Characterization of the First Secreted Sorting Nexin Identified in the Leishmania Protists
Abstract
:1. Introduction
2. Results
2.1. The LdBPK_352470.1 Gene Product Is Highly Conserved in Leishmania spp. and Has Orthologs in Trypanosoma spp.
2.2. Structure Prediction of A0A504X805 Reveals Domains That Classify It to the BAR Subfamily of Sorting Nexins
2.3. Cloning of the LdBPK_352470 Gene from L. donovani Genomic DNA and Generation of Specific Molecular Reagents
2.4. LdSNXi Is Expressed in L. donovani as a Soluble Cytosolic and as Secreted Protein
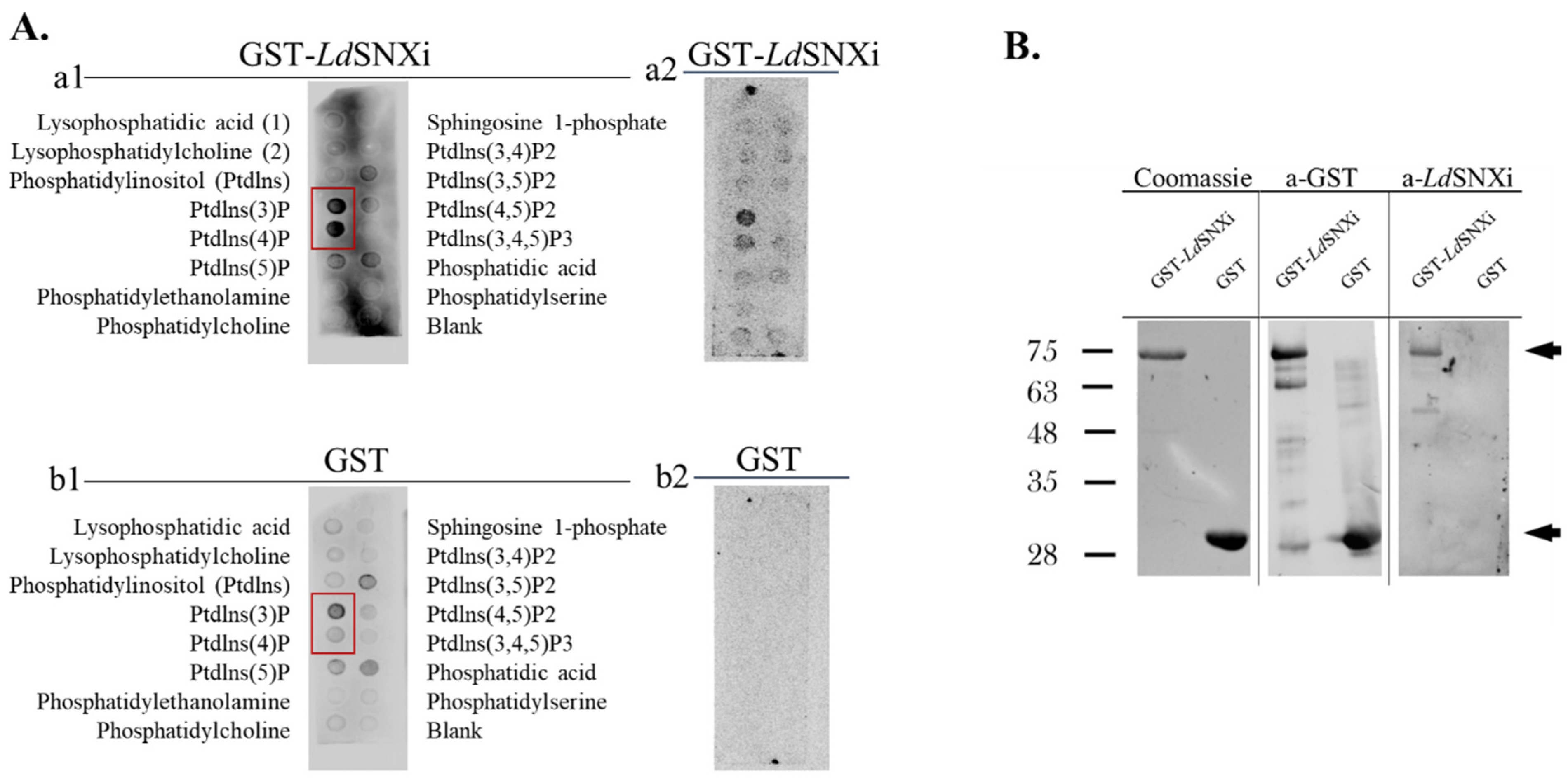
2.5. LdSNXi Binds Preferentially to PtdIns3P and PtdIns4P
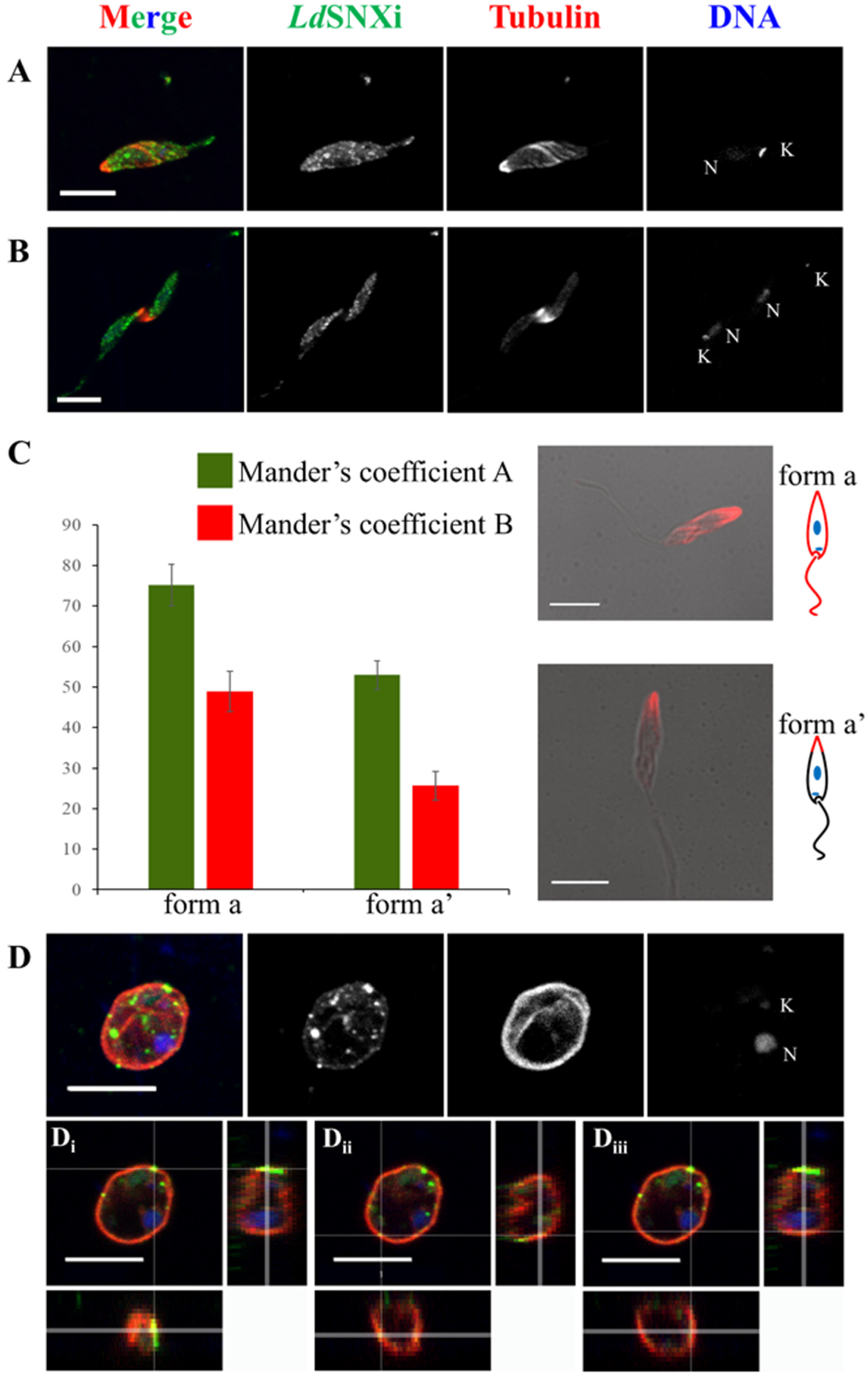
2.6. Endogenous LdSNXi Shows a Vesicular Localization Pattern in L. donovani Promastigotes and Axenic Amastigote-like Cells and Partially Co-Localizes with Microtubules
2.7. LdSNXi Is Predicted as the Evolutionary Structural Homologue of Human SNX1 and SNX2
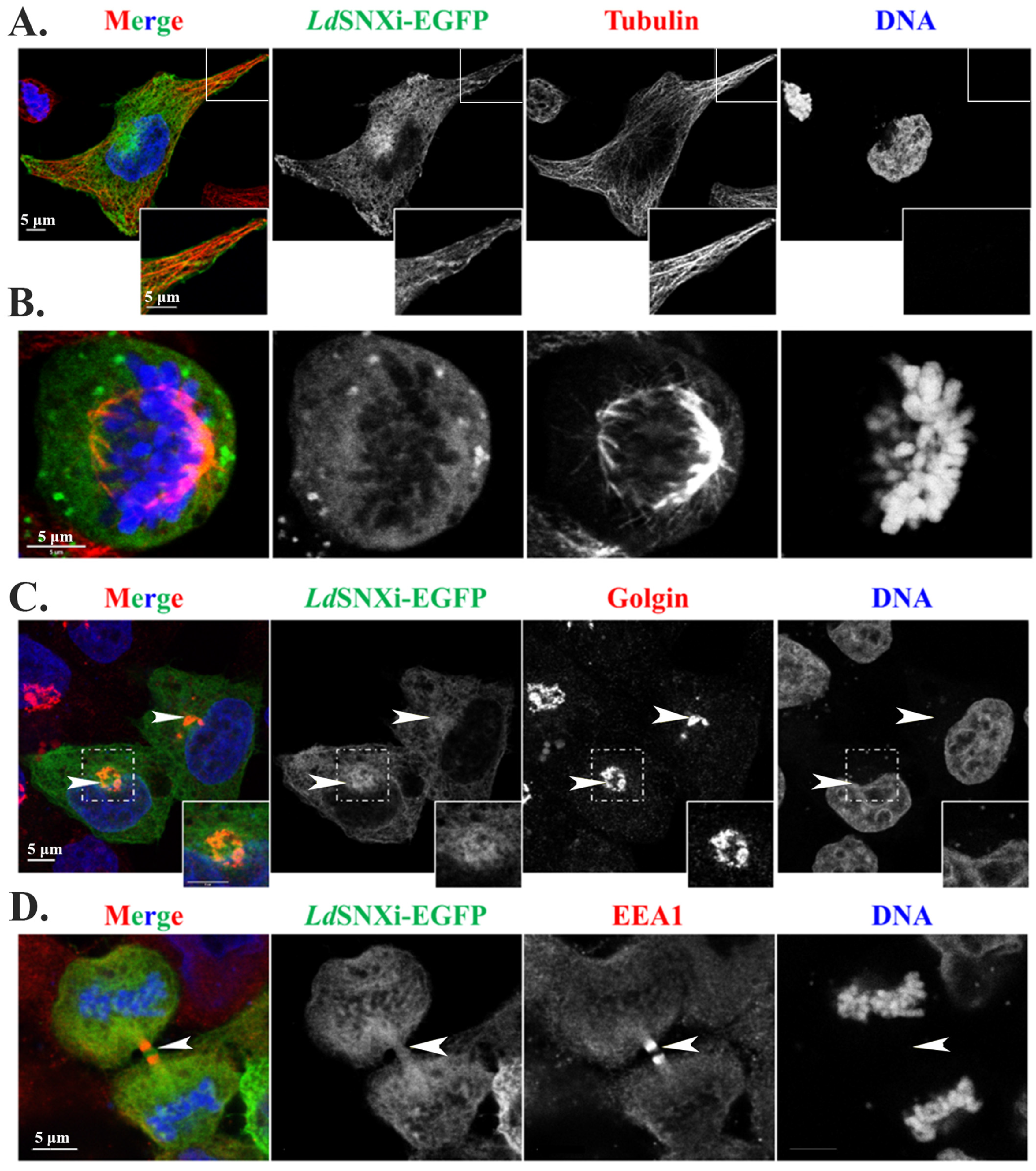
2.8. Multiple Localizations of the LdSNXi-GFP Heterologously Expressed in Mammalian Cells in Golgi, Microtubules, and the Vicinity of Mitotic Chromosomes
2.9. In Silico Analysis Reveals That LdSNXi Is Functionally Homologous to Human SNX1 and SNX2
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Cloning and DNA Constructs
4.3. Overexpression and Purification of Recombinant LdSNXi Forms
4.4. Generation and Purification of Polyclonal Antibodies
4.5. Phosphoinositide Binding Assay
4.6. Detergent-Based Protein Fractionation
4.7. Protein Analysis by SDS-PAGE and Western Blot
4.8. Immunofluorescence
4.9. Transfection of Mammalian Cells
4.10. Imaging by Confocal Microscopy
4.11. Co-localization Analysis
4.12. Secretion Assay
4.13. Sequence Data Retrieval, Sequence Alignment, and Phylogenetic Tree Construction
4.14. Structure and Functional Predictionss
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Teasdale, R.D.; Collins, B.M. Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: Structures, functions and roles in disease. Biochem. J. 2012, 441, 39–59. [Google Scholar] [CrossRef] [PubMed]
- Hanley, S.E.; Cooper, K.F. Sorting Nexins in Protein Homeostasis. Cells 2020, 10, 17. [Google Scholar] [CrossRef] [PubMed]
- Haft, C.R.; Sierra, M.d.l.L.; Barr, V.A.; Haft, D.H.; Taylor, S.I. Identification of a family of sorting nexin molecules and characterization of their association with receptors. Mol. Cell. Biol. 1998, 18, 7278–7287. [Google Scholar] [CrossRef] [PubMed]
- Peter, B.J.; Kent, H.M.; Mills, I.G.; Vallis, Y.; Butler, P.J.G.; Evans, P.R.; McMahon, H.T. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science 2004, 303, 495–499. [Google Scholar] [CrossRef] [PubMed]
- Chishti, A.H.; Kim, A.C.; Marfatia, S.M.; Lutchman, M.; Hanspal, M.; Jindal, H.; Liu, S.-C.; Low, P.S.; Rouleau, G.A.; Mohandas, N.; et al. The FERM domain: A unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends Biochem. Sci. 1998, 23, 281–282. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, E.F.; Katzenell, S.; Andhare, D.; Bauer, K.M.; Ragusa, M.J. A Comparative Analysis of the Membrane Binding and Remodeling Properties of Two Related Sorting Nexin Complexes Involved in Autophagy. Biochemistry 2023, 62, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Chandra, M.; Chin, Y.K.-Y.; Mas, C.; Feathers, J.R.; Paul, B.; Datta, S.; Chen, K.-E.; Jia, X.; Yang, Z.; Norwood, S.J.; et al. Classification of the human phox homology (PX) domains based on their phosphoinositide binding specificities. Nat. Commun. 2019, 10, 1528. [Google Scholar] [CrossRef] [PubMed]
- Petsana, M.; Roumia, A.F.; Bagos, P.G.; Boleti, H.; Braliou, G.G. In Silico Identification and Analysis of Proteins Containing the Phox Homology Phosphoinositide-Binding Domain in Kinetoplastea Protists: Evolutionary Conservation and Uniqueness of Phox-Homology-Domain-Containing Protein Architectures. Int. J. Mol. Sci. 2023, 24, 11521. [Google Scholar] [CrossRef] [PubMed]
- Chandra, M.; Collins, B.M. The Phox Homology (PX) Domain. In Protein Reviews–Purinergic Recepto; Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2019; Volume 1111, pp. 1–17. [Google Scholar]
- Shewan, A.; Eastburn, D.J.; Mostov, K. Phosphoinositides in cell architecture. Cold Spring Harb. Perspect. Biol. 2011, 3, a004796. [Google Scholar] [CrossRef] [PubMed]
- Carlton, J.; Bujny, M.; Peter, B.J.; Oorschot, V.M.J.; Rutherford, A.; Mellor, H.; Klumperman, J.; McMahon, H.T.; Cullen, P.J. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high-curvature membranes and 3-phosphoinositides. Curr. Biol. 2004, 14, 1791–1800. [Google Scholar] [CrossRef] [PubMed]
- Cockcroft, S. Biology of Phosphoinositides; Oxford University Press: Oxford, UK, 2000. [Google Scholar]
- Balla, T. Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol. Rev. 2013, 93, 1019–1137. [Google Scholar] [CrossRef] [PubMed]
- Di Paolo, G.; De Camilli, P. Phosphoinositides in cell regulation and membrane dynamics. Nature 2006, 443, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, M.A. Phosphoinositide recognition domains. Traffic 2003, 4, 201–213. [Google Scholar] [CrossRef] [PubMed]
- DiNitto, J.P.; Cronin, T.C.; Lambright, D.G. Membrane recognition and targeting by lipid-binding domains. Sci. STKE 2003, 2003, re16. [Google Scholar] [CrossRef] [PubMed]
- Cestari, I.; Stuart, K. The phosphoinositide regulatory network in Trypanosoma brucei: Implications for cell-wide regulation in eukaryotes. PLoS Negl. Trop. Dis. 2020, 14, e0008689. [Google Scholar] [CrossRef] [PubMed]
- van Weering, J.R.; Verkade, P.; Cullen, P.J. SNX-BAR proteins in phosphoinositide-mediated, tubular-based endosomal sorting. Semin. Cell Dev. Biol. 2010, 21, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Yong, X.; Zhao, L.; Deng, W.; Sun, H.; Zhou, X.; Mao, L.; Hu, W.; Shen, X.; Sun, Q.; Billadeau, D.D.; et al. Mechanism of cargo recognition by retromer-linked SNX-BAR proteins. PLoS Biol. 2020, 18, e3000631. [Google Scholar] [CrossRef] [PubMed]
- van Weering, J.R.; Sessions, R.B.; Traer, C.J.; Kloer, D.P.; Bhatia, V.K.; Stamou, D.; Carlsson, S.R.; Hurley, J.H.; Cullen, P.J. Molecular basis for SNX-BAR-mediated assembly of distinct endosomal sorting tubules. EMBO J. 2012, 31, 4466–4480. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Burd, C.G. Retrograde trafficking and plasma membrane recycling pathways of the budding yeast Saccharomyces cerevisiae. Traffic 2020, 21, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Anton, Z.; Betin, V.M.S.; Simonetti, B.; Traer, C.J.; Attar, N.; Cullen, P.J.; Lane, J.D. A heterodimeric SNX4--SNX7 SNX-BAR autophagy complex coordinates ATG9A trafficking for efficient autophagosome assembly. J. Cell Sci. 2020, 133, jcs246306. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, F.; Gallon, M.; Winfield, M.; Thomas, E.C.; Bell, A.J.; Heesom, K.J.; Tavaré, J.M.; Cullen, P.J. A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat. Cell Biol. 2013, 15, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Kocmar, T.; Çağlayan, E.; Rayaman, E.; Nagata, K.; Turan, K. Human sorting nexin 2 protein interacts with Influenza A virus PA protein and has a negative regulatory effect on the virus replication. Mol. Biol. Rep. 2022, 49, 497–510. [Google Scholar] [CrossRef] [PubMed]
- Li, D.Y.; Wen, J.-H.; Liang, S.; Tang, J.-X. The Essential Role of Sorting Nexin 5 in Virus-Induced Autophagy. Front. Immunol. 2022, 13, 947384. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; He, P.; Zhang, Y.; Ren, Y.; Zhang, L. The emerging roles of retromer and sorting nexins in the life cycle of viruses. Virol. Sin. 2022, 37, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Liebl, D.; Qi, X.; Zhe, Y.; Barnett, T.C.; Teasdale, R.D. SopB-Mediated Recruitment of SNX18 Facilitates Salmonella Typhimurium Internalization by the Host Cell. Front. Cell. Infect. Microbiol. 2017, 7, 257. [Google Scholar] [CrossRef] [PubMed]
- Paul, B.; Kim, H.S.; Kerr, M.C.; Huston, W.M.; Teasdale, R.D.; Collins, B.M. Structural basis for the hijacking of endosomal sorting nexin proteins by Chlamydia trachomatis. eLife 2017, 6, e22311. [Google Scholar] [CrossRef] [PubMed]
- Braun, V.; Wong, A.; Landekic, M.; Hong, W.J.; Grinstein, S.; Brumell, J.H. Sorting nexin 3 (SNX3) is a component of a tubular endosomal network induced by Salmonella and involved in maturation of the Salmonella-containing vacuole. Cell. Microbiol. 2010, 12, 1352–1367. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Yong, X.; Sun, X.; Yang, F.; Dai, Z.; Gong, Y.; Zhou, L.; Zhang, X.; Niu, D.; Dai, L.; et al. Structural and functional insights into sorting nexin 5/6 interaction with bacterial effector IncE. Signal Transduct. Target. Ther. 2017, 2, 17030. [Google Scholar] [CrossRef]
- Finsel, I.; Ragaz, C.; Hoffmann, C.; Harrison, C.F.; Weber, S.; van Rahden, V.A.; Johannes, L.; Hilbi, H. The Legionella Effector RidL Inhibits Retrograde Trafficking to Promote Intracellular Replication. Cell Host Microbe 2013, 14, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Cernikova, L.; Faso, C.; Hehl, A.B. Roles of Phosphoinositides and Their binding Proteins in Parasitic Protozoa. Trends Parasitol. 2019, 35, 996–1008. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.H.Y.; Stahelin, R.V.; Bhattacharjee, S.; Haldar, K. Eukaryotic virulence determinants utilize phosphoinositides at the ER and host cell surface. Trends Microbiol. 2013, 21, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Koumandou, V.L.; Klute, M.J.; Herman, E.K.; Nunez-Miguel, R.; Dacks, J.B.; Field, M.C. Evolutionary reconstruction of the retromer complex and its function in Trypanosoma brucei. J. Cell Sci. 2011, 124 Pt 9, 1496–1509. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, D.; Zhang, N.; Zoltner, M.; del Pino, R.C.; Field, M.C. Evolution of protein trafficking in kinetoplastid parasites: Complexity and pathogenesis. Traffic 2018, 19, 803–812. [Google Scholar] [CrossRef]
- O’Malley, M.A.; Leger, M.M.; Wideman, J.G.; Ruiz-Trillo, I. Concepts of the last eukaryotic common ancestor. Nat. Ecol. Evol. 2019, 3, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Kostygov, A.Y.; Karnkowska, A.; Votýpka, J.; Tashyreva, D.; Maciszewski, K.; Yurchenko, V.; Lukeš, J. Euglenozoa: Taxonomy, diversity and ecology, symbioses and viruses. Open Biol. 2021, 11, 200407. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.R.; Auger, K.R. Phylogenomics of phosphoinositide lipid kinases: Perspectives on the evolution of second messenger signaling and drug discovery. BMC Evolut. Biol. 2011, 11, 4. [Google Scholar] [CrossRef]
- Demmel, L.; Schmidt, K.; Lucast, L.; Havlicek, K.; Zankel, A.; Koestler, T.; Reithofer, V.; de Camilli, P.; Warren, G. The endocytic activity of the flagellar pocket in Trypanosoma brucei is regulated by an adjacent phosphatidylinositol phosphate kinase. J. Cell Sci. 2014, 127 Pt 10, 2351–2364. [Google Scholar] [CrossRef] [PubMed]
- Sudhahar, C.G.; Haney, R.M.; Xue, Y.; Stahelin, R.V. Cellular membranes and lipid-binding domains as attractive targets for drug development. Curr. Drug Targets 2008, 9, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Lukes, J.; Butenko, A.; Hashimi, H.; Maslov, D.A.; Votýpka, J.; Yurchenko, V. Trypanosomatids Are Much More than Just Trypanosomes: Clues from the Expanded Family Tree. Trends Parasitol. 2018, 34, 466–480. [Google Scholar] [CrossRef] [PubMed]
- Silverman, J.M.; Chan, S.K.; Robinson, D.P.; Dwyer, D.M.; Nandan, D.; Foster, L.J.; E Reiner, N. Proteomic analysis of the secretome of Leishmania donovani. Genome Biol. 2008, 9, R35. [Google Scholar] [CrossRef] [PubMed]
- Seaman, M.N.; Williams, H.P. Identification of the functional domains of yeast sorting nexins Vps5p and Vps17p. Mol. Biol. Cell 2002, 13, 2826–2840. [Google Scholar] [CrossRef] [PubMed]
- Doukas, A.; Karena, E.; Botou, M.; Papakostas, K.; Papadaki, A.; Tziouvara, O.; Xingi, E.; Frillingos, S.; Boleti, H. Heterologous expression of the mammalian sodium-nucleobase transporter rSNBT1 in Leishmania tarentolae. Biochim. Biophys. Acta Biomembr. 2019, 1861, 1546–1557. [Google Scholar] [CrossRef] [PubMed]
- Foucher, A.L.; Papadopoulou, B.; Ouellette, M. Prefractionation by digitonin extraction increases representation of the cytosolic and intracellular proteome of Leishmania infantum. J. Proteome Res. 2006, 5, 1741–1750. [Google Scholar] [CrossRef] [PubMed]
- Papadaki, A.; Tziouvara, O.; Kotopouli, A.; Koumarianou, P.; Doukas, A.; Rios, P.; Tardieux, I.; Köhn, M.; Boleti, H. The Leishmania donovani LDBPK_220120.1 Gene Encodes for an Atypical Dual Specificity Lipid-Like Phosphatase Expressed in Promastigotes and Amastigotes; Substrate Specificity, Intracellular Localizations, and Putative Role(s). Front. Cell. Infect. Microbiol. 2021, 11, 591868. [Google Scholar] [CrossRef] [PubMed]
- Ambit, A.; Woods, K.L.; Cull, B.; Coombs, G.H.; Mottram, J.C. Morphological events during the cell cycle of Leishmania major. Eukaryot. Cell 2011, 10, 1429–1438. [Google Scholar] [CrossRef]
- Shortill, S.P.; Frier, M.S.; Conibear, E. You can go your own way: SNX-BAR coat complexes direct traffic at late endosomes. Curr. Opin. Cell Biol. 2022, 76, 102087. [Google Scholar] [CrossRef] [PubMed]
- Okada, H.; Zhang, W.; Peterhoff, C.; Hwang, J.C.; Nixon, R.A.; Ryu, S.H.; Kim, T.-W. Proteomic identification of sorting nexin 6 as a negative regulator of BACE1-mediated APP processing. FASEB J. 2010, 24, 2783–2794. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Huang, T.; Hong, Y.; Yang, W.; Zhang, X.; Luo, H.; Xu, H.; Wang, X. The Retromer Complex and Sorting Nexins in Neurodegenerative Diseases. Front. Aging Neurosci. 2018, 10, 79. [Google Scholar] [CrossRef] [PubMed]
- Frémont, S.; Echard, A. Membrane Traffic in the Late Steps of Cytokinesis. Curr. Biol. 2018, 28, R458–R470. [Google Scholar] [CrossRef] [PubMed]
- Kendall, A.K.; Xie, B.; Xu, P.; Wang, J.; Burcham, R.; Frazier, M.N.; Binshtein, E.; Wei, H.; Graham, T.R.; Nakagawa, T.; et al. Mammalian Retromer Is an Adaptable Scaffold for Cargo Sorting from Endosomes. Structure 2020, 28, 393–405.e4. [Google Scholar] [CrossRef] [PubMed]
- Fuse, A.; Furuya, N.; Kakuta, S.; Inose, A.; Sato, M.; Koike, M.; Saiki, S.; Hattori, N. VPS29–VPS35 intermediate of retromer is stable and may be involved in the retromer complex assembly process. FEBS Lett. 2015, 589, 1430–1436. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Shibata, Y.; Yun, C.; Ron, D.; Rapoport, T.A. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 2004, 429, 841–847. [Google Scholar] [CrossRef] [PubMed]
- Rao, B.; Li, S.; Yao, D.; Wang, Q.; Xia, Y.; Jia, Y.; Shen, Y.; Cao, Y. The cryo-EM structure of an ERAD protein channel formed by tetrameric human Derlin-1. Sci. Adv. 2021, 7, eabe8591. [Google Scholar] [CrossRef] [PubMed]
- Kervin, T.A.; Wiseman, B.C.; Overduin, M. Phosphoinositide Recognition Sites Are Blocked by Metabolite Attachment. Front. Cell Dev. Biol. 2021, 9, 690461. [Google Scholar] [CrossRef] [PubMed]
- Posor, Y.; Jang, W.; Haucke, V. Phosphoinositides as membrane organizers. Nat. Rev. Mol. Cell Biol. 2022, 23, 797–816. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.W.; Lemmon, M.A. All Phox Homology (PX) Domains from Saccharomyces cerevisiae Specifically Recognize Phosphatidylinositol 3-Phosphate. J. Biol. Chem. 2001, 276, 44179–44184. [Google Scholar] [CrossRef] [PubMed]
- Carlton, J.G.; Bujny, M.V.; Peter, B.J.; Oorschot, V.M.J.; Rutherford, A.; Arkell, R.S.; Klumperman, J.; McMahon, H.T.; Cullen, P.J. Sorting nexin-2 is associated with tubular elements of the early endosome, but is not essential for retromer-mediated endosome-to-TGN transport. J. Cell Sci. 2005, 118 Pt 19, 4527–4539. [Google Scholar] [CrossRef] [PubMed]
- Wassmer, T.; Attar, N.; Bujny, M.V.; Oakley, J.; Traer, C.J.; Cullen, P.J. A loss-of-function screen reveals SNX5 and SNX6 as potential components of the mammalian retromer. J. Cell Sci. 2007, 120, 45–54. [Google Scholar] [CrossRef] [PubMed]
- McGough, I.J.; Cullen, P.J. Recent Advances in Retromer Biology. Traffic 2011, 12, 963–971. [Google Scholar] [CrossRef] [PubMed]
- Wassmer, T.; Attar, N.; Harterink, M.; van Weering, J.R.; Traer, C.J.; Oakley, J.; Goud, B.; Stephens, D.J.; Verkade, P.; Korswagen, H.C.; et al. The retromer coat complex coordinates endosomal sorting and dynein-mediated transport, with carrier recognition by the trans-Golgi network. Dev. Cell 2009, 17, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Yang, Y.; Zhang, C.; Niu, Y.; Li, K.; Zhao, X.; Liu, J.-J. The retromer component SNX6 interacts with dynactin p150(Glued) and mediates endosome-to-TGN transport. Cell Res. 2009, 19, 1334–1349. [Google Scholar] [CrossRef] [PubMed]
- Schlacht, A.; Herman, E.K.; Klute, M.J.; Field, M.C.; Dacks, J.B. Missing pieces of an ancient puzzle: Evolution of the eukaryotic membrane-trafficking system. Cold Spring Harb. Perspect. Biol. 2014, 6, a016048. [Google Scholar] [CrossRef] [PubMed]
- Seaman, M.N.; McCaffery, J.M.; Emr, S.D. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 1998, 142, 665–681. [Google Scholar] [CrossRef] [PubMed]
- Horazdovsky, B.F.; Davies, B.A.; McLaughlin, S.A.; Yoon, S.; Shi, Y.; Stefan, C.J.; Rue, S.M.; Teis, D.; Emr, S.D.; Schmid, M.E.S.L.; et al. A sorting nexin-1 homologue, Vps5p, forms a complex with Vps17p and is required for recycling the vacuolar protein-sorting receptor. Mol. Biol. Cell 1997, 8, 1529–1541. [Google Scholar] [CrossRef] [PubMed]
- Simonetti, B.; Cullen, P.J. Endosomal Sorting: Architecture of the Retromer Coat. Curr. Biol. 2018, 28, R1350–R1352. [Google Scholar] [CrossRef] [PubMed]
- Rojas, R.; Kametaka, S.; Haft, C.R.; Bonifacino, J.S. Interchangeable but essential functions of SNX1 and SNX2 in the association of retromer with endosomes and the trafficking of mannose 6-phosphate receptors. Mol. Cell. Biol. 2007, 27, 1112–1124. [Google Scholar] [CrossRef] [PubMed]
- Carlton, J.G.; Jones, H.; Eggert, U.S. Membrane and organelle dynamics during cell division. Nat. Rev. Mol. Cell Biol. 2020, 21, 151–166. [Google Scholar] [CrossRef] [PubMed]
- Fraschini, R. Cytokinesis in Eukaryotic Cells: The Furrow Complexity at a Glance. Cells 2020, 9, 271. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.P.; Chircop, M. SNX9, SNX18 and SNX33 are required for progression through and completion of mitosis. J. Cell Sci. 2012, 125 Pt 18, 4372–4382. [Google Scholar] [CrossRef] [PubMed]
- Pissarra, J.; Pagniez, J.; Petitdidier, E.; Séveno, M.; Vigy, O.; Bras-Gonçalves, R.; Lemesre, J.-L.; Holzmuller, P. Proteomic Analysis of the Promastigote Secretome of Seven Leishmania Species. J. Proteome Res. 2022, 21, 30–48. [Google Scholar] [CrossRef] [PubMed]
- Forrest, D.M.; Batista, M.; Marchini, F.K.; Tempone, A.J.; Traub-Csekö, Y.M. Proteomic analysis of exosomes derived from procyclic and metacyclic-like cultured Leishmania infantum chagasi. J. Proteom. 2020, 227, 103902. [Google Scholar] [CrossRef] [PubMed]
- Boleti, H.; Smirlis, D.; Dalagiorgou, G.; Meurs, E.F.; Christoforidis, S.; Mavromara, P. ER targeting and retention of the HCV NS4B protein relies on the concerted action of multiple structural features including its transmembrane domains. Mol. Membr. Biol. 2010, 27, 45–62. [Google Scholar] [CrossRef] [PubMed]
- Papadaki, A.; Politou, A.S.; Smirlis, D.; Kotini, M.P.; Kourou, K.; Papamarcaki, T.; Boleti, H. The Leishmania donovani histidine acid ecto-phosphatase LdMAcP: Insight into its structure and function. Biochem. J. 2015, 467, 473–486. [Google Scholar] [CrossRef] [PubMed]
- Smirlis, D.; Bisti, S.N.; Xingi, E.; Konidou, G.; Thiakaki, M.; Soteriadou, K.P. Leishmania histone H1 overexpression delays parasite cell-cycle progression, parasite differentiation and reduces Leishmania infectivity in vivo. Mol. Microbiol. 2006, 60, 1457–1473. [Google Scholar] [CrossRef] [PubMed]
- Costes, S.V.; Daelemans, D.; Cho, E.H.; Dobbin, Z.; Pavlakis, G.; Lockett, S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 2004, 86, 3993–4003. [Google Scholar] [CrossRef] [PubMed]
- Amos, B.; Aurrecoechea, C.; Barba, M.; Barreto, A.; Basenko, E.Y.; Bażant, W.; Belnap, R.; Blevins, A.S.; Böhme, U.; Brestelli, J.; et al. VEuPathDB: The eukaryotic pathogen, vector and host bioinformatics resource center. Nucleic Acids Res. 2021, 50, D898–D911. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2. 0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Higgins, D.G. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. 2014, 1079, 105–116. [Google Scholar] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Coudert, E.; Gehant, S.; de Castro, E.; Pozzato, M.; Baratin, D.; Neto, T.; Sigrist, C.J.A.; Redaschi, N.; Bridge, A.; Aimo, L.; et al. Annotation of biologically relevant ligands in UniProtKB using ChEBI. Bioinformatics 2022, 39, btac793. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- McGuffin, L.J.; Bryson, K.; Jones, D.T. The PSIPRED protein structure prediction server. Bioinformatics 2000, 16, 404–405. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2021, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
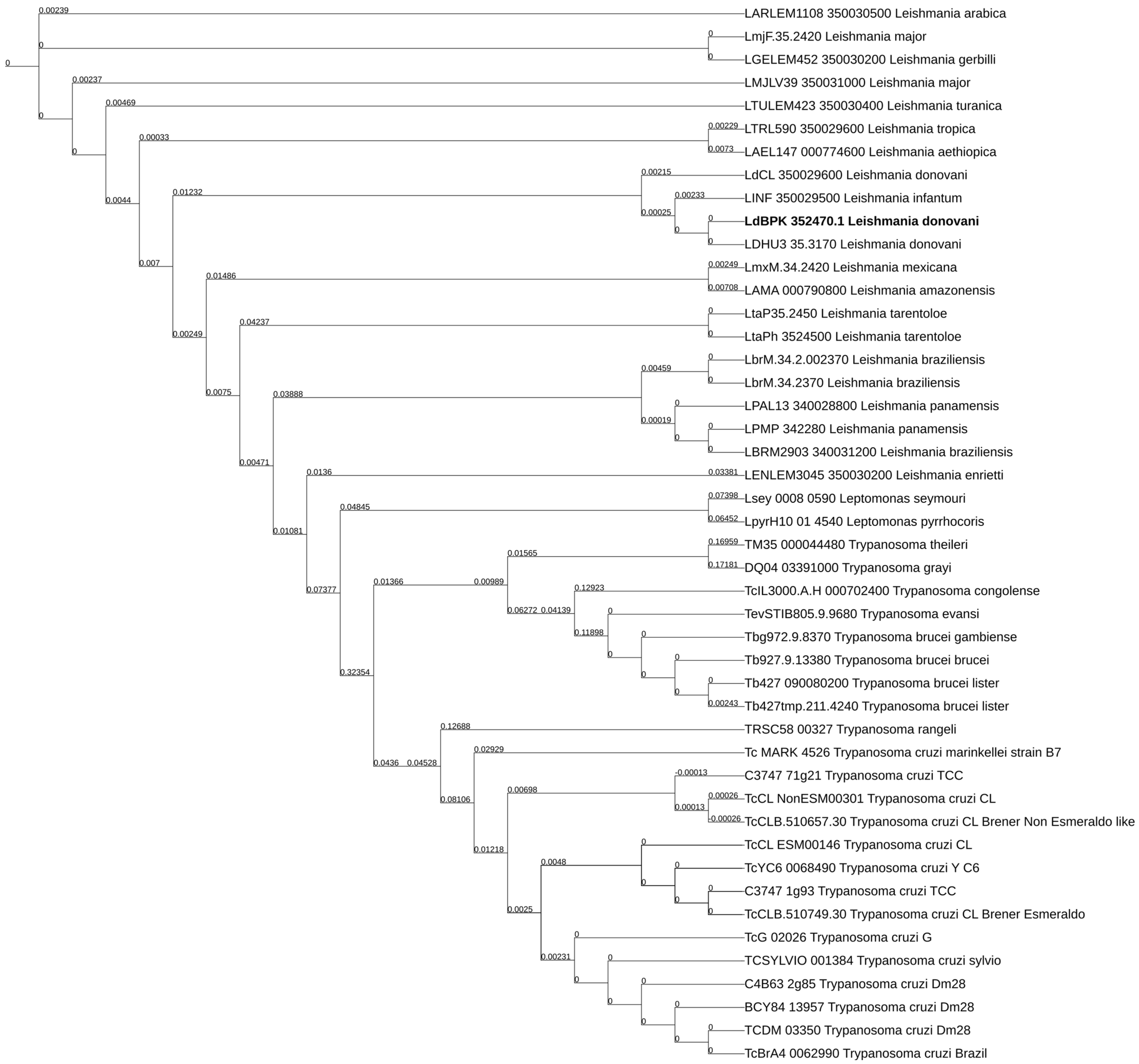



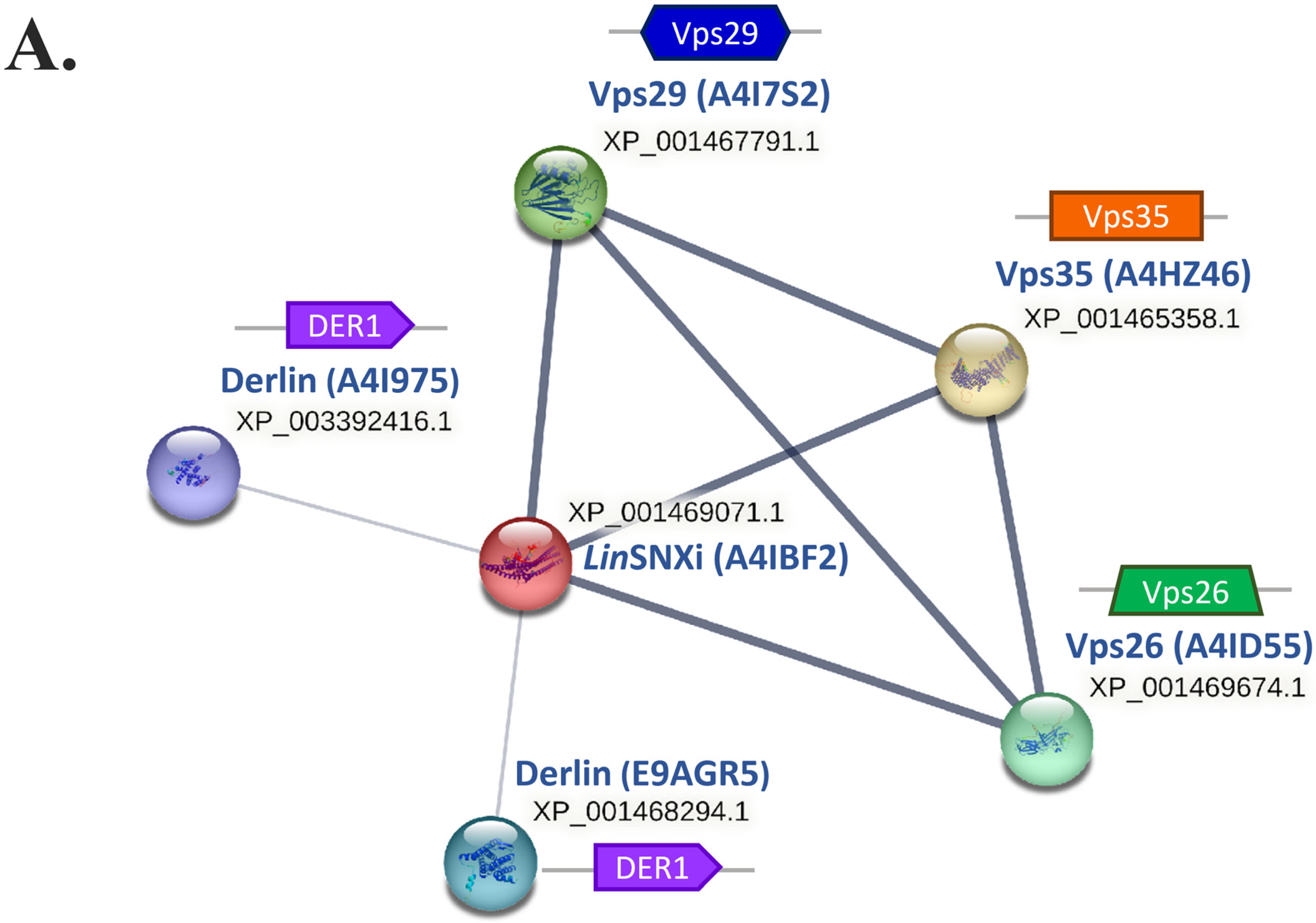
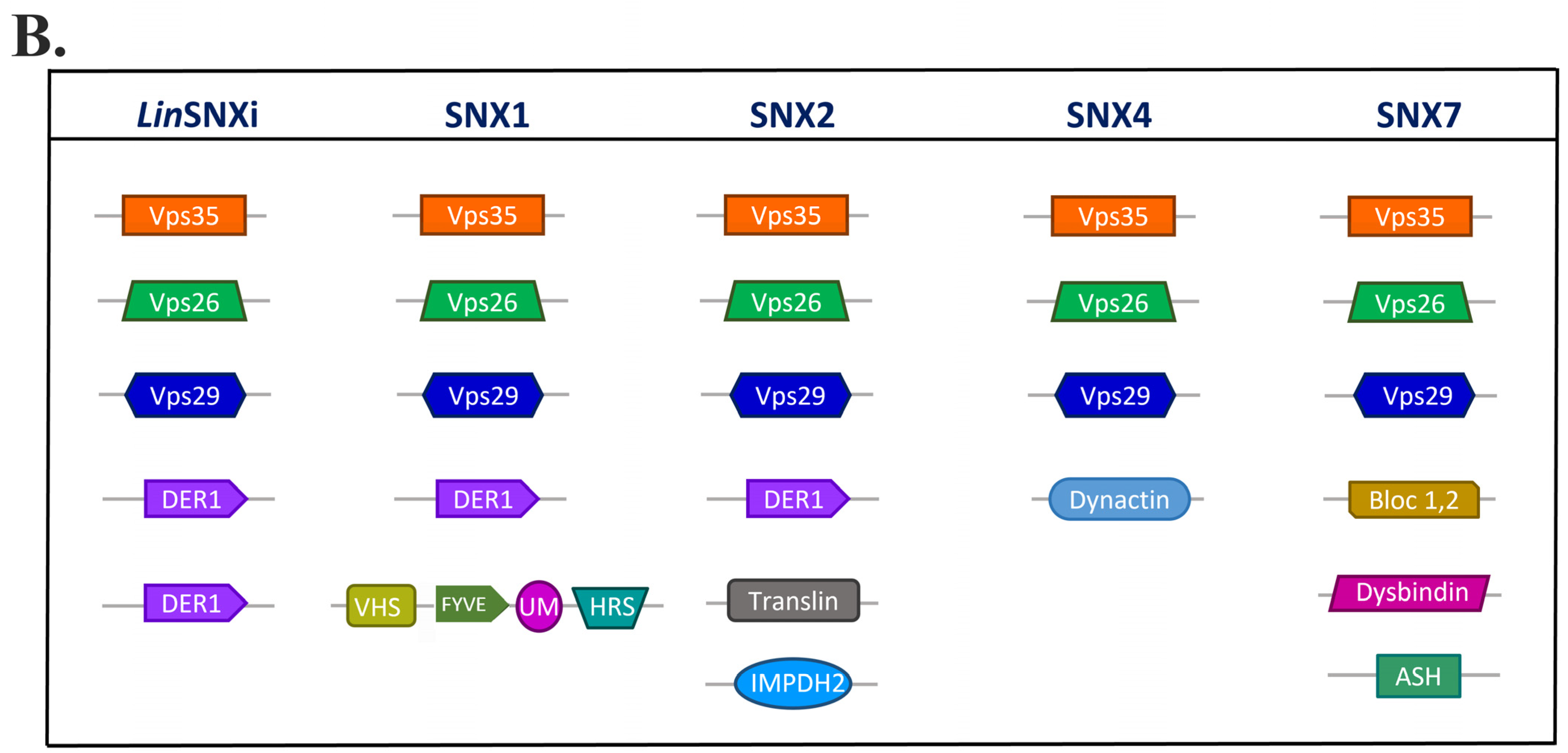
| Protein ID | Query Cover | E-Value | Per. Ident. | Acc. Len |
|---|---|---|---|---|
| LARLEM1108_350030500 (Leishmania arabica) | 100% | 0.0 | 97.36% | 417 |
| LmjF.35.2420 (Leishmania major) | 100% | 0.0 | 97.60% | 417 |
| LGELEM452_350030200 (Leishmania gerbilli) | 100% | 0.0 | 97.60% | 417 |
| LMJLV39_350031000 (Leishmania major) | 100% | 0.0 | 97.36% | 417 |
| LTULEM423_350030400 (Leishmania turanica) | 100% | 0.0 | 97.12% | 417 |
| LTRL590_350029600 (Leishmania tropica) | 100% | 0.0 | 97.84% | 417 |
| LAEL147_000774600 (Leishmania aethiopica) | 100% | 0.0 | 97.12% | 417 |
| LdCL_350029600 (Leishmania donovani) | 100% | 0.0 | 99.76% | 417 |
| LINF_350029500 (Leishmania infantum) | 100% | 0.0 | 99.76% | 417 |
| LDHU3_35.3170 (Leishmania donovani) | 100% | 0.0 | 100.00% | 417 |
| LmxM.34.2420 (Leishmania mexicana) | 99% | 0.0 | 97.12% | 418 |
| LAMA_000790800 (Leishmania amazonensis) | 99% | 0.0 | 96.40% | 418 |
| LtaP35.2450 (Leishmania tarentoloe) | 100% | 0.0 | 92.11% | 418 |
| LtaPh_3524500 (Leishmania tarentoloe) | 100% | 0.0 | 92.11% | 418 |
| LbrM.34.2.002370 (Leishmania braziliensis) | 100% | 0.0 | 92.34% | 418 |
| LbrM.34.2370 (Leishmania braziliensis) | 100% | 0.0 | 92.34% | 418 |
| LPAL13_340028800 (Leishmania panamensis) | 100% | 0.0 | 92.82% | 418 |
| LPMP_342280 (Leishmania panamensis) | 100% | 0.0 | 92.82% | 418 |
| LBRM2903_340031200 (Leishmania braziliensis) | 100% | 0.0 | 92.82% | 418 |
| LENLEM3045_350030200 (Leishmania enrietti) | 100% | 0.0 | 90.91% | 418 |
| Lsey_0008_0590 (Leptomonas seymouri) | 99% | 0.0 | 77.18% | 426 |
| LpyrH10_01_4540 (Leptomonas pyrrhocoris) | 99% | 0.0 | 75.59% | 427 |
| TM35_000044480 (Trypanosoma theileri) | 98% | 1 × 10−68 | 33.88% | 415 |
| DQ04_03391000 (Trypanosoma grayi) | 96% | 1 × 10−71 | 34.13% | 426 |
| TcIL3000.A.H_000702400 (Trypanosoma congolense) | 98% | 8 × 10−58 | 31.24% | 419 |
| TevSTIB805.9.9680 (Trypanosoma evansi) | 97% | 9 × 10−67 | 31.24% | 419 |
| Tbg972.9.8370 (Trypanosoma brucei gambiense) | 97% | 9 × 10−67 | 31.24% | 419 |
| Tb927.9.13380 (Trypanosoma brucei brucei) | 97% | 9 × 10−67 | 31.24% | 419 |
| Tb427_090080200 (Trypanosoma brucei lister) | 97% | 9 × 10−67 | 31.24% | 419 |
| Tb427tmp.211.4240 (Trypanosoma brucei lister) | 97% | 7 × 10−66 | 31.00% | 419 |
| TRSC58_00327 (Trypanosoma rangeli) | 98% | 1 × 10−67 | 33.18% | 422 |
| Tc_MARK_4526 (Trypanosoma cruzi marinkellei strain B7) | 97% | 1 × 10−70 | 33.25% | 422 |
| C3747_71g21 (Trypanosoma cruzi TCC) | 97% | 4 × 10−73 | 34.43% | 422 |
| TcCL_NonESM00301 (Trypanosoma cruzi CL) | 97% | 4 × 10−73 | 34.43% | 429 |
| TcCLB.510657.30 (Trypanosoma cruzi CL Brener Non-Esmeraldo) | 97% | 4 × 10−73 | 34.43% | 422 |
| TcCL_ESM00146 (Trypanosoma cruzi CL) | 97% | 2 × 10−74 | 34.43% | 422 |
| TcYC6_0068490 (Trypanosoma cruzi Y C6) | 97% | 2 × 10−74 | 34.43% | 422 |
| C3747_1g93 (Trypanosoma cruzi TCC) | 97% | 2 × 10−74 | 34.43% | 422 |
| TcCLB.510749.30 (Trypanosoma cruzi CL Brener Esmeraldo) | 97% | 2 × 10−74 | 34.43% | 422 |
| TcG_02026 (Trypanosoma cruzi G) | 97% | 8 × 10−75 | 34.67% | 422 |
| TCSYLVIO_001384 (Trypanosoma cruzi sylvio) | 97% | 8 × 10−75 | 34.67% | 422 |
| C4B63_2g85 (Trypanosoma cruzi Dm28) | 97% | 8 × 10−75 | 34.67% | 422 |
| BCY84_13957 (Trypanosoma cruzi Dm28) | 97% | 8 × 10−75 | 34.67% | 422 |
| TCDM_03350 (Trypanosoma cruzi Dm28) | 97% | 8 × 10−75 | 34.67% | 422 |
| TcBrA4_0062990 (Trypanosoma cruzi) | 97% | 8 × 10−75 | 34.67% | 422 |
| Protein ID | Query Cover | E-Value | Per. Ident. | Acc. Length |
|---|---|---|---|---|
| SNX2 | 96% | 2 × 10−19 | 23.56% | 519 |
| SNX1 | 96% | 4 × 10−20 | 24.11% | 522 |
| SNX5 | 28% | 2 × 10−6 | 28.03% | 404 |
| SNX32 | 13% | 1 × 10−2 | 33.85% | 403 |
| SNX4 | 23% | 1 × 10−1 | 36.00% | 450 |
| SNX7 | 30% | 3 × 10−11 | 32.31% | 387 |
| SNX3 | 31% | 1 × 10−8 | 26.72% | 162 |
| SNX9 | 23% | 9 × 10−69 | 23.76% | 595 |
| SNX33 | 31% | 2 × 10−62 | 24.44% | 574 |
| SNX18 | 31% | 4 × 10−94 | 26.43% | 628 |
| Human SNX | Query Cover | E-Value | Per. Ident. | |
|---|---|---|---|---|
| PX | ||||
| PX_SNX4 (61-187) | 68% | 4 × 10−15 | 38.30% | |
| PX_SNX7 (30-151) | 97% | 8 × 10−14 | 32.31% | |
| PX_SNX2 (140-269) | 84% | 2 × 10−13 | 31.67% | |
| PX_SNX30 (89-210) | 93% | 6 × 10−11 | 26.02% | |
| PX_SNX1 (143-272) | 84% | 1 × 10−10 | 30.83% | |
| PX_SNX18 (276-386) | 91% | 8 × 10−10 | 26.83% | |
| PX_SNX8 (73-181) | 65% | 5 × 10−9 | 28.74% | |
| PX_SNX5 (25-172) | 88% | 1 × 10−8 | 28.03% | |
| PX_SNX9 (250-361) | 66% | 2 × 10−6 | 23.60% | |
| PX_SNX33 (230-340) | 90% | 2 × 10−6 | 24.79% | |
| PX_SNX32 (20-168) | 41% | 2 × 10−4 | 34.38% | |
| PX_SNX6 (26-173) | 46% | 5 × 10−3 | 30.99% | |
| BAR | ||||
| BAR_SNX2_human (299-519) | 91% | 2 × 10−8 | 21.72% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tziouvara, O.; Petsana, M.; Kourounis, D.; Papadaki, A.; Basdra, E.; Braliou, G.G.; Boleti, H. Characterization of the First Secreted Sorting Nexin Identified in the Leishmania Protists. Int. J. Mol. Sci. 2024, 25, 4095. https://doi.org/10.3390/ijms25074095
Tziouvara O, Petsana M, Kourounis D, Papadaki A, Basdra E, Braliou GG, Boleti H. Characterization of the First Secreted Sorting Nexin Identified in the Leishmania Protists. International Journal of Molecular Sciences. 2024; 25(7):4095. https://doi.org/10.3390/ijms25074095
Chicago/Turabian StyleTziouvara, Olympia, Marina Petsana, Drosos Kourounis, Amalia Papadaki, Efthimia Basdra, Georgia G. Braliou, and Haralabia Boleti. 2024. "Characterization of the First Secreted Sorting Nexin Identified in the Leishmania Protists" International Journal of Molecular Sciences 25, no. 7: 4095. https://doi.org/10.3390/ijms25074095





