway in ovarian cancer by dephosphorylating and inactivating β-catenin. J. Biol. Chem. 2019, 294, 18306–18323. [Google Scholar] [CrossRef] [PubMed]
- Owens, D.M.; Keyse, S.M. Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene 2007, 26, 3203–3213. [Google Scholar] [CrossRef] [PubMed]
- Krysan, K.; Reckamp, K.L.; Dalwadi, H.; Sharma, S.; Rozengurt, E.; Dohadwala, M.; Dubinett, S.M. Prostaglandin E2 activates mitogen-activated protein kinase/Erk pathway signaling and cell proliferation in non-small cell lung cancer cells in an epidermal growth factor receptor-independent manner. Cancer Res. 2005, 65, 6275–6281. [Google Scholar] [CrossRef] [PubMed]
- Kanai, M.; Konda, Y.; Nakajima, T.; Izumi, Y.; Kanda, N.; Nanakin, A.; Kubohara, Y.; Chiba, T. Differentiation-inducing factor-1 (DIF-1) inhibits STAT3 activity involved in gastric cancer cell proliferation via MEK-ERK-dependent pathway. Oncogene 2003, 22, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Keyse, S.M. Dual-specificity MAP kinase phosphatases (MKPs) and cancer. Cancer Metastasis Rev. 2008, 27, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Calvisi, D.F.; Pinna, F.; Meloni, F.; Ladu, S.; Pellegrino, R.; Sini, M.; Daino, L.; Simile, M.M.; De Miglio, M.R.; Virdis, P.; et al. Dual-specificity phosphatase 1 ubiquitination in extracellular signal-regulated kinase-mediated control of growth in human hepatocellular carcinoma. Cancer Res. 2008, 68, 4192–4200. [Google Scholar] [CrossRef] [PubMed]
- Moncho-Amor, V.; Ibañez de Cáceres, I.; Bandres, E.; Martínez-Poveda, B.; Orgaz, J.L.; Sánchez-Pérez, I.; Zazo, S.; Rovira, A.; Albanell, J.; Jiménez, B.; et al. DUSP1/MKP1 promotes angiogenesis, invasion and metastasis in non-small-cell lung cancer. Oncogene 2011, 30, 668–678. [Google Scholar] [CrossRef] [PubMed]
- Magi-Galluzzi, C.; Mishra, R.; Fiorentino, M.; Montironi, R.; Yao, H.; Capodieci, P.; Wishnow, K.; Kaplan, I.; Stork, P.J.; Loda, M. Mitogen-activated protein kinase phosphatase 1 is overexpressed in prostate cancers and is inversely related to apoptosis. Lab. Investig. 1997, 76, 37–51. [Google Scholar]
- Srikanth, S.; Franklin, C.C.; Duke, R.C.; Kraft, R.S. Human DU145 prostate cancer cells overexpressing mitogen-activated protein kinase phosphatase-1 are resistant to Fas ligand-induced mitochondrial perturbations and cellular apoptosis. Mol. Cell. Biochem. 1999, 199, 169–178. [Google Scholar] [CrossRef]
- Loda, M.; Capodieci, P.; Mishra, R.; Yao, H.; Corless, C.; Grigioni, W.; Wang, Y.; Magi-Galluzzi, C.; Stork, P.J. Expression of mitogen-activated protein kinase phosphatase-1 in the early phases of human epithelial carcinogenesis. Am. J. Pathol. 1996, 149, 1553–1564. [Google Scholar]
- Shen, J.; Zhang, Y.; Yu, H.; Shen, B.; Liang, Y.; Jin, R.; Liu, X.; Shi, L.; Cai, X. Role of DUSP1/MKP1 in tumorigenesis, tumor progression and therapy. Cancer Med. 2016, 5, 2061–2068. [Google Scholar] [CrossRef] [PubMed]
- Bhore, N.; Wang, B.J.; Chen, Y.W.; Liao, Y.F. Critical Roles of Dual-Specificity Phosphatases in Neuronal Proteostasis and Neurological Diseases. Int. J. Mol. Sci. 2017, 18, 1963. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.P.; Qi, X.W.; Sun, N.; Sun, Y.Y.; Zhang, Y.; Tan, X.N.; Ding, J.; Han, F.; Zhang, Y. The emerging roles of dual-specificity phosphatases and their specific characteristics in human cancer. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188562. [Google Scholar] [CrossRef]
- Lee, C.; Rhee, I. Important roles of protein tyrosine phosphatase PTPN12 in tumor progression. Pharmacol. Res. 2019, 144, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Nair, A.; Chung, H.C.; Sun, T.; Tyagi, S.; Dobrolecki, L.E.; Dominguez-Vidana, R.; Kurley, S.J.; Orellana, M.; Renwick, A.; Henke, D.M.; et al. Combinatorial inhibition of PTPN12-regulated receptors leads to a broadly effective therapeutic strategy in triple-negative breast cancer. Nat. Med. 2018, 24, 505–511. [Google Scholar] [CrossRef]
- Rhee, I.; Zhong, M.C.; Reizis, B.; Cheong, C.; Veillette, A. Control of dendritic cell migration, T cell-dependent immunity, and autoimmunity by protein tyrosine phosphatase PTPN12 expressed in dendritic cells. Mol. Cell. Biol. 2014, 34, 888–899. [Google Scholar] [CrossRef]
- Souza, C.M.; Davidson, D.; Rhee, I.; Gratton, J.P.; Davis, E.C.; Veillette, A. The phosphatase PTP-PEST/PTPN12 regulates endothelial cell migration and adhesion, but not permeability, and controls vascular development and embryonic viability. J. Biol. Chem. 2012, 287, 43180–43190. [Google Scholar] [CrossRef]
- Doench, J.G.; Fusi, N.; Sullender, M.; Hegde, M.; Vaimberg, E.W.; Donovan, K.F.; Smith, I.; Tothova, Z.; Wilen, C.; Orchard, R. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016, 34, 184–191. [Google Scholar] [CrossRef]
- Lai, D.; Ho, K.C.; Hao, Y.; Yang, X. Taxol resistance in breast cancer cells is mediated by the hippo pathway component TAZ and its downstream transcriptional targets Cyr61 and CTGF. Cancer Res. 2011, 71, 2728–2738. [Google Scholar] [CrossRef]
- Janse van Rensburg, H.J.; Azad, T.; Ling, M.; Hao, Y.; Snetsinger, B.; Khanal, P.; Minassian, L.M.; Graham, C.H.; Rauh, M.J.; Yang, X. The Hippo pathway component TAZ promotes immune evasion in human cancer through PD-L1. Cancer Res. 2018, 78, 1457–1470. [Google Scholar] [CrossRef] [PubMed]


| Construct | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| PTPN12-pTRIPZ | 5′-CGACCGGTGCCACCATGGAAG ACTACAAAGACGATGACGACAA GATGGAGCAAGTGGAGATCCTG-3′ | 5′-ACGACGCGTTCATGTCCATTC TGAAGGTG-3′ |
| PTPRR-pTRIPZ | 5′-CGACCGGTGCCACCATGGACTA CAAAGACGATGACGACAAGATGA TTCTTCACAGATTAAAAGAAAG-3′ | 5′-ACGACGCGTTCACTGGACAGT CTCTGCTG-3′ |
| DUSP1-pTRIPZ | 5′-CGACCGGTGCCACCATGGACTA CAAAGACGATGACGACAAGATGG TCATGGAAGTGGGCAC-3′ | 5′-ACGACGCGTTCAGCAGCTGGG AGAGGTCGTAATG-3′ |
| PTPDC1-pTRIPZ | 5′-CGACCGGTGCCACCATGGACTA CAAAGACGATGACGACAAGATGG CTGCAGGAGTCTTGCC-3′ | 5′-ACGACGCGTCTAGAGGCCAGG CTTAGGGC-3′ |
| Primary or Secondary | Protein | Antibody | Dilution | Company |
|---|---|---|---|---|
| Primary | PTPs | α-FLAG (M2) | 1:1000 | Sigma-Aldrich |
| Primary | ꞵ-actin | α-ꞵ-actin | 1:10,000 | Sigma-Aldrich |
| Primary | Cas9 | Cas9 (7A9-3A3) Mouse mAb | 1:1000 | Cell Signaling (Danvers, MA, USA) |
| Primary | PTPN12 | PTP-PEST Rabbit mAb | 1:1000 | Cell Signaling |
| Primary | Phospho-YAP | Phospho-YAP (Ser127)-mAb | 1:1000 | Cell Signaling |
| Primary | YAP/TAZ | YAP/TAZ Rabbit mAb | 1:1000 | Cell Signaling |
| Secondary | - | HRP goat α-mouse | 1:10,000 | Cell Signaling |
| Secondary | - | HRP goat α-rabbit | 1:10,000 | Cell Signaling |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarmasti Emami, S.; Ge, A.; Zhang, D.; Hao, Y.; Ling, M.; Rubino, R.; Nicol, C.J.B.; Wang, W.; Yang, X. Identification of PTPN12 Phosphatase as a Novel Negative Regulator of Hippo Pathway Effectors YAP/TAZ in Breast Cancer. Int. J. Mol. Sci. 2024, 25, 4064. https://doi.org/10.3390/ijms25074064
Sarmasti Emami S, Ge A, Zhang D, Hao Y, Ling M, Rubino R, Nicol CJB, Wang W, Yang X. Identification of PTPN12 Phosphatase as a Novel Negative Regulator of Hippo Pathway Effectors YAP/TAZ in Breast Cancer. International Journal of Molecular Sciences. 2024; 25(7):4064. https://doi.org/10.3390/ijms25074064
Chicago/Turabian StyleSarmasti Emami, Sahar, Anni Ge, Derek Zhang, Yawei Hao, Min Ling, Rachel Rubino, Christopher J. B. Nicol, Wenqi Wang, and Xiaolong Yang. 2024. "Identification of PTPN12 Phosphatase as a Novel Negative Regulator of Hippo Pathway Effectors YAP/TAZ in Breast Cancer" International Journal of Molecular Sciences 25, no. 7: 4064. https://doi.org/10.3390/ijms25074064







