High NaCl Concentrations in Water Are Associated with Developmental Abnormalities and Altered Gene Expression in Zebrafish
Abstract
1. Introduction
2. Results
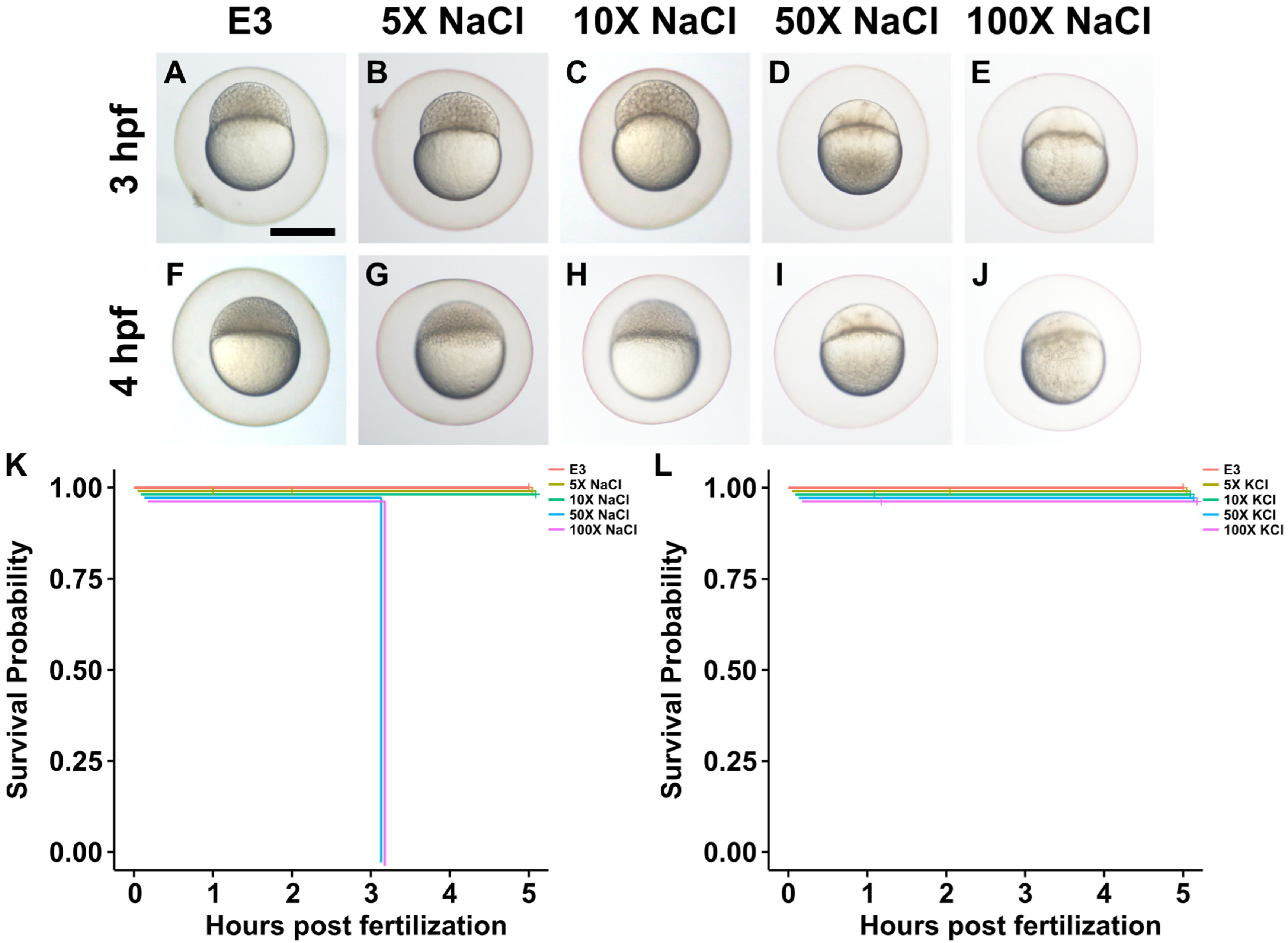
2.1. Impact of Increased Salt Concentrations on Zebrafish Development from the Blastocyst to Larval Stage
2.2. Impact of Increased Salt Concentrations on Zebrafish Development from the Two-Cell to Blastocyst Stage
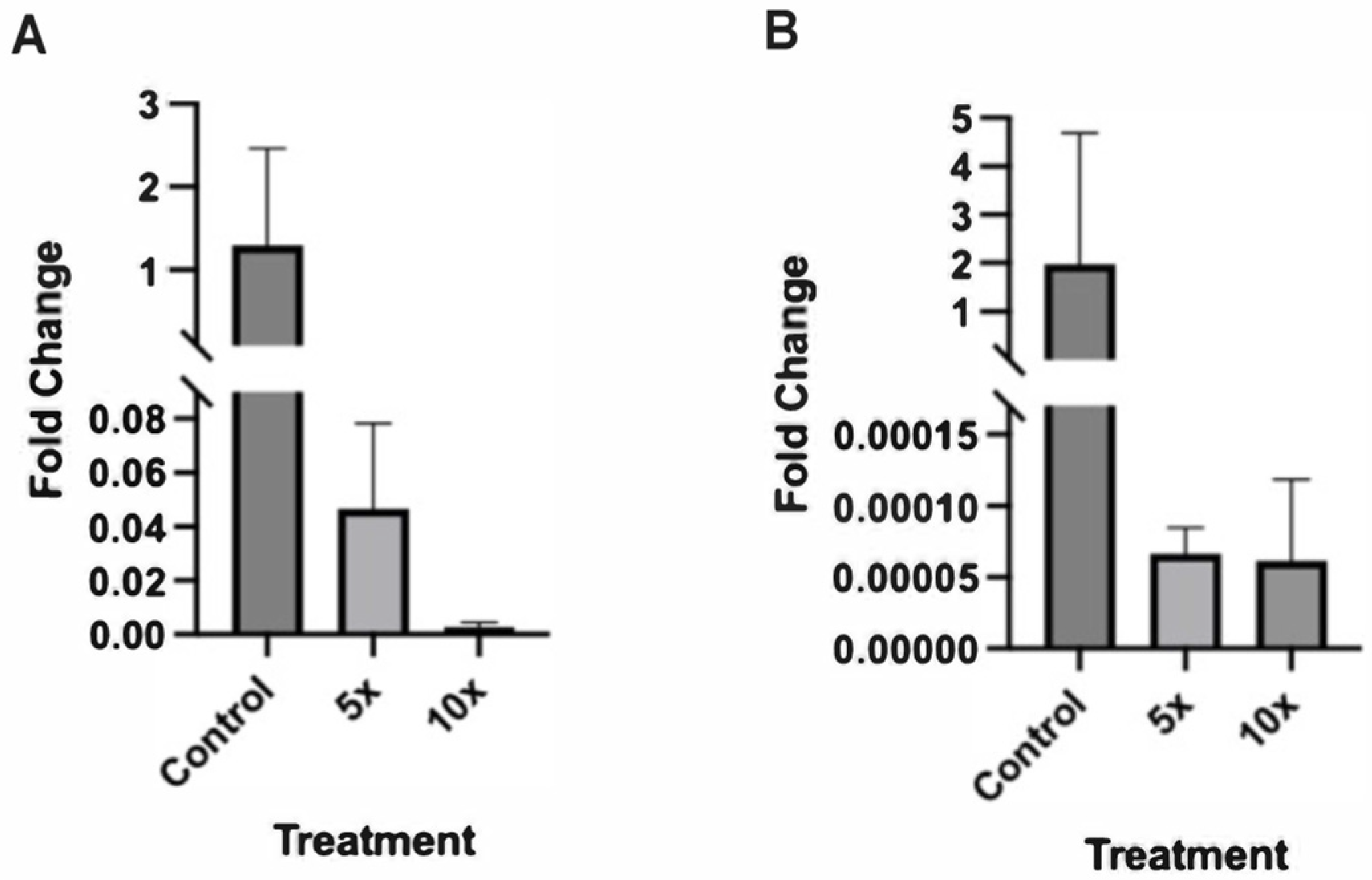
2.3. Altered Expression of Sonic Hedgehog Pathway Genes in Zebrafish Embryos Treated with Higher NaCl Concentrations
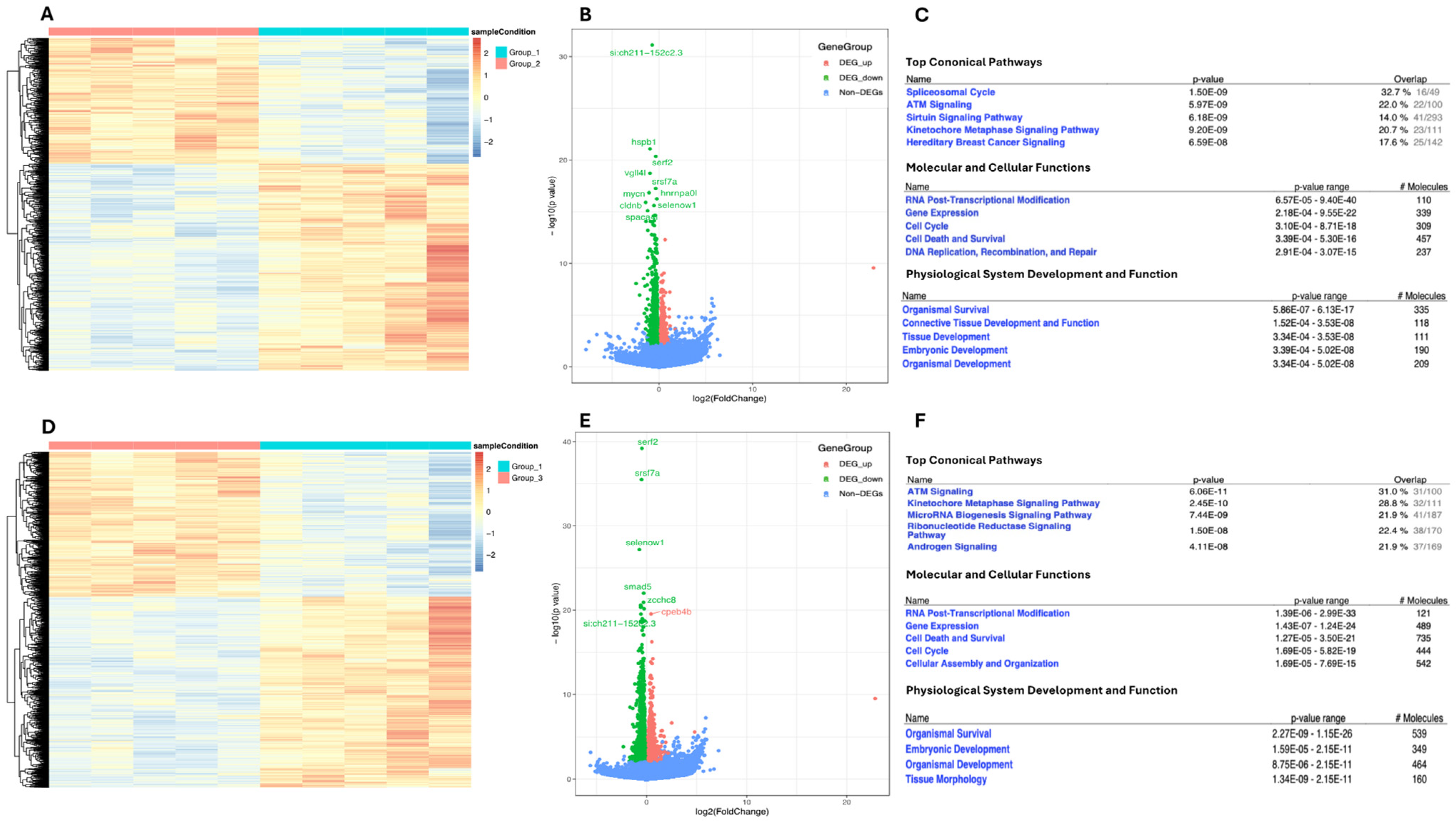
2.4. Transcriptomic Landscape of Zebrafish Embryos Treated with Higher NaCl Concentrations
3. Discussion
4. Materials and Methods
4.1. Zebrafish Culture
4.2. Quantitative Reverse-Transcription Polymerase Chain Reaction (qRT-PCR)
4.3. Statistical Analyses
4.4. RNA Extraction and Sequencing
4.5. RNA Sequencing
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pieper, K.J.; Tang, M.; Jones, C.N.; Weiss, S.; Greene, A.; Mohsin, H.; Parks, J.; Edwards, M.A. Impact of Road Salt on Drinking Water Quality and Infrastructure Corrosion in Private Wells. Environ. Sci. Technol. 2018, 52, 14078–14087. [Google Scholar] [CrossRef] [PubMed]
- Hintz, W.D.; Fay, L.; Relyea, R.A. Road salts, human safety, and the rising salinity of our fresh waters. Front. Ecol. Environ. 2022, 20, 22–30. [Google Scholar] [CrossRef]
- Nasrin, S.; Mahmood, H.; Siddique, M.; Masudur, M. Adaptive responses to salinity: Seed germination traits of Sonneratia apetala along salinity gradient in sundarbans, Bangladesh. J. Trop. For. Sci. 2020, 32, 179–186. [Google Scholar] [CrossRef]
- Wilson, B.J.; Servais, S.; Charles, S.P.; Mazzei, V.; Gaiser, E.E.; Kominoski, J.S.; Richards, J.H.; Troxler, T.G. Phosphorus Alleviation of Salinity Stress: Effects of Saltwater Intrusion on an Everglades Freshwater Peat Marsh. Ecology 2019, 100, e02672. [Google Scholar] [CrossRef] [PubMed]
- Witteveen, J. The Impact of the Salinity of Soil-Water and Food on the Physiology, Behaviour and Ecology of Salt-Marsh Collembola. Funct. Ecol. 1988, 2, 49–55. [Google Scholar] [CrossRef]
- Kujawa, R.; Piech, P. Influence of Water Salinity on the Growth and Survivability of Asp Larvae Leuciscus aspius (Linnaeus, 1758) under Controlled Conditions. Animals 2022, 12, 2299. [Google Scholar] [CrossRef]
- Cañedo-Argüelles, M.; Kefford, B.; Schäfer, R. Salt in freshwaters: Causes, effects and prospects-introduction to the theme issue. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 374, 20180002. [Google Scholar] [CrossRef]
- Engeszer, R.; Patterson, L.; Rao, A.; Parichy, D. Zebrafish in the wild: A review of natural history and new notes from the field. Spring 2007, 4, 21–40. [Google Scholar] [CrossRef]
- Azevedo, R.D.S.; Falcão, K.V.G.; Amaral, I.P.G.; Leite, A.C.R.; Bezerra, R.S. Mitochondria as targets for toxicity and metabolism research using zebrafish. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2020, 1864, 129634. [Google Scholar] [CrossRef]
- Keatinge, M.; Bui, H.; Menke, A.; Chen, Y.; Sokol, A.M.; Bai, Q.; Ellett, F.; Costa, M.D.; Burke, D.; Gegg, M.; et al. Glucocerebrosidase 1 deficient Danio rerio mirror key pathological aspects of human Gaucher disease and provide evidence of early microglial activation preceding alpha-synuclein-independent neuronal cell death. Hum. Mol. Genet. 2015, 24, 6640–6652. [Google Scholar] [CrossRef]
- Pelster, B.; Burggren, W.W. Disruption of hemoglobin oxygen transport does not impact oxygen-dependent physiological processes in developing embryos of zebra fish (Danio rerio). Circ. Res. 1996, 79, 358–362. [Google Scholar] [CrossRef] [PubMed]
- Gould, A.; Missailidis, S. Targeting the hedgehog pathway: The development of cyclopamine and the development of anti-cancer drugs targeting the hedgehog pathway. Mini Rev. Med. Chem. 2011, 11, 200–213. [Google Scholar] [CrossRef] [PubMed]
- Winata, C.L.; Korzh, S.; Kondrychyn, I.; Zheng, W.; Korzh, V.; Gong, Z. Development of zebrafish swimbladder: The requirement of the hedgehog signaling specification and organization of the three tissue layers. Dev. Biol. 2009, 331, 222–236. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, W.; Yuan, M.; Liu, X. The synthetic pyrethroid deltamethrin impairs zebrafish (Danio rerio) swim bladder development. Sci. Total Environ. 2020, 701, 134870. [Google Scholar] [CrossRef] [PubMed]
- Jackson, J.K.; Funk, D.H. Temperature affects acute mayfly responses to elevated salinity: Implications for toxicity of road de-icing salts. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180081. [Google Scholar] [CrossRef] [PubMed]
- Arreguin Rebolledo, U.; Nandini, S.; Sarma, S.S.S.; Escobar-Sánchez, O. Effect of salinity and temperature on the acute and chronic toxicity of arsenic to the marine rotifers Proales similis and Brachionus ibericus. Mar. Pollut. Bull. 2020, 157, 111341. [Google Scholar] [CrossRef] [PubMed]
- Sheng, T.; Li, C.; Zhang, X.; Chi, S.; He, N.; Chen, K.; McCormick, F.; Gatalica, Z.; Xie, J. Activation of the hedgehog pathway in advanced prostate cancer. Mol. Cancer 2004, 3, 29. [Google Scholar] [CrossRef] [PubMed]
- Nüsslein-Volhard, C.; Wieschaus, E. Mutations affecting segment number and polarity in Drosophila. Nature 1980, 287, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Xie, G.; Fan, Q.; Xie, J. Activation of the hedgehog-signaling pathway in human cancer and the clinical implications. Oncogene 2010, 29, 469–481. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, C.; Zhang, J.; Lin, J.; Song, D.; Zhang, P.; Liu, Y.; Chen, L.; Zhang, L. Cadmium impairs zebrafish swim bladder development via ROS mediated inhibition of the Wnt/Hedgehog pathway. Aquat. Toxicol. 2022, 247, 106180. [Google Scholar] [CrossRef]
- Carballo, G.B.; Honorato, J.R.; Lopes, G.P.F.D.; Spohr, T.C.L.D.S. A highlight on Sonic hedgehog pathway. Cell Commun. Signal. 2018, 16, 11. [Google Scholar] [CrossRef] [PubMed]
- Kurz, E.U.; Lees-Miller, S.P. DNA damage-induced activation of ATM and ATM-dependent signaling pathways. DNA Repair. 2004, 3, 889–900. [Google Scholar] [CrossRef] [PubMed]
- Barzilai, A.; Yamamoto, K. DNA damage response to oxidative stress. DNA Repair. 2004, 3, 1109–1115. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.L.; Kremer, P.; Cravotta, C.A.; Scheirer, K.E.; Goldsmith, S.T. Long-term impacts of impervious surface cover change and roadway deicing agent application on chloride concentrations in exurban and suburban watersheds. Sci. Total Environ. 2022, 851, 157933. [Google Scholar] [CrossRef] [PubMed]
- Hintz, W.D.; Relyea, R.A. Impacts of road deicing salts on the early-life growth and development of a stream salmonid: Salt type matters. Environ. Pollut. 2017, 223, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Leite, T.; Santos, J.M.; Ferreira, T.M.; Canhoto, C.; Branco, P. Does short-term salinization of freshwater alter the behaviour of the Iberian barbel (Luciobarbus bocagei, Steindachner 1864)? Sci. Total Environ. 2019, 15, 648–655. [Google Scholar] [CrossRef]
- Velasco, J.; Gutiérrez-Cánovas, C.; Botella-Cruz, M.; Sánchez-Fernández, D.; Arribas, P.; Carbonell, J.A.; Millán, A.; Pallarés, S. Effects of salinity changes on aquatic organisms in a multiple stressor context. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180011. [Google Scholar] [CrossRef] [PubMed]
- Cañedo-Argüelles, M.; Hawkins, C.P.; Kefford, B.J.; Schäfer, R.B.; Dyack, B.J.; Brucet, S.; Buchwalter, D.; Dunlop, J.; Frör, O.; Lazorchak, J.; et al. Saving freshwater from salts. Science 2016, 351, 914–916. [Google Scholar] [CrossRef]
- Jackson, M.C.; Loewen, C.J.; Vinebrooke, R.D.; Chimimba, C.T. Net effects of multiple stressors in freshwater ecosystems: A meta-analysis. Glob. Chang. Biol. 2016, 22, 180–189. [Google Scholar] [CrossRef]
- Lawrence, C. Advances in zebrafish husbandry and management. Methods Cell Biol. 2011, 104, 429–451. [Google Scholar]
- Lawrence, C. New frontiers for zebrafish management. Methods Cell Biol. 2016, 135, 483–508. [Google Scholar] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seli, D.A.; Prendergast, A.; Ergun, Y.; Tyagi, A.; Taylor, H.S. High NaCl Concentrations in Water Are Associated with Developmental Abnormalities and Altered Gene Expression in Zebrafish. Int. J. Mol. Sci. 2024, 25, 4104. https://doi.org/10.3390/ijms25074104
Seli DA, Prendergast A, Ergun Y, Tyagi A, Taylor HS. High NaCl Concentrations in Water Are Associated with Developmental Abnormalities and Altered Gene Expression in Zebrafish. International Journal of Molecular Sciences. 2024; 25(7):4104. https://doi.org/10.3390/ijms25074104
Chicago/Turabian StyleSeli, Denis A., Andrew Prendergast, Yagmur Ergun, Antariksh Tyagi, and Hugh S. Taylor. 2024. "High NaCl Concentrations in Water Are Associated with Developmental Abnormalities and Altered Gene Expression in Zebrafish" International Journal of Molecular Sciences 25, no. 7: 4104. https://doi.org/10.3390/ijms25074104
APA StyleSeli, D. A., Prendergast, A., Ergun, Y., Tyagi, A., & Taylor, H. S. (2024). High NaCl Concentrations in Water Are Associated with Developmental Abnormalities and Altered Gene Expression in Zebrafish. International Journal of Molecular Sciences, 25(7), 4104. https://doi.org/10.3390/ijms25074104





