Domains in Action: Understanding Ddi1’s Diverse Functions in the Ubiquitin-Proteasome System
Abstract
:1. Introduction
2. Ddi1, Rad23, and Dsk2: Shared Characteristics and Contrasts
3. Ddi1’s Atypical UBL Domain
4. Ubiquitin Recognition: Yeast UBA Domains and Human UIM Motifs
5. Combined Role of UBL and UBA
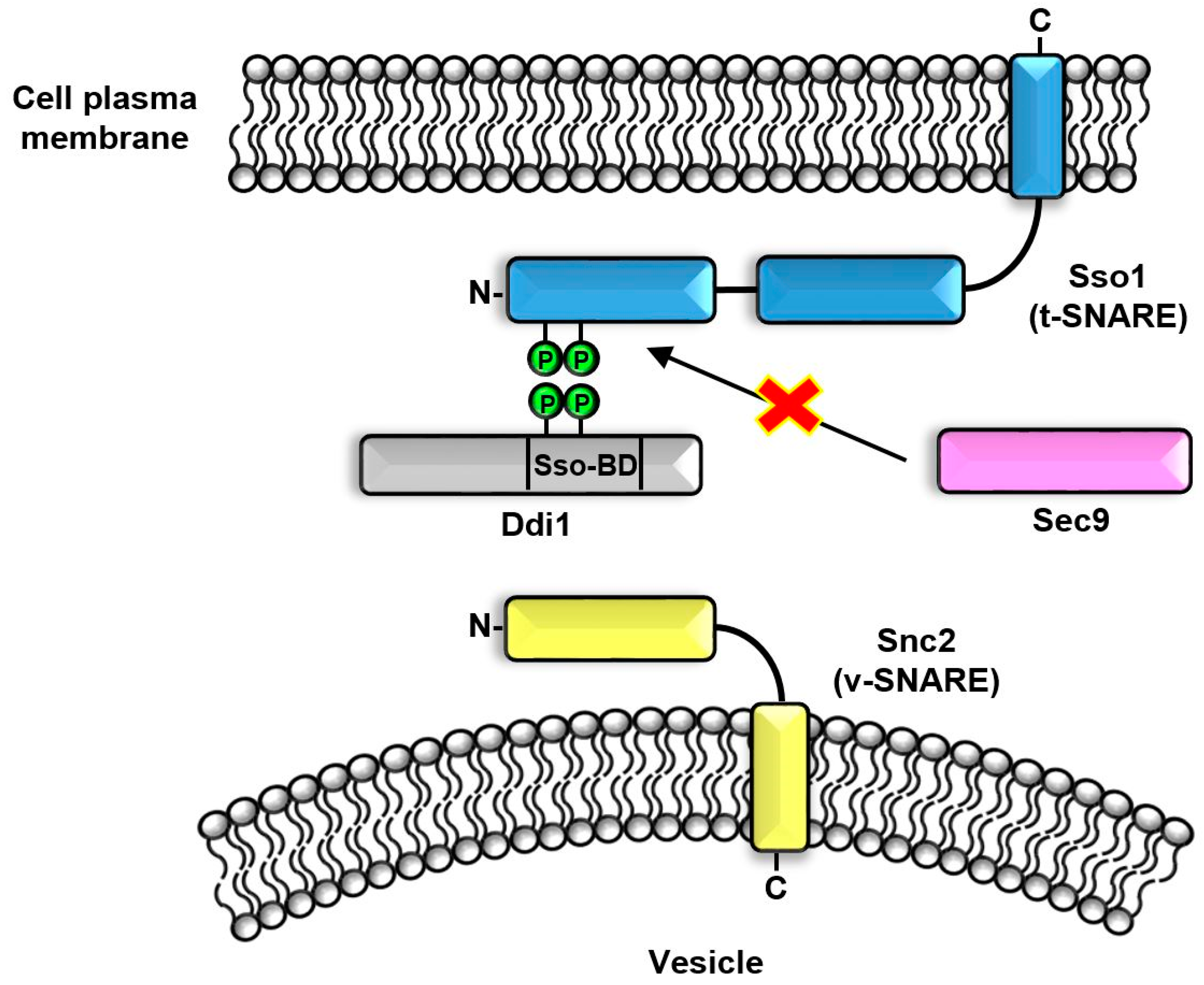
6. SSO-BD Domain
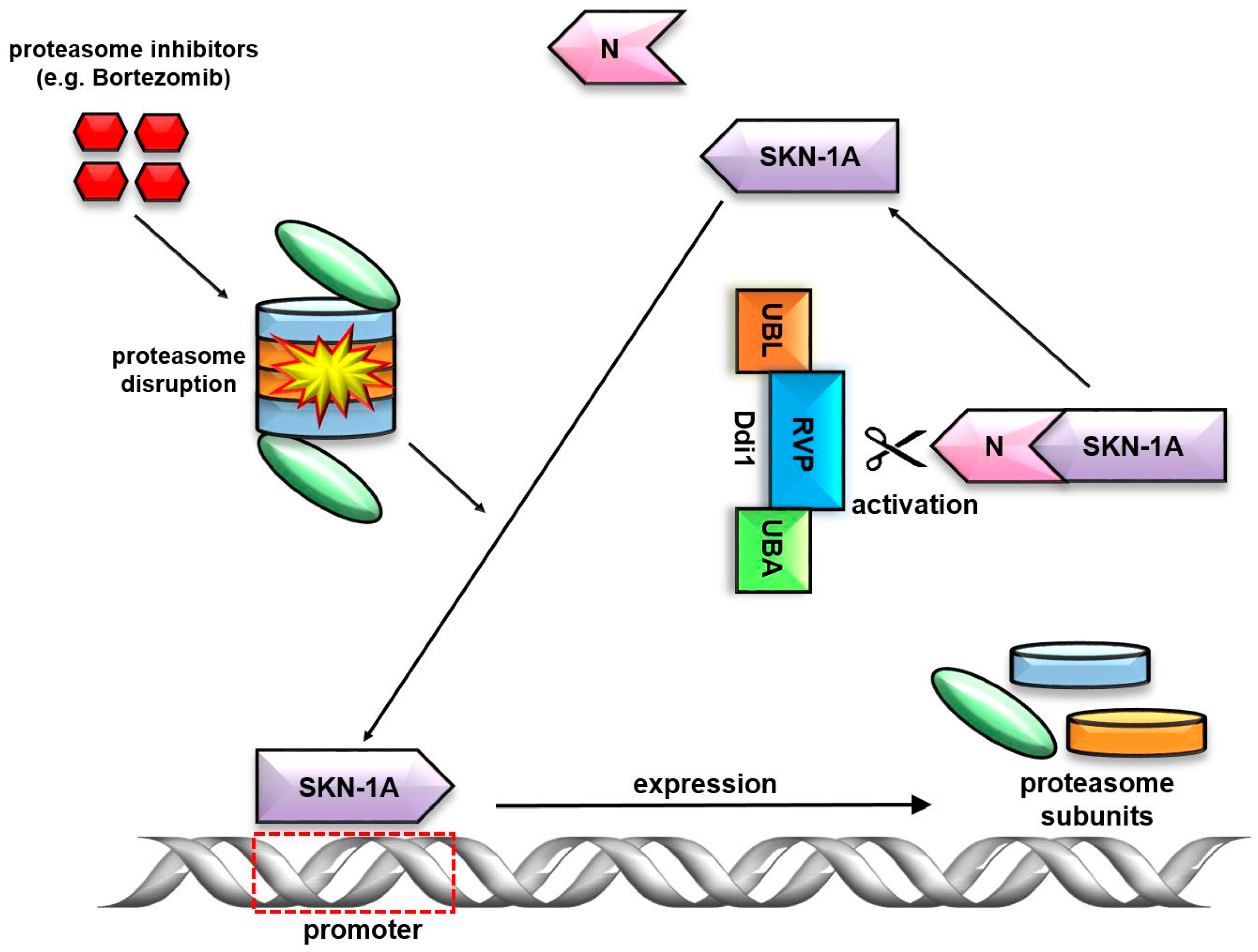
7. Ddi1’s Unique RVP Domain
8. The Helical Domain Ddi1 (HDD)
9. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef] [PubMed]
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef] [PubMed]
- Schwertman, P.; Bekker-Jensen, S.; Mailand, N. Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat. Rev. Mol. Cell Biol. 2016, 17, 379–394. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.M.; Finley, D.; Vogel, C. K63 polyubiquitination is a new modulator of the oxidative stress response. Nat. Struct. Mol. Biol. 2015, 22, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Lauwers, E.; Jacob, C.; André, B. K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J. Cell Biol. 2009, 185, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Erpapazoglou, Z.; Walker, O.; Haguenauer-Tsapis, R. Versatile roles of k63-linked ubiquitin chains in trafficking. Cells 2014, 3, 1027–1088. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M. Targeting of substrates to the 26S proteasome. FASEB J. 1997, 11, 1055–1066. [Google Scholar] [CrossRef] [PubMed]
- Kravtsova-Ivantsiv, Y.; Sommer, T.; Ciechanover, A. The lysine48-based polyubiquitin chain proteasomal signal: Not a single child anymore. Angew. Chem. Int. Ed. Engl. 2013, 52, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Husnjak, K.; Dikic, I. Ubiquitin-binding proteins: Decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 2012, 81, 291–322. [Google Scholar] [CrossRef]
- Wilkinson, C.R.; Seeger, M.; Hartmann-Petersen, R.; Stone, M.; Wallace, M.; Semple, C.; Gordon, C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol. 2001, 3, 939–943. [Google Scholar] [CrossRef]
- Saeki, Y.; Saitoh, A.; Toh-e, A.; Yokosawa, H. Ubiquitin-like proteins and Rpn10 play cooperative roles in ubiquitin-dependent proteolysis. Biochem. Biophys. Res. Commun. 2002, 293, 986–992. [Google Scholar] [CrossRef] [PubMed]
- Rao, H.; Sastry, A. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J. Biol. Chem. 2002, 277, 11691–11695. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Chen, X.; Elsasser, S.; Stocks, B.B.; Tian, G.; Lee, B.-H.; Shi, Y.; Zhang, N.; de Poot, S.A.H.; Tuebing, F.; et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 2016, 351, aad9421. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Chen, T.; Ziv, I.; Rosenzweig, R.; Matiuhin, Y.; Bronner, V.; Glickman, M.H.; Fushman, D. Together, Rpn10 and Dsk2 can serve as a polyubiquitin chain-length sensor. Mol. Cell 2009, 36, 1018–1033. [Google Scholar] [CrossRef] [PubMed]
- Husnjak, K.; Elsasser, S.; Zhang, N.; Chen, X.; Randles, L.; Shi, Y.; Hofmann, K.; Walters, K.J.; Finley, D.; Dikic, I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 2008, 453, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Elsasser, S.; Gali, R.R.; Schwickart, M.; Larsen, C.N.; Leggett, D.S.; Müller, B.; Feng, M.T.; Tübing, F.; Dittmar, G.A.; Finley, D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 2002, 4, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Lam, Y.A.; Lawson, T.G.; Velayutham, M.; Zweier, J.L.; Pickart, C.M. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 2002, 416, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Gomez, T.A.; Kolawa, N.; Gee, M.; Sweredoski, M.J.; Deshaies, R.J. Identification of a functional docking site in the Rpn1 LRR domain for the UBA-UBL domain protein Ddi1. BMC Biol. 2011, 9, 33. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, R.; Bronner, V.; Zhang, D.; Fushman, D.; Glickman, M.H. Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. J. Biol. Chem. 2012, 287, 14659–14671. [Google Scholar] [CrossRef]
- Liu, Y.; Xiao, W. Bidirectional regulation of two DNA-damage-inducible genes, MAG1 and DDI1, from Saccharomyces cerevisiae. Mol. Microbiol. 1997, 23, 777–789. [Google Scholar] [CrossRef]
- Gabriely, G.; Kama, R.; Gelin-Licht, R.; Gerst, J.E. Different domains of the UBL-UBA ubiquitin receptor, Ddi1/Vsm1, are involved in its multiple cellular roles. Mol. Biol. Cell 2008, 19, 3625–3637. [Google Scholar] [CrossRef] [PubMed]
- Lehrbach, N.J.; Ruvkun, G. Proteasome dysfunction triggers activation of SKN-1A/Nrf1 by the aspartic protease DDI-1. eLife 2016, 5, e17721. [Google Scholar] [CrossRef] [PubMed]
- Raasi, S.; Varadan, R.; Fushman, D.; Pickart, C.M. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat. Struct. Mol. Biol. 2005, 12, 708–714. [Google Scholar] [CrossRef]
- Varadan, R.; Assfalg, M.; Raasi, S.; Pickart, C.; Fushman, D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol. Cell 2005, 18, 687–698. [Google Scholar] [CrossRef]
- Hiyama, H.; Yokoi, M.; Masutani, C.; Sugasawa, K.; Maekawa, T.; Tanaka, K.; Hoeijmakers, J.H.J.; Hanaoka, F. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J. Biol. Chem. 1999, 274, 28019–28025. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Liu, S.; Zhang, Y.; Wang, Z. Roles of Rad23 protein in yeast nucleotide excision repair. Nucleic Acids Res. 2004, 32, 5981–5990. [Google Scholar] [CrossRef]
- Masutani, C.; Araki, M.; Sugasawa, K.; van der Spek, P.J.; Yamada, A.; Uchida, A.; Maekawa, T.; Bootsma, D.; Hoeijmakers, J.H.J.; Hanaoka, F. Identification and characterization of XPC-binding domain of hHR23B. Mol. Cell Biol. 1997, 17, 6915–6923. [Google Scholar] [CrossRef]
- Sugasawa, K.; Ng, J.M.Y.; Masutani, C.; Maekawa, T.; Uchida, A.; van der Spek, P.J.; Eker, A.P.M.; Rademakers, S.; Visser, C.; Aboussekhra, A.; et al. Two human homologs of Rad23 are functionally interchangeable in complex formation and stimulation of XPC repair activity. Mol. Cell Biol. 1997, 17, 6924–6931. [Google Scholar] [CrossRef]
- Cheon, N.Y.; Kim, H.S.; Yeo, J.E.; Schärer, O.D.; Lee, J.Y. Single-molecule visualization reveals the damage search mechanism for the human NER protein XPC-RAD23B. Nucleic Acids Res. 2019, 47, 8337–8347. [Google Scholar] [CrossRef]
- Bergink, S.; Toussaint, W.; Luijsterburg, M.S.; Dinant, C.; Alekseev, S.; Hoeijmakers, J.H.; Dantuma, N.P.; Houtsmuller, A.B.; Vermeulen, W. Recognition of DNA damage by XPC coincides with disruption of the XPC-RAD23 complex. J. Cell Biol. 2012, 196, 681–688. [Google Scholar] [CrossRef]
- Ng, J.M.; Vermeulen, W.; van der Horst, G.T.; Bergink, S.; Sugasawa, K.; Vrieling, H.; Hoeijmakers, J.H. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Genes Dev. 2003, 17, 1630–1645. [Google Scholar] [CrossRef] [PubMed]
- Medicherla, B.; Kostova, Z.; Schaefer, A.; Wolf, D.H. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 2004, 5, 692–697. [Google Scholar] [CrossRef] [PubMed]
- Berner, N.; Reutter, K.R.; Wolf, D.H. Protein Quality Control of the Endoplasmic Reticulum and Ubiquitin-Proteasome-Triggered Degradation of Aberrant Proteins: Yeast Pioneers the Path. Annu. Rev. Biochem. 2018, 87, 751–782. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.S.; Uehara, T.; Tsuruma, K.; Nomura, Y. Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett. 2004, 566, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Hartmann-Petersen, R.; Hendil, K.B.; Gordon, C. Ubiquitin binding proteins protect ubiquitin conjugates from disassembly. FEBS Lett. 2003, 535, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.; Mi, K.; Rao, H. Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol. Biol. Cell 2004, 15, 3357–3365. [Google Scholar] [CrossRef] [PubMed]
- Kaplun, L.; Tzirkin, R.; Bakhrat, A.; Shabek, N.; Ivantsiv, Y.; Raveh, D. The DNA damage-inducible UbL-UbA protein Ddi1 participates in Mec1-mediated degradation of Ho endonuclease. Mol. Cell Biol. 2005, 25, 5355–5362. [Google Scholar] [CrossRef] [PubMed]
- Lassmann, T.; Frings, O.; Sonnhammer, E.L. Kalign2: High-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res. 2009, 37, 858–865. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Nowicka, U.; Zhang, D.; Walker, O.; Krutauz, D.; Castañeda, C.A.; Chaturvedi, A.; Chen, T.Y.; Reis, N.; Glickman, M.H.; Fushman, D. DNA-damage-inducible 1 protein (Ddi1) contains an uncharacteristic ubiquitin-like domain that binds ubiquitin. Structure 2015, 23, 542–557. [Google Scholar] [CrossRef]
- Sivá, M.; Svoboda, M.; Veverka, V.; Trempe, J.-F.; Hofmann, K.; Kožíšek, M.; Hexnerová, R.; Sedlák, F.; Belza, J.; Brynda, J.; et al. Human DNA-Damage-Inducible 2 Protein Is Structurally and Functionally Distinct from Its Yeast Ortholog. Sci. Rep. 2016, 6, 30443. [Google Scholar] [CrossRef] [PubMed]
- Bertolaet, B.L.; Clarke, D.J.; Wolff, M.; Watson, M.H.; Henze, M.; Divita, G.; Reed, S.I. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Biol. 2001, 8, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Zhang, N.; Koepp, D.M.; Walters, K.J. Ubiquitin receptor proteins hHR23a and hPLIC2 interact. J. Mol. Biol. 2007, 365, 1093–1101. [Google Scholar] [CrossRef] [PubMed]
- Trempe, J.-F.; Brown, N.R.; Lowe, E.D.; Gordon, C.; Campbell, I.D.; Noble, M.E.M.; Endicott, J.A. Mechanism of Lys48-linked polyubiquitin chain recognition by the Mud1 UBA domain. EMBO J. 2005, 24, 3178–3189. [Google Scholar] [CrossRef] [PubMed]
- Heinen, C.; Acs, K.; Hoogstraten, D.; Dantuma, N.P. C-terminal UBA domains protect ubiquitin receptors by preventing initiation of protein degradation. Nat. Commun. 2011, 2, 191. [Google Scholar] [CrossRef]
- Kang, Y.; Vossler, R.A.; Diaz-Martinez, L.A.; Winter, N.S.; Clarke, D.J.; Walters, K.J. UBL/UBA ubiquitin receptor proteins bind a common tetraubiquitin chain. J. Mol. Biol. 2006, 356, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Dang, F.; Nie, L.; Wei, W. Ubiquitin signaling in cell cycle control and tumorigenesis. Cell Death Differ. 2021, 28, 427–438. [Google Scholar] [CrossRef]
- Kaplun, L.; Ivantsiv, Y.; Bakhrat, A.; Raveh, D. DNA damage response-mediated degradation of Ho endonuclease via the ubiquitin system involves its nuclear export. J. Biol. Chem. 2003, 278, 48727–48734. [Google Scholar] [CrossRef]
- Kaplun, L.; Ivantsiv, Y.; Bakhrat, A.; Tzirkin, R.; Baranes, K.; Shabek, N.; Raveh, D. The F-box protein, Ufo1, maintains genome stability by recruiting the yeast mating switch endonuclease, Ho, for rapid proteasome degradation. Isr. Med. Assoc. J. 2006, 8, 246–248. [Google Scholar] [PubMed]
- Ivantsiv, Y.; Kaplun, L.; Tzirkin-Goldin, R.; Shabek, N.; Raveh, D. Unique role for the UbL-UbA protein Ddi1 in turnover of SCFUfo1 complexes. Mol. Cell Biol. 2006, 26, 1579–1588. [Google Scholar] [CrossRef]
- Voloshin, O.; Bakhrat, A.; Herrmann, S.; Raveh, D. Transfer of Ho endonuclease and Ufo1 to the proteasome by the UbL-UbA shuttle protein, Ddi1, analysed by complex formation in vitro. PLoS ONE 2012, 7, e39210. [Google Scholar] [CrossRef]
- Kama, R.; Gabriely, G.; Kanneganti, V.; Gerst, J.E. Cdc48 and ubiquilins confer selective anterograde protein sorting and entry into the multivesicular body in yeast. Mol. Biol. Cell 2018, 29, 948–963. [Google Scholar] [CrossRef] [PubMed]
- Lustgarten, V.; Gerst, J.E. Yeast VSM1 encodes a v-SNARE binding protein that may act as a negative regulator of constitutive exocytosis. Mol. Cell Biol. 1999, 19, 4480–4494. [Google Scholar] [CrossRef]
- Marash, M.; Gerst, J.E. Phosphorylation of the autoinhibitory domain of the Sso t-SNAREs promotes binding of the Vsm1 SNARE regulator in yeast. Mol. Biol. Cell 2003, 14, 3114–3125. [Google Scholar] [CrossRef] [PubMed]
- Krylov, D.M.; Koonin, E.V. A novel family of predicted retroviral-like aspartyl proteases with a possible key role in eukaryotic cell cycle control. Curr. Biol. 2001, 11, R584–R587. [Google Scholar] [CrossRef]
- Trempe, J.-F.; Šašková, K.G.; Sivá, M.; Ratcliffe, C.D.H.; Veverka, V.; Hoegl, A.; Ménade, M.; Feng, X.; Shenker, S.; Svoboda, M.; et al. Structural studies of the yeast DNA damage-inducible protein Ddi1 reveal domain architecture of this eukaryotic protein family. Sci. Rep. 2016, 6, 33671. [Google Scholar] [CrossRef] [PubMed]
- White, R.E.; Powell, D.J.; Berry, C. HIV proteinase inhibitors target the Ddi1-like protein of Leishmania parasites. FASEB J. 2011, 25, 1729–1736. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Martínez, L.A.; Kang, Y.; Walters, K.J.; Clarke, D.J. Yeast UBL-UBA proteins have partially redundant functions in cell cycle control. Cell Div. 2006, 1, 28. [Google Scholar] [CrossRef]
- Clarke, D.J.; Mondesert, G.; Segal, M.; Bertolaet, B.L.; Jensen, S.; Wolff, M.; Henze, M.; Reed, S.I. Dosage suppressors of pds1 implicate ubiquitin-associated domains in checkpoint control. Mol. Cell Biol. 2001, 21, 1997–2007. [Google Scholar] [CrossRef]
- Ciosk, R.; Zachariae, W.; Michaelis, C.; Shevchenko, A.; Mann, M.; Nasmyth, K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 1998, 93, 1067–1076. [Google Scholar] [CrossRef]
- Sha, Z.; Goldberg, A.L. Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Curr. Biol. 2014, 24, 1573–1583. [Google Scholar] [CrossRef] [PubMed]
- Vangala, J.R.; Sotzny, F.; Krüger, E.; Deshaies, R.J.; Radhakrishnan, S.K. Nrf1 can be processed and activated in a proteasome-independent manner. Curr. Biol. 2016, 26, R834–R835. [Google Scholar] [CrossRef] [PubMed]
- Dirac-Svejstrup, A.B.; Walker, J.; Faull, P.; Encheva, V.; Akimov, V.; Puglia, M.; Perkins, D.; Kümper, S.; Hunjan, S.S.; Blagoev, B.; et al. DDI2 Is a Ubiquitin-Directed Endoprotease Responsible for Cleavage of Transcription Factor NRF1. Mol. Cell 2020, 79, 332–341.e7. [Google Scholar] [CrossRef] [PubMed]
- Northrop, A.; Vangala, J.R.; Feygin, A.; Radhakrishnan, S.K. Disabling the Protease DDI2 Attenuates the Transcriptional Activity of NRF1 and Potentiates Proteasome Inhibitor Cytotoxicity. Int. J. Mol. Sci. 2020, 21, 327. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, S.; Irie, T.; Hirayama, S.; Sakurai, Y.; Yashiroda, H.; Naguro, I.; Ichijo, H.; Hamazaki, J.; Murata, S. The aspartyl protease DDI2 activates Nrf1 to compensate for proteasome dysfunction. eLife 2016, 5, e18357. [Google Scholar] [CrossRef] [PubMed]
- Yip, M.C.J.; Bodnar, N.O.; Rapoport, T.A. Ddi1 is a ubiquitin-dependent protease. Proc. Natl. Acad. Sci. USA 2020, 117, 7776–7781. [Google Scholar] [CrossRef] [PubMed]
- Kottemann, M.C.; Conti, B.A.; Lach, F.P.; Smogorzewska, A. Removal of RTF2 from Stalled Replisomes Promotes Maintenance of Genome Integrity. Mol. Cell 2018, 69, 24–35.e5. [Google Scholar] [CrossRef] [PubMed]
- Conti, B.A.; Smogorzewska, A. Mechanisms of direct replication restart at stressed replisomes. DNA Repair. 2020, 95, 102947. [Google Scholar] [CrossRef]
- Conti, B.A.; Ruiz, P.D.; Broton, C.; Blobel, N.J.; Kottemann, M.C.; Sridhar, S.; Lach, F.P.; Wiley, T.F.; Sasi, N.K.; Carroll, T.; et al. RTF2 controls replication repriming and ribonucleotide excision at the replisome. bioRxiv 2023. [Google Scholar] [CrossRef]
- Svoboda, M.; Konvalinka, J.; Trempe, J.F.; Grantz Saskova, K. The yeast proteases Ddi1 and Wss1 are both involved in the DNA replication stress response. DNA Repair. 2019, 80, 45–51. [Google Scholar] [CrossRef]
- Serbyn, N.; Noireterre, A.; Bagdiul, I.; Plank, M.; Michel, A.H.; Loewith, R.; Kornmann, B.; Stutz, F. The Aspartic Protease Ddi1 Contributes to DNA-Protein Crosslink Repair in Yeast. Mol. Cell 2020, 77, 1066–1079.e9. [Google Scholar] [CrossRef] [PubMed]
- Noireterre, A.; Serbyn, N.; Bagdiul, I.; Stutz, F. Ubx5-Cdc48 assists the protease Wss1 at DNA-protein crosslink sites in yeast. EMBO J. 2023, 42, e113609. [Google Scholar] [CrossRef] [PubMed]
- Waite, K.A.; Vontz, G.; Lee, S.Y.; Roelofs, J. Proteasome condensate formation is driven by multivalent interactions with shuttle factors and K48-linked ubiquitin chains. Preprint. bioRxiv 2023. [Google Scholar] [CrossRef]
- Liu, Z.; Guo, X.; Guo, A.; Zhang, S.; Zou, Y.; Wang, Y.; Li, X.; He, W.; Pu, L.; Zhang, S.; et al. HIV protease inhibitor nelfinavir is a potent drug candidate against echinococcosis by targeting Ddi1-like protein. EBioMedicine 2022, 82, 104177. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fabijan, A.; Polis, B.; Zawadzka-Fabijan, A.; Korabiewska, I.; Zakrzewski, K.; Nowosławska, E.; Chojnacki, M. Domains in Action: Understanding Ddi1’s Diverse Functions in the Ubiquitin-Proteasome System. Int. J. Mol. Sci. 2024, 25, 4080. https://doi.org/10.3390/ijms25074080
Fabijan A, Polis B, Zawadzka-Fabijan A, Korabiewska I, Zakrzewski K, Nowosławska E, Chojnacki M. Domains in Action: Understanding Ddi1’s Diverse Functions in the Ubiquitin-Proteasome System. International Journal of Molecular Sciences. 2024; 25(7):4080. https://doi.org/10.3390/ijms25074080
Chicago/Turabian StyleFabijan, Artur, Bartosz Polis, Agnieszka Zawadzka-Fabijan, Izabela Korabiewska, Krzysztof Zakrzewski, Emilia Nowosławska, and Michał Chojnacki. 2024. "Domains in Action: Understanding Ddi1’s Diverse Functions in the Ubiquitin-Proteasome System" International Journal of Molecular Sciences 25, no. 7: 4080. https://doi.org/10.3390/ijms25074080




