Fibrocyte Phenotype of ENTPD1+CD55+ Cells and Its Association with Pain in Osteoarthritic Synovium
Abstract
:1. Introduction
2. Results
2.1. CD55+ Population in the Osteoarthritic Synovium
2.2. Characterization of ENTPD1+CD55+ and ENTPD1−CD55+ Using LC/MS and qPCR
2.3. Correlation between Proportion of ENTPD1+CD55+ and OA Pathology
3. Discussion
4. Materials and Methods
4.1. Recruitment of Patients and Study Criteria
4.2. Cell Sorting
4.3. LC/MS Analysis
Quantitative Real-Time PCR (qRT-PCR) Analysis
4.4. Correlation between Proportion of ENTPD1+CD55+ Cells and OA Pathology
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perruccio, A.V.; Young, J.J.; Wilfong, J.M.; Denise Power, J.; Canizares, M.; Badley, E.M. Osteoarthritis year in review 2023: Epidemiology & therapy. Osteoarthr. Cartil. 2024, 32, 159–165. [Google Scholar]
- Salman, L.A.; Ahmed, G.; Dakin, S.G.; Kendrick, B.; Price, A. Osteoarthritis: A narrative review of molecular approaches to disease management. Arthritis Res. Ther. 2023, 25, 27. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, Y.; Uchida, K.; Fukushima, K.; Inoue, G.; Takaso, M. Mechanisms of Peripheral and Central Sensitization in Osteoarthritis Pain. Cureus 2023, 15, e35331. [Google Scholar] [CrossRef] [PubMed]
- Perrot, S.; Anne-Priscille, T. Pain in osteoarthritis from a symptom to a disease. Best. Pract. Res. Clin. Rheumatol. 2023, 37, 101825. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Hu, Y.; Zhang, H.; Wang, S.; Xu, K.; Su, J. Single-cell RNA sequencing in osteoarthritis. Cell Prolif. 2023, 56, e13517. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Bhutani, N. Profiling joint tissues at single-cell resolution: Advances and insights. Nat. Rev. Rheumatol. 2024, 20, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Karpus, O.N.; Kiener, H.P.; Niederreiter, B.; Yilmaz-Elis, A.S.; van der Kaa, J.; Ramaglia, V.; Arens, R.; Smolen, J.S.; Botto, M.; Tak, P.P.; et al. CD55 deposited on synovial collagen fibers protects from immune complex-mediated arthritis. Arthritis Res. Ther. 2015, 17, 6. [Google Scholar] [CrossRef] [PubMed]
- McHugh, J. Functionally distinct fibroblast subsets in RA. Nat. Rev. Rheumatol. 2019, 15, 449. [Google Scholar] [CrossRef]
- Nygaard, G.; Firestein, G.S. Restoring synovial homeostasis in rheumatoid arthritis by targeting fibroblast-like synoviocytes. Nat. Rev. Rheumatol. 2020, 16, 316–333. [Google Scholar] [CrossRef]
- Vassallo, V.; Stellavato, A.; Russo, R.; Cimini, D.; Valletta, M.; Alfano, A.; Pedone, P.V.; Chambery, A.; Schiraldi, C. Molecular Fingerprint of Human Pathological Synoviocytes in Response to Extractive Sulfated and Biofermentative Unsulfated Chondroitins. Int. J. Mol. Sci. 2022, 23, 5865. [Google Scholar] [CrossRef]
- Chen, H.W.; Huang, C.H.; Huang, C.F.; Chang, C.H.; Liao, H.J. Distinct subsets of synovial fibroblasts control cartilage destruction in joint diseases. Clin. Exp. Rheumatol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, M.; Ohashi, Y.; Kodera, Y.; Satoh, M.; Matsui, T.; Fukushima, K.; Iwase, D.; Aikawa, J.; Mukai, M.; Inoue, G.; et al. CD39+CD55− Fb Subset Exhibits Myofibroblast-Like Phenotype and Is Associated with Pain in Osteoarthritis of the Knee. Biomedicines 2023, 11, 3047. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, C.; Wang, T.; Li, S.; Bai, Y.; Pan, F.; Wang, J.; Han, J.; Luo, R.; Wan, X.; et al. Single-cell communication patterns and their intracellular information flow in synovial fibroblastic osteoarthritis and rheumatoid arthritis. Immunol. Lett. 2023, 263, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, J.W.; Breuer, C.K. Fibrocytes: A Critical Review and Practical Guide. Front. Immunol. 2021, 12, 784401. [Google Scholar] [CrossRef] [PubMed]
- Elhaj Mahmoud, D.; Kaabachi, W.; Sassi, N.; Mokhtar, A.; Ben Ammar, L.; Rekik, S.; Tarhouni, L.; Kallel-Sellami, M.; Cheour, E.; Laadhar, L. Expression of extracellular matrix components and cytokine receptors in human fibrocytes during rheumatoid arthritis. Connect. Tissue Res. 2021, 62, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Galligan, C.L.; Fish, E.N. The role of circulating fibrocytes in inflammation and autoimmunity. J. Leukoc. Biol. 2013, 93, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Heukels, P.; van Hulst, J.A.C.; van Nimwegen, M.; Boorsma, C.E.; Melgert, B.N.; van den Toorn, L.M.; Boomars, K.A.T.; Wijsenbeek, M.S.; Hoogsteden, H.; von der Thusen, J.H.; et al. Fibrocytes are increased in lung and peripheral blood of patients with idiopathic pulmonary fibrosis. Respir. Res. 2018, 19, 90. [Google Scholar] [CrossRef] [PubMed]
- Quan, T.E.; Cowper, S.E.; Bucala, R. The role of circulating fibrocytes in fibrosis. Curr. Rheumatol. Rep. 2006, 8, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Allard, B.; Longhi, M.S.; Robson, S.C.; Stagg, J. The ectonucleotidases CD39 and CD73: Novel checkpoint inhibitor targets. Immunol. Rev. 2017, 276, 121–144. [Google Scholar] [CrossRef]
- Jiang, X.; Wu, X.; Xiao, Y.; Wang, P.; Zheng, J.; Wu, X.; Jin, Z. The ectonucleotidases CD39 and CD73 on T cells: The new pillar of hematological malignancy. Front. Immunol. 2023, 14, 1110325. [Google Scholar] [CrossRef]
- Huang, X.; Gu, S.; Liu, C.; Zhang, L.; Zhang, Z.; Zhao, Y.; Khoong, Y.; Li, H.; Gao, Y.; Liu, Y.; et al. CD39+ Fibroblasts Enhance Myofibroblast Activation by Promoting IL-11 Secretion in Hypertrophic Scars. J. Investig. Dermatol. 2022, 142, 1065–1076.e19. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, M.A.; Diaz-Torne, C.; De Agustin, J.J.; Estrada, P.; Reina, D.; Hernandez, M.V.; Sang, H.Y.; Zamora, C.; Canto, E.; Corominas, H.; et al. Altered CD39 and CD73 Expression in Rheumatoid Arthritis: Implications for Disease Activity and Treatment Response. Biomolecules 2024, 14, 1. [Google Scholar] [CrossRef]
- Chen, N.; Fan, B.; He, Z.; Yu, X.; Wang, J. Identification of HBEGF+ fibroblasts in the remission of rheumatoid arthritis by integrating single-cell RNA sequencing datasets and bulk RNA sequencing datasets. Arthritis Res. Ther. 2022, 24, 215. [Google Scholar] [CrossRef] [PubMed]
- Ji, Q.; Zheng, Y.; Zhang, G.; Hu, Y.; Fan, X.; Hou, Y.; Wen, L.; Li, L.; Xu, Y.; Wang, Y.; et al. Single-cell RNA-seq analysis reveals the progression of human osteoarthritis. Ann. Rheum. Dis. 2019, 78, 100–110. [Google Scholar] [CrossRef]
- Zhang, F.; Wei, K.; Slowikowski, K.; Fonseka, C.Y.; Rao, D.A.; Kelly, S.; Goodman, S.M.; Tabechian, D.; Hughes, L.B.; Salomon-Escoto, K.; et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 2019, 20, 928–942. [Google Scholar] [CrossRef]
- Pilling, D.; Fan, T.; Huang, D.; Kaul, B.; Gomer, R.H. Identification of markers that distinguish monocyte-derived fibrocytes from monocytes, macrophages, and fibroblasts. PLoS ONE 2009, 4, e7475. [Google Scholar] [CrossRef]
- Smith, T.J. Potential role for bone marrow-derived fibrocytes in the orbital fibroblast heterogeneity associated with thyroid-associated ophthalmopathy. Clin. Exp. Immunol. 2010, 162, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Just, S.A.; Nielsen, C.; Werlinrud, J.C.; Larsen, P.V.; Hejbol, E.K.; Tenstad, H.B.; Daa Schroder, H.; Barington, T.; Torfing, T.; Humby, F.; et al. Fibrocytes in early and long-standing rheumatoid arthritis: A 6-month trial with repeated synovial biopsy, imaging and lung function test. RMD Open 2021, 7, e001494. [Google Scholar] [CrossRef] [PubMed]
- Knights, A.J.; Redding, S.J.; Maerz, T. Inflammation in osteoarthritis: The latest progress and ongoing challenges. Curr. Opin. Rheumatol. 2023, 35, 128–134. [Google Scholar] [CrossRef]
- van den Bosch, M.H.J.; Blom, A.B.; van der Kraan, P.M. Inflammation in osteoarthritis: Our view on its presence and involvement in disease development over the years. Osteoarthr. Cartil. 2023, 32, 355–364. [Google Scholar] [CrossRef]
- Lee, D.G.; Woo, J.W.; Kwok, S.K.; Cho, M.L.; Park, S.H. MRP8 promotes Th17 differentiation via upregulation of IL-6 production by fibroblast-like synoviocytes in rheumatoid arthritis. Exp. Mol. Med. 2013, 45, e20. [Google Scholar] [CrossRef] [PubMed]
- Cesaro, A.; Anceriz, N.; Plante, A.; Page, N.; Tardif, M.R.; Tessier, P.A. An inflammation loop orchestrated by S100A9 and calprotectin is critical for development of arthritis. PLoS ONE 2012, 7, e45478. [Google Scholar] [CrossRef] [PubMed]
- Austermann, J.; Zenker, S.; Roth, J. S100-alarmins: Potential therapeutic targets for arthritis. Expert Opin. Ther. Targets 2017, 21, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Kambayashi, T.; Laufer, T.M. Atypical MHC class II-expressing antigen-presenting cells: Can anything replace a dendritic cell? Nat. Rev. Immunol. 2014, 14, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Unanue, E.R.; Turk, V.; Neefjes, J. Variations in MHC Class II Antigen Processing and Presentation in Health and Disease. Annu. Rev. Immunol. 2016, 34, 265–297. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Grieshaber-Bouyer, R.; Rao, D.A.; Kolb, P.; Chen, H.; Andreeva, I.; Tretter, T.; Lorenz, H.M.; Watzl, C.; Wabnitz, G.; et al. Effect of JAK Inhibition on the Induction of Proinflammatory HLA-DR+CD90+ Rheumatoid Arthritis Synovial Fibroblasts by Interferon-gamma. Arthritis Rheumatol. 2022, 74, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Layton, T.B.; Williams, L.; McCann, F.; Zhang, M.; Fritzsche, M.; Colin-York, H.; Cabrita, M.; Ng, M.T.H.; Feldmann, M.; Sansom, S.N.; et al. Cellular census of human fibrosis defines functionally distinct stromal cell types and states. Nat. Commun. 2020, 11, 2768. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Chen, Z.; Guo, H.; Wang, Z.; Sun, K.; Yang, X.; Zhao, X.; Ma, L.; Wang, J.; Meng, Z.; et al. Extensive cytokine analysis in synovial fluid of osteoarthritis patients. Cytokine 2021, 143, 155546. [Google Scholar] [CrossRef] [PubMed]
- Ciregia, F.; Deroyer, C.; Cobraiville, G.; Plener, Z.; Malaise, O.; Gillet, P.; Fillet, M.; Malaise, M.G.; de Seny, D. Modulation of alphaVbeta6 integrin in osteoarthritis-related synovitis and the interaction with VTN(381–397 a.a.) competing for TGF-beta1 activation. Exp. Mol. Med. 2021, 53, 210–222. [Google Scholar] [CrossRef]
- Lundblad, H.; Kreicbergs, A.; Jansson, K.A. Prediction of persistent pain after total knee replacement for osteoarthritis. J. Bone Jt. Surg. Br. 2008, 90, 166–171. [Google Scholar] [CrossRef]
- Ohashi, Y.; Fukushima, K.; Inoue, G.; Uchida, K.; Koyama, T.; Tsuchiya, M.; Uchiyama, K.; Takahira, N.; Takaso, M. Central sensitization inventory scores correlate with pain at rest in patients with hip osteoarthritis: A retrospective study. BMC Musculoskelet. Disord. 2020, 21, 595. [Google Scholar] [CrossRef] [PubMed]
- Power, J.D.; Perruccio, A.V.; Gandhi, R.; Veillette, C.; Davey, J.R.; Syed, K.; Mahomed, N.N.; Rampersaud, Y.R. Neuropathic pain in end-stage hip and knee osteoarthritis: Differential associations with patient-reported pain at rest and pain on activity. Osteoarthr. Cartil. 2018, 26, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xing, R.; Huang, Z.; Ding, L.; Zhang, L.; Li, M.; Li, X.; Wang, P.; Mao, J. Synovial Fibrosis Involvement in Osteoarthritis. Front. Med. 2021, 8, 684389. [Google Scholar] [CrossRef] [PubMed]
- Barabas, K.; Bakos, J.; Zeitler, Z.; Balint, G.; Nagy, E.; Lakatos, T.; Kekesi, A.K.; Gaspar, L.; Szekanecz, Z. Effects of laser treatment on the expression of cytosolic proteins in the synovium of patients with osteoarthritis. Lasers Surg. Med. 2014, 46, 644–649. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Ruiz, R.; Avila, J.; Lopez-Bote, J.P.; Bernabeu, C.; Larraga, V. Decreased tubulin synthesis in synoviocytes from adjuvant-induced arthritic rats. Biochim. Biophys. Acta 1992, 1138, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Yuan, Z.; Ma, Z.; Song, J.; Xie, X.; Chen, Y. KEGG-PATH: Kyoto encyclopedia of genes and genomes-based pathway analysis using a path analysis model. Mol. Biosyst. 2014, 10, 2441–2447. [Google Scholar] [CrossRef] [PubMed]
- Kellgren, J.H.; Lawrence, J.S. Radiological assessment of osteo-arthrosis. Ann. Rheum. Dis. 1957, 16, 494–502. [Google Scholar] [CrossRef] [PubMed]
- Chan, E.F.; Cockman, M.D.; Goel, P.; Newman, P.S.; Hipp, J.A. Characterization of the mid-coronal plane method for measurement of radiographic change in knee joint space width across different levels of image parallax. Osteoarthr. Cartil. 2021, 29, 1306–1313. [Google Scholar] [CrossRef]
- Ohashi, Y.; Uchida, K.; Fukushima, K.; Satoh, M.; Koyama, T.; Tsuchiya, M.; Saito, H.; Uchiyama, K.; Takahira, N.; Inoue, G.; et al. Increased Synovial CD14 mRNA Expression and Proportion of CD14(high) Subsets in Early-Stage Hip Osteoarthritis: Propensity Matched Score Analysis. Int. J. Mol. Sci. 2022, 23, 3622. [Google Scholar] [CrossRef]

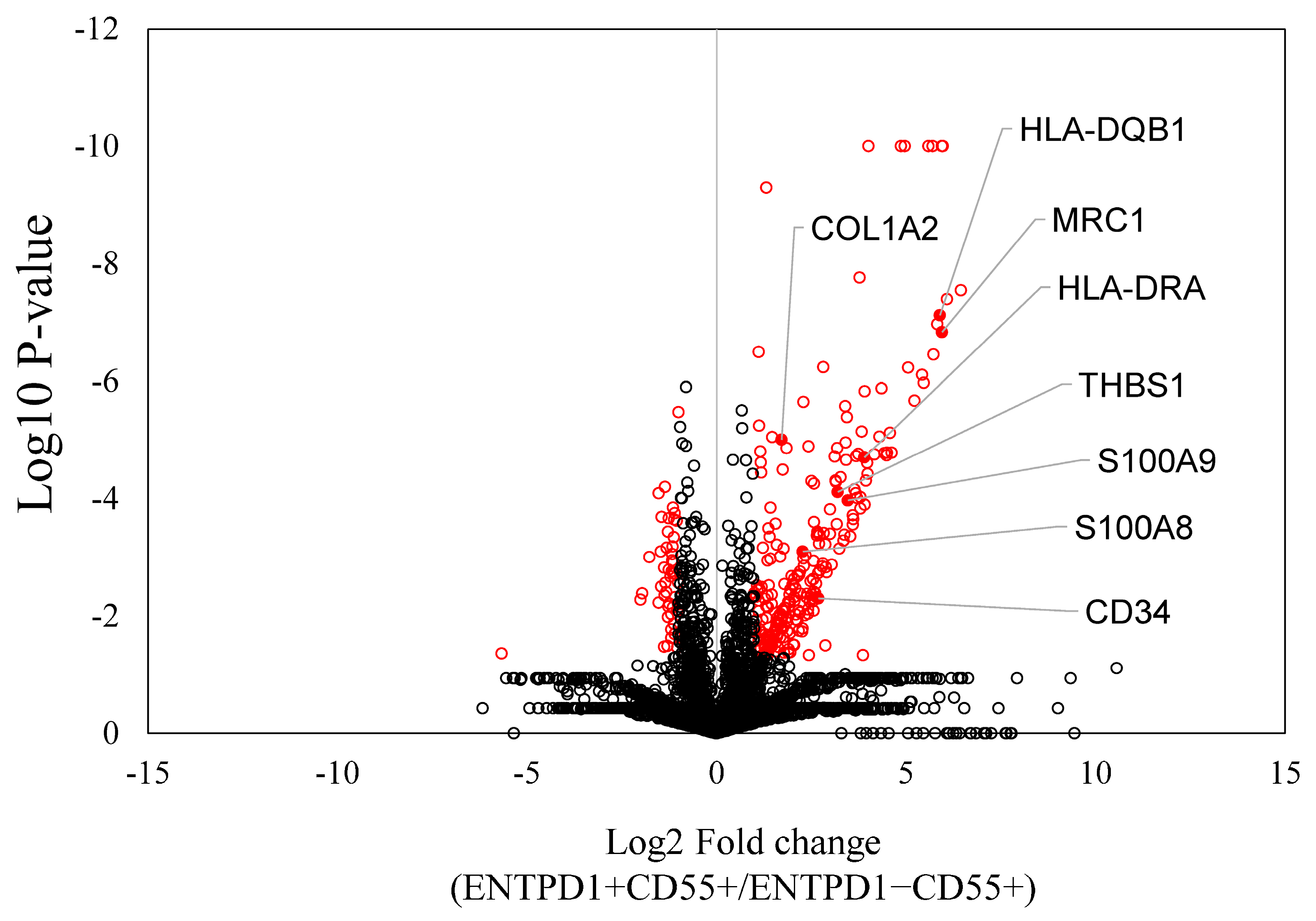
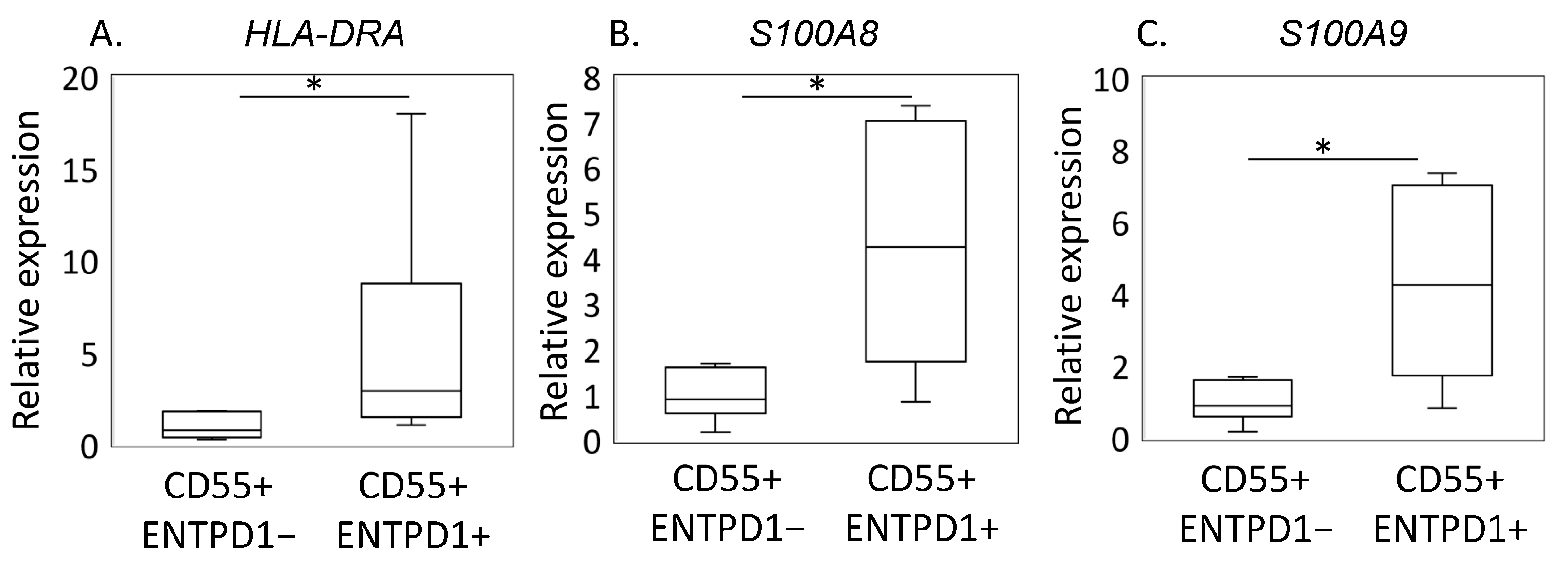

| Protein ID | Protein Name | Log2FC | p-Value | Pathway |
|---|---|---|---|---|
| P07711 | Cathepsin L | 1.596 | 0.001 | Rheum |
| P16070 | CD44 | 1.689 | 0.001 | ECM |
| P08123 | Collagen alpha-2(I) chain | 1.720 | 0.000 | ECM |
| P02458 | Collagen alpha-1(II) chain | 2.544 | 0.006 | ECM |
| P49747 | Cartilage oligomeric matrix protein | 2.673 | 0.000 | ECM |
| P01920 | HLA-DQB1 | 5.901 | 0.000 | Rheum |
| P01903 | HLA-DRA | 3.904 | 0.000 | Rheum |
| P05362 | Intercellular adhesion molecule 1 | 1.425 | 0.001 | Rheum |
| P23229 | Integrin alpha-6 | 1.122 | 0.008 | ECM |
| P16144 | Integrin beta-4 | 5.714 | 0.000 | ECM |
| O15230 | Laminin subunit alpha-5 | 3.868 | 0.047 | ECM |
| P61421 | V-type proton ATPase subunit d 1 | 1.331 | 0.011 | Rheum |
| O75348 | V-type proton ATPase subunit G 1 | 1.869 | 0.011 | Rheum |
| Q9UI12 | V-type proton ATPase subunit H | 1.372 | 0.040 | Rheum |
| Q93050 | V-type proton ATPase 116 kDa subunit a1 | 2.142 | 0.002 | Rheum |
| P04004 | Vitronectin | 2.557 | 0.002 | ECM |
| Pathway | Number of Genes | p-Value |
|---|---|---|
| Systemic lupus erythematosus | 21 | 7.0 × 10−12 |
| Alcoholism | 22 | 3.6 × 10−10 |
| Lysosome | 18 | 2.1 × 10−9 |
| Shigellosis | 22 | 5.0 × 10−8 |
| Phagosome | 16 | 6.9 × 10−7 |
| Neutrophil extracellular trap formation | 17 | 2.6 × 10−6 |
| Transcriptional misregulation in cancer | 17 | 3.0 × 10−6 |
| Rheumatoid arthritis | 10 | 1.5 × 10−4 |
| Tuberculosis | 13 | 4.2 × 10−4 |
| Valine, leucine, and isoleucine degradation | 7 | 4.9 × 10−4 |
| Toxoplasmosis | 10 | 5.5 × 10−4 |
| ECM–receptor interaction | 9 | 5.7 × 10−4 |
| Complement and coagulation cascades | 8 | 2.2 × 10−3 |
| Metabolic pathways | 49 | 2.9 × 10−3 |
| Staphylococcus aureus infection | 8 | 4.2 × 10−3 |
| Pathway | Number of Genes | p-Value |
|---|---|---|
| Carbon metabolism | 8 | 1.6 × 10−7 |
| Biosynthesis of amino acids | 6 | 7.2 × 10−6 |
| Metabolic pathways | 17 | 3.4 × 10−5 |
| Glycolysis/glucogenesis | 5 | 1.0 × 10−4 |
| HIF-1 signaling pathway | 4 | 7.4 × 10−3 |
| Biosynthesis of nucleotide sugars | 3 | 8.2 × 10−3 |
| Amino sugar and nucleotide sugar metabolism | 3 | 1.4 × 10−2 |
| Cysteine and methionine metabolism | 3 | 1.6 × 10−2 |
| RNA degradation | 3 | 3.4 × 10−2 |
| Pathogenic Escherichia coli infection | 4 | 3.6 × 10−2 |
| Gap Junction | 3 | 4.2 × 10−2 |
| Salmonella infection | 4 | 6.3 × 10−4 |
| Prion disease | 4 | 2.2 × 10−3 |
| Age (years) | 74.1 ± 9.2 |
| Sex, male/female, n | 10/30 |
| BMI (kg/m2) | 27.2 ± 4.9 |
| KL grade (2/3/4), n | 1/10/29 |
| JSW (mm) | 2.7 ± 1.8 |
| VAS resting pain (mm) | 14.0 ± 22.7 |
| VAS at active pain (mm) | 59.2 ± 27.8 |
| Gene | Direction | Primer Sequence (5′–3′) | Product Size (bp) |
|---|---|---|---|
| HLA-DRA | F | ATCCTGACCAATCAGGCGAG | 124 |
| R | GCCTCAAAGCTGGCAAATCG | ||
| S100A8 | F | GACGTCTGGTTCAAAGAGTTGG | 89 |
| R | GCCACGCCCATCTTTATCAC | ||
| S100A9 | F | GACTTGCAAAATGTCGCAGC | 80 |
| R | GCCCCAGCTTCACAGAGTAT | ||
| GAPDH | F | TGTTGCCATCAATGACCCCTT | 202 |
| R | CTCCACGACGTACTCAGCG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsuchiya, M.; Ohashi, Y.; Fukushima, K.; Okuda, Y.; Suto, A.; Matsui, T.; Kodera, Y.; Sato, M.; Tsukada, A.; Inoue, G.; et al. Fibrocyte Phenotype of ENTPD1+CD55+ Cells and Its Association with Pain in Osteoarthritic Synovium. Int. J. Mol. Sci. 2024, 25, 4085. https://doi.org/10.3390/ijms25074085
Tsuchiya M, Ohashi Y, Fukushima K, Okuda Y, Suto A, Matsui T, Kodera Y, Sato M, Tsukada A, Inoue G, et al. Fibrocyte Phenotype of ENTPD1+CD55+ Cells and Its Association with Pain in Osteoarthritic Synovium. International Journal of Molecular Sciences. 2024; 25(7):4085. https://doi.org/10.3390/ijms25074085
Chicago/Turabian StyleTsuchiya, Maho, Yoshihisa Ohashi, Kensuke Fukushima, Yusei Okuda, Arisa Suto, Takashi Matsui, Yoshio Kodera, Masashi Sato, Ayumi Tsukada, Gen Inoue, and et al. 2024. "Fibrocyte Phenotype of ENTPD1+CD55+ Cells and Its Association with Pain in Osteoarthritic Synovium" International Journal of Molecular Sciences 25, no. 7: 4085. https://doi.org/10.3390/ijms25074085
APA StyleTsuchiya, M., Ohashi, Y., Fukushima, K., Okuda, Y., Suto, A., Matsui, T., Kodera, Y., Sato, M., Tsukada, A., Inoue, G., Takaso, M., & Uchida, K. (2024). Fibrocyte Phenotype of ENTPD1+CD55+ Cells and Its Association with Pain in Osteoarthritic Synovium. International Journal of Molecular Sciences, 25(7), 4085. https://doi.org/10.3390/ijms25074085





