Research Progress of Small Plant Peptides on the Regulation of Plant Growth, Development, and Abiotic Stress
Abstract
1. Introduction
2. Types and Sources of Small Peptides in Plants
2.1. Post-Translationally Modified Peptides
2.2. Cysteine-Rich Peptides
2.3. The Relationship between Non-Secretory Small Peptides, Post-Translationally Modified Peptides, and Cysteine-Rich Peptides
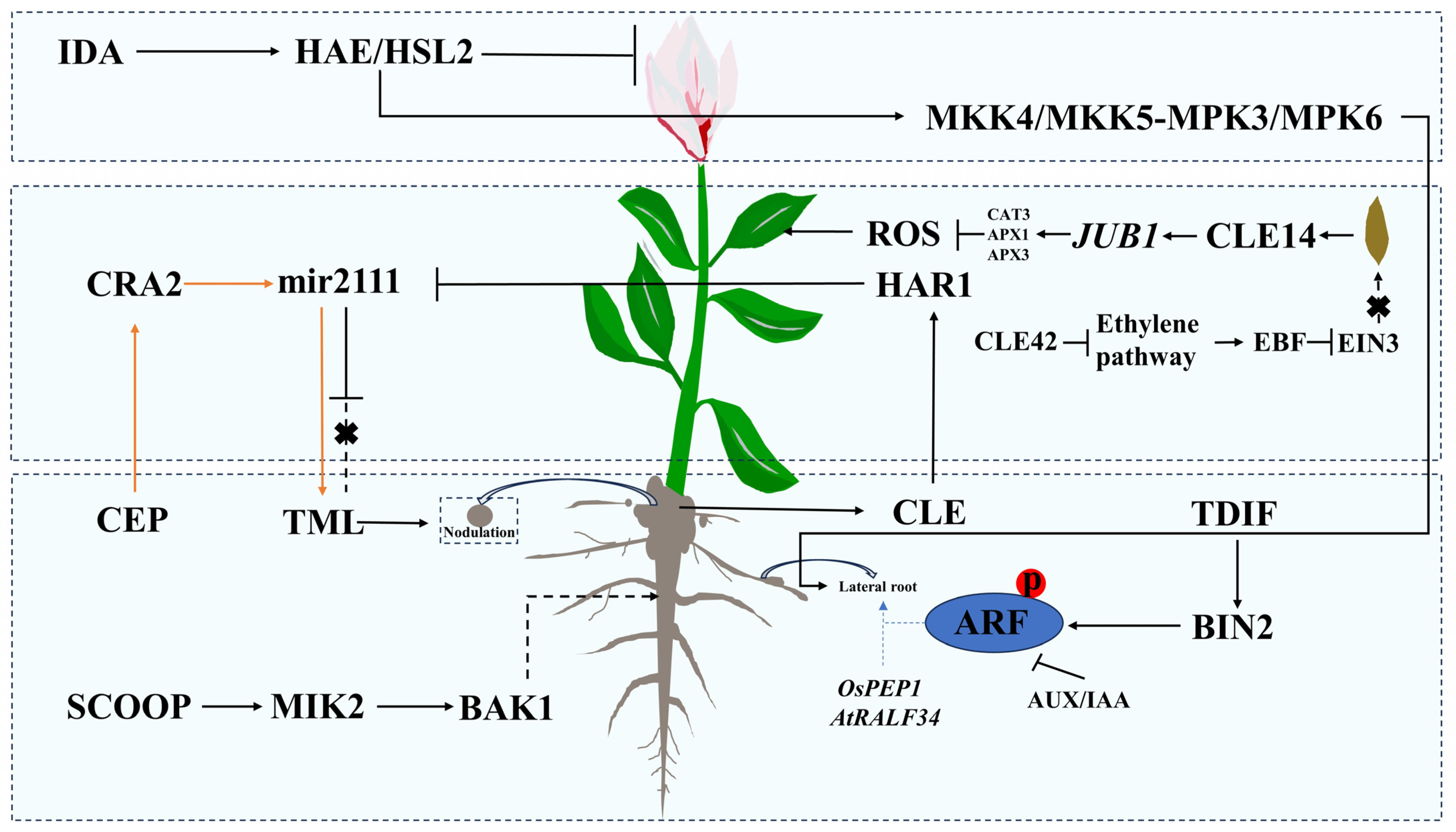
3. The Role of Small Peptides in Plant Growth and Development
3.1. The Role of Small Peptides in Root Development
3.2. The Role of Small Peptides in Leaf Senescence
3.3. Other Functions of Small Peptides in Plant Growth and Development
4. The Role of Small Peptides in Plant under Abiotic Stress
4.1. The Role of Small Plant Peptides under Drought Stress
4.2. The Role of Small Peptides under Salt Stress in Plants
4.3. The Role of Small Plant Peptides under Temperature Stress
4.4. The Role of Small Peptides under Other Stress Conditions and Biological Stress in Plants
5. Long-Distance Signal Transduction Mediated by Small Peptides under Stress
6. Plant Peptides Control the Biosynthesis of Hormones
7. Discussion and Prospects
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Djordjevic, M.A.; Mohd-Radzman, N.A.; Imin, N. Small-peptide signals that control root nodule number, development, and symbiosis. J. Exp. Bot. 2015, 17, 5171–5181. [Google Scholar] [CrossRef] [PubMed]
- Matsubayashi, Y.; Sakagami, Y. Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc. Natl. Acad. Sci. USA 1996, 15, 7623–7627. [Google Scholar] [CrossRef] [PubMed]
- Tabata, R.; Sumida, K.; Yoshii, T.; Ohyama, K.; Shinohara, H.; Matsubayashi, Y. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 2014, 6207, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Seo, P.J.; Kim, J. Signaling Peptides and Receptors Coordinating Plant Root Development. Trends Plant Sci. 2018, 4, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.C.; Katinakis, P.; Hendriks, P.; Smolders, A.; de Vries, F.; Spee, J.; van Kammen, A.; Bisseling, T.; Franssen, H. Characterization of GmENOD40, a gene showing novel patterns of cell-specific expression during soybean nodule development. Plant J. 1993, 4, 573–585. [Google Scholar] [CrossRef] [PubMed]
- Frueauf, J.B.; Dolata, M.; Leykam, J.F.; Lloyd, E.A.; Gonzales, M.; VandenBosch, K.; Kieliszewski, M.J. Peptides isolated from cell walls of Medicago truncatula nodules and uninfected root. Phytochemistry 2000, 5, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, P.; Roy, D.; Das, T.; Kundu, A.; Cartieaux, F.; Ghosh, Z.; DasGupta, M. The Natural Antisense Transcript DONE40 Derived from the lncRNA ENOD40 Locus Interacts with SET Domain Protein ASHR3 During Inception of Symbiosis in Arachis hypogaea. Mol. Plant-Microbe Interact. 2021, 9, 1057–1070. [Google Scholar] [CrossRef] [PubMed]
- Crespi, M.D.; Jurkevitch, E.; Poiret, M.; d’Aubenton-Carafa, Y.; Petrovics, G.; Kondorosi, E.; Kondorosi, A. enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 1994, 21, 5099–5112. [Google Scholar] [CrossRef] [PubMed]
- Campalans, A.; Kondorosi, A.; Crespi, M. Enod40, a short open reading frame-containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant Cell 2004, 4, 1047–1059. [Google Scholar] [CrossRef]
- Xie, H.P.; Zhao, W.; Li, W.L.; Zhang, Y.Z.; Hajny, J.; Han, H.B. Small signaling peptides mediate plant adaptions to abiotic environmental stress. Planta 2022, 4, 12. [Google Scholar] [CrossRef]
- Li, W.; Ye, T.; Ye, W.; Liang, J.; Wang, W.; Han, D.; Liu, X.; Huang, L.; Ouyang, Y.; Liao, J.; et al. S-acylation of a non-secreted peptide controls plant immunity via secreted-peptide signal activation. EMBO Rep. 2024, 25, 489–505. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.H.; Bai, J.P.; Wang, J.H. TDIF peptides regulate root growth by affecting auxin homeostasis and PINs expression in Arabidopsis thaliana. Planta 2020, 6, 109. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, Y.C.; Yamada, M. The Roles of Peptide Hormones and Their Receptors during Plant Root Development. Genes 2021, 1, 22. [Google Scholar] [CrossRef]
- Horak, H. As above, so below: CLE peptide signaling in shoot and root apical meristems. Plant Cell 2022, 4, 1159–1160. [Google Scholar] [CrossRef]
- Altmann, M.; Altmann, S.; Rodriguez, P.A.; Weller, B.; Vergara, L.E.; Palme, J.; la Rosa, N.M.D.; Sauer, M.; Wenig, M.; Villaécija-Aguilar, J.A.; et al. Extensive signal integration by the phytohormone protein network. Nature 2020, 583, 271–276. [Google Scholar] [CrossRef]
- Luo, Z.P.; Wang, J.; Li, F.Y.; Lu, Y.T.; Fang, Z.J.; Fu, M.D.; Mysore, K.S.; Wen, J.Q.; Gong, J.M.; Murray, J.D.; et al. The small peptide CEP1 and the NIN-like protein NLP1 regulate NRT2.1 to mediate root nodule formation across nitrate concentrations. Plant Cell 2023, 2, 776–794. [Google Scholar] [CrossRef] [PubMed]
- Couzigou, J.-M.; Andre, O.; Guillotin, B.; Alexandre, M.; Combier, J.-P. Use of microRNA-encoded peptide miPEP172c to stimulate nodulation in soybean. New Phytol. 2016, 2, 379–381. [Google Scholar] [CrossRef]
- Shinohara, H. Root meristem growth factor RGF, a sulfated peptide hormone in plants. Peptides 2021, 142, 170556. [Google Scholar] [CrossRef]
- Geng, R.; Shan, Y.; Li, L.; Shi, C.L.; Zhang, W.; Wang, J.; Sarwar, R.; Xue, Y.X.; Li, Y.L.; Zhu, K.M.; et al. CRISPR-mediated BnaIDA editing prevents silique shattering, floral organ abscission, and spreading of Sclerotinia sclerotiorum in Brassica napus. Plant Commun. 2022, 6, 4. [Google Scholar] [CrossRef]
- Zhou, H.P.; Xiao, F.; Zheng, Y.; Liu, G.Y.; Zhuang, Y.F.; Wang, Z.Y.; Zhang, Y.Y.; He, J.X.; Fu, C.X.; Lin, H.H. PAMP-INDUCED SECRETED PEPTIDE 3 modulates salt tolerance through RECEPTOR-LIKE KINASE 7 in plants. Plant Cell 2022, 2, 927–944. [Google Scholar] [CrossRef]
- Sin, W.C.; Lam, H.M.; Ngai, S.M. Identification of Diverse Stress-Responsive Xylem Sap Peptides in Soybean. Int. J. Mol. Sci. 2022, 15, 8641. [Google Scholar] [CrossRef]
- Stegmann, M.; Monaghan, J.; Smakowska-Luzan, E.; Rovenich, H.; Lehner, A.; Holton, N.; Belkhadir, Y.; Zipfel, C. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 2017, 6322, 287–289. [Google Scholar] [CrossRef]
- Matsubayashi, Y. Recent progress in research on small post-translationally modified peptide signals in plants. Genes. Cells 2012, 1, 1–10. [Google Scholar] [CrossRef]
- Onrubia, M.; Pollier, J.; Vanden Bossche, R.; Goethals, M.; Gevaert, K.; Moyano, E.; Vidal-Limon, H.; Cusido, R.M.; Palazon, J.; Goossens, A. Taximin, a conserved plant-specific peptide is involved in the modulation of plant-specialized metabolism. Plant Biotechnol. J. 2015, 12, 971–983. [Google Scholar] [CrossRef]
- Tang, W.H.; Ezcurra, I.; Muschietti, J.; McCormick, S. A cysteine-rich extracellular protein, LAT52, interacts with the extracellular domain of the pollen receptor kinase LePRK2. Plant Cell 2002, 9, 2277–2287. [Google Scholar] [CrossRef] [PubMed]
- Guillou, M.C.; Balliau, T.; Vergne, E.; Canut, H.; Chourre, J.; Herrera-Leon, C.; Ramos-Martin, F.; Ahmadi-Afzadi, M.; D’Amelio, N.; Ruelland, E.; et al. The PROSCOOP10 Gene Encodes Two Extracellular Hydroxylated Peptides and Impacts Flowering Time in Arabidopsis. Plants 2022, 24, 3554. [Google Scholar] [CrossRef] [PubMed]
- Guillou, M.C.; Vergne, E.; Aligon, S.; Pelletier, S.; Simonneau, F.; Rolland, A.; Chabout, S.; Mouille, G.; Gully, K.; Grappin, P.; et al. The peptide SCOOP12 acts on reactive oxygen species homeostasis to modulate cell division and elongation in Arabidopsis primary root. J. Exp. Bot. 2022, 18, 6115–6132. [Google Scholar] [CrossRef]
- Venancio, T.M.; Aravind, L. CYSTM, a novel cysteine-rich transmembrane module with a role in stress tolerance across eukaryotes. Bioinformatics 2010, 2, 149–152. [Google Scholar] [CrossRef]
- Matsubayashi, Y. Posttranslationally Modified Small-Peptide Signals in Plants. Annu. Rev. Plant Biol. 2014, 65, 385–413. [Google Scholar] [CrossRef] [PubMed]
- Stuhrwohldt, N.; Schaller, A. Regulation of plant peptide hormones and growth factors by post-translational modification. Plant Biol. 2019, 21, 49–63. [Google Scholar] [CrossRef]
- Whitewoods, C.D. Evolution of CLE peptide signalling. Semin. Cell Dev. Biol. 2021, 109, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.D.; Fiers, M. CLE peptide signaling during plant development. Protoplasma 2010, 240, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Willoughby, A.C.; Nimchuk, Z.L. WOX going on: CLE peptides in plant development. Curr. Opin. Plant Biol. 2021, 63, 102056. [Google Scholar] [CrossRef] [PubMed]
- Kucukoglu, M.; Nilsson, O. CLE peptide signaling in plants-the power of moving around. Physiol. Plant 2015, 1, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, Y. CLAVATA3, a plant peptide controlling stem cell fate in the meristem. Peptides 2021, 142, 170579. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.Y.; Schmidt, W.; Aalen, R.B.; Xu, C.; Takahashi, F. Editorial: Peptide Signaling in Plants. Front. Plant Sci. 2022, 13, 3918. [Google Scholar] [CrossRef]
- Yamaguchi, Y.L.; Ishida, T.; Sawa, S. CLE peptides and their signaling pathways in plant development. J. Exp. Bot. 2016, 16, 4813–4826. [Google Scholar] [CrossRef]
- Fletcher, J.C. Recent Advances in Arabidopsis CLE Peptide Signaling. Trends Plant Sci. 2020, 10, 1005–1016. [Google Scholar] [CrossRef]
- Stuhrwohldt, N.; Buhler, E.; Sauter, M.; Schaller, A. Phytosulfokine (PSK) precursor processing by subtilase SBT3.8 and PSK signaling improve drought stress tolerance in Arabidopsis. J. Exp. Bot. 2021, 9, 3427–3440. [Google Scholar] [CrossRef]
- Frederick, R.O.; Haruta, M.; Tonelli, M.; Lee, W.; Cornilescu, G.; Cornilescu, C.C.; Sussman, M.R.; Markley, J.L. Function and solution structure of the Arabidopsis thaliana RALF8 peptide. Protein Sci. 2019, 6, 1115–1126. [Google Scholar] [CrossRef]
- Gao, Q.F.; Wang, C.; Xi, Y.S.; Shao, Q.L.; Hou, C.C.; Li, L.G.; Luan, S. RALF signaling pathway activates MLO calcium channels to maintain pollen tube integrity. Cell Res. 2023, 33, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.R.; Pacheco, J.M.; Estevez, J.M.; Yu, F. Autocrine regulation of root hair size by the RALF-FERONIA-RSL4 signaling pathway. New Phytol. 2020, 1, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Li, L.; Wang, Z.J.; Ge, Z.X.; Li, Q.Y.; Bleckmann, A.; Wang, J.Z.; Song, Z.H.; Shi, Y.H.; Liu, T.X.; et al. RALF peptide signaling controls the polytubey block in Arabidopsis. Science 2022, 6578, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Mamaeva, A.; Lyapina, I.; Knyazev, A.; Golub, N.; Mollaev, T.; Chudinova, E.; Elansky, S.; Babenko, V.V.; Veselovsky, V.A.; Klimina, K.M.; et al. RALF peptides modulate immune response in the moss Physcomitrium patens. Front. Plant Sci. 2023, 14, 1077301. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.Y.; Kittur, F.S.; Wharton, K.N.; Umstead, M.L.; Burwell, D.B.; Thomas, M.; Qi, Q.; Zhang, J.H.; Oldham, C.E.; Burkey, K.O.; et al. A Rapid Alkalinization Factor-like Peptide EaF82 Impairs Tapetum Degeneration during Pollen Development through Induced ATP Deficiency. Cells 2023, 11, 1542. [Google Scholar] [CrossRef] [PubMed]
- Breiden, M.; Olsson, V.; Blumke, P.; Schlegel, J.; Gustavo-Pinto, K.; Dietrich, P.; Butenko, M.A.; Simon, R. The Cell Fate Controlling CLE40 Peptide Requires CNGCs to Trigger Highly Localized Ca2+ Transients in Arabidopsis thaliana Root Meristems. Plant Cell Physiol. 2021, 8, 1290–1301. [Google Scholar] [CrossRef]
- Fu, B.L.; Xu, Z.P.; Lei, Y.T.; Dong, R.; Wang, Y.A.; Guo, X.L.; Zhu, H.; Cao, Y.R.; Yan, Z. A novel secreted protein, NISP1, is phosphorylated by soybean Nodulation Receptor Kinase to promote nodule symbiosis. J. Integr. Plant Biol. 2023, 5, 1297–1311. [Google Scholar] [CrossRef] [PubMed]
- Solís-Miranda, J.; Juárez-Verdayes, M.A.; Nava, N.; Rosas, P.; Leija-Salas, A.; Cárdenas, L.; Quinto, C. The Phaseolus vulgaris Receptor-Like Kinase PvFER1 and the Small Peptides PvRALF1 and PvRALF6 Regulate Nodule Number as a Function of Nitrate Availability. Int. J. Mol. Sci. 2023, 6, 5230. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.X.; Wang, C.; Zhang, F.R.; Chen, L.; Sun, Z.X.; Cai, Y.P.; Luo, Y.Q.; Liao, J.W.; Wang, Y.L.; Cha, Y.Y.; et al. A nitrogen fixing symbiosis-specific pathway required for legume flowering. Sci. Adv. 2023, 2, eade1150. [Google Scholar] [CrossRef]
- Zhang, M.B.; Su, H.N.; Gresshoff, P.M.; Ferguson, B.J. Shoot-derived miR2111 controls legume root and nodule development. Plant Cell Environ. 2021, 5, 1627–1641. [Google Scholar] [CrossRef]
- Moreau, C.; Gautrat, P.; Frugier, F. Nitrate-induced CLE35 signaling peptides inhibit nodulation through the SUNN receptor and miR2111 repression. Plant Physiol. 2021, 3, 1216–1228. [Google Scholar] [CrossRef]
- Tsikou, D.; Yan, Z.; Holt, D.B.; Abel, N.B.; Reid, D.E.; Madsen, L.H.; Bhasin, H.; Sexauer, M.; Stougaard, J.; Markmann, K. Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science 2018, 6411, 233–235. [Google Scholar] [CrossRef]
- Gautrat, P.; Laffont, C.; Frugier, F. Compact Root Architecture 2 Promotes Root Competence for Nodulation through the miR2111 Systemic Effector. Curr. Biol. 2020, 7, 1339–1345. [Google Scholar] [CrossRef]
- Nanjo, Y.; Maruyama, K.; Yasue, H.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Komatsu, S. Transcriptional responses to flooding stress in roots including hypocotyl of soybean seedlings. Plant Mol. Biol. 2011, 77, 129–144. [Google Scholar] [CrossRef]
- Liu, Y.T.; Zuo, T.T.; Qiu, Z.W.; Zhuang, K.Q.; Hu, S.P.; Han, H.B. Genome-wide identification reveals the function of CEP peptide in cucumber root development. Plant Physiol. Biochem. 2021, 169, 119–126. [Google Scholar] [CrossRef]
- Huang, A.X.; Cui, T.T.; Zhang, Y.; Ren, X.F.; Wang, M.F.; Jia, L.Y.; Zhang, Y.H.; Wang, G.D. CRISPR/Cas9-Engineered Large Fragment Deletion Mutations in Arabidopsis CEP Peptide-Encoding Genes Reveal Their Role in Primary and Lateral Root Formation. Plant Cell Physiol. 2023, 1, 19–26. [Google Scholar] [CrossRef]
- Qiu, Z.W.; Zhuang, K.Q.; Liu, Y.T.; Ge, X.M.; Chen, C.; Hu, S.P.; Han, H.B. Functional characterization of C-TERMINALLY ENCODED PEPTIDE (CEP) family in Brassica rapa L. Plant Signal. Behav. 2022, 1, 10. [Google Scholar] [CrossRef]
- Xiang, D.; Meng, F.N.; Wang, A.D.; Wu, Y.R.; Wang, Z.Y.; Zheng, S.J.; Mao, C.A.Z. Root-secreted peptide OsPEP1 regulates primary root elongation in rice. Plant J. 2021, 2, 480–492. [Google Scholar] [CrossRef]
- De Rybel, B.; Vassileva, V.; Parizot, B.; Demeulenaere, M.; Grunewald, W.; Audenaert, D.; Van Campenhout, J.; Overvoorde, P.; Jansen, L.; Vanneste, S.; et al. A Novel Aux/IAA28 Signaling Cascade Activates GATA23-Dependent Specification of Lateral Root Founder Cell Identity. Curr. Biol. 2010, 19, 1697–1706. [Google Scholar] [CrossRef]
- Shumilina, J.; Kiryushkin, A.S.; Frolova, N.; Mashkina, V.; Ilina, E.L.; Puchkova, V.A.; Danko, K.; Silinskaya, S.; Serebryakov, E.B.; Soboleva, A.; et al. Integrative Proteomics and Metabolomics Analysis Reveals the Role of Small Signaling Peptide Rapid Alkalinization Factor 34 (RALF34) in Cucumber Roots. Int. J. Mol. Sci. 2023, 8, 7654. [Google Scholar] [CrossRef]
- Zhang, B.L.; Xin, B.N.; Sun, X.Q.; Chao, D.; Zheng, H.W.; Peng, L.Y.; Chen, X.X.; Zhang, L.; Yu, J.Y.; Ma, D.; et al. Small peptide signaling via OsCIF1/2 mediates Casparian strip formation at the root endodermal and nonendodermal cell layers in rice. Plant Cell 2023, 36, 383–403. [Google Scholar] [CrossRef]
- Zhang, Z.L.; Liu, C.; Li, K.; Li, X.X.; Xu, M.M.; Guo, Y.F. CLE14 functions as a “brake signal”to suppress age-dependent and stress-induced leaf senescence by promoting JUB1-mediated ROS scavenging in Arabidopsis. Mol. Plant 2022, 1, 179–188. [Google Scholar] [CrossRef]
- Zhang, Y.; Tan, S.Y.; Gao, Y.H.; Kan, C.C.; Wang, H.L.; Yang, Q.; Xia, X.L.; Ishida, T.; Sawa, S.; Guo, H.W.; et al. CLE42 delays leaf senescence by antagonizing ethylene pathway in Arabidopsis. New Phytol. 2022, 2, 550–562. [Google Scholar] [CrossRef]
- Czyzewicz, N.; Yue, K.; Beeckman, T.; De Smet, I. Message in a bottle: Small signalling peptide outputs during growth and development. J. Exp. Bot. 2013, 17, 5281–5296. [Google Scholar] [CrossRef]
- Djami-Tchatchou, A.T.; Tetorya, M.; Godwin, J.; Codjoe, J.M.; Li, H.; Shah, D.M. Small Cationic Cysteine-Rich Defensin-Derived Antifungal Peptide Controls White Mold in Soybean. J. Fungi 2023, 9, 873. [Google Scholar] [CrossRef]
- Wang, P.Y.; Jia, H.M.; Guo, T.; Zhang, Y.Y.; Wang, W.Q.; Nishimura, H.; Li, Z.G.; Kawano, Y. The secreted immune response peptide 1 functions as a phytocytokine in rice immunity. J. Exp. Bot. 2023, 3, 1059–1073. [Google Scholar] [CrossRef]
- Wang, J.; Wang, D.; Ji, X.B.; Wang, J.; Klosterman, S.J.; Dai, X.F.; Chen, J.Y.; Subbarao, K.V.; Hao, X.J.; Zhang, D.D. The Verticillium dahliae Small Cysteine-Rich Protein VdSCP23 Manipulates Host Immunity. Int. J. Mol. Sci. 2023, 11, 9403. [Google Scholar] [CrossRef]
- Wu, Q.Q.; Kuang, K.Y.; Lyu, M.H.; Zhao, Y.; Li, Y.; Li, J.; Pan, Y.; Shi, H.; Zhong, S.W. Allosteric deactivation of PIFs and EIN3 by microproteins in light control of plant development. Proc. Natl. Acad. Sci. USA 2020, 31, 18858–18868. [Google Scholar] [CrossRef]
- Roy, S.; Griffiths, M.; Torres-Jerez, I.; Sanchez, B.; Antonelli, E.; Jain, D.; Krom, N.; Zhang, S.L.; York, L.M.; Scheible, W.R.; et al. Application of Synthetic Peptide CEP1 Increases Nutrient Uptake Rates Along Plant Roots. Front. Plant Sci. 2022, 12, 793145. [Google Scholar] [CrossRef]
- Su, Q.; Wang, K.; Zhang, Z.Y. Ecotopic Expression of the Antimicrobial Peptide DmAMP1W Improves Resistance of Transgenic Wheat to Two Diseases: Sharp Eyespot and Common Root Rot. Int. J. Mol. Sci. 2020, 2, 647. [Google Scholar] [CrossRef]
- Dong, J.H.; Wang, Y.; Xu, L.; Li, B.S.; Wang, K.; Ying, J.L.; He, Q.; Liu, L.W. RsCLE22a regulates taproot growth through an auxin signaling-related pathway in radish (Raphanus sativus L.). J. Exp. Bot. 2023, 1, 233–250. [Google Scholar] [CrossRef]
- Liu, P.; Shi, C.; Liu, S.; Lei, J.; Lu, Q.; Hu, H.; Ren, Y.; Zhang, N.; Sun, C.; Chen, L.; et al. A papain-like cysteine protease-released small signal peptide confers wheat resistance to wheat yellow mosaic virus. Nat. Commun. 2023, 1, 7773. [Google Scholar] [CrossRef]
- Niu, X.Y.; Yamamoto, N.; Yang, G.J.; Lin, H.; Jiang, L.J.; Liu, Y.; Zheng, A.P. A small secreted protein, RsMf8HN, in Rhizoctonia solani triggers plant immune response, which interacts with rice OsHIPP28. Microbiol. Res. 2023, 266, 127219. [Google Scholar] [CrossRef]
- Tong, X.; Zhao, J.J.; Feng, Y.L.; Wang, X.B. The small peptide VISP1 acts as a selective autophagy receptor regulating plant-virus interactions. Autophagy 2023, 12, 3246–3247. [Google Scholar] [CrossRef]
- Lan, Z.J.; Song, Z.H.; Wang, Z.J.; Li, L.; Liu, Y.Q.; Zhi, S.H.; Wang, R.H.; Wang, J.Z.; Li, Q.Y.; Bleckmann, A.; et al. Antagonistic RALF peptides control an intergeneric hybridization barrier on Brassicaceae stigmas. Cell 2023, 22, 4773–4787. [Google Scholar] [CrossRef]
- Takahashi, F.; Hanada, K.; Kondo, T.; Shinozaki, K. Hormone-like peptides and small coding genes in plant stress signaling and development. Curr. Opin. Plant Biol. 2019, 51, 88–95. [Google Scholar] [CrossRef]
- Kim, S.I.; Lee, K.H.; Kwak, J.S.; Kwon, D.H.; Song, J.T.; Seo, H.S. Overexpression of Rice OsS1Fa1 Gene Confers Drought Tolerance in Arabidopsis. Plants 2021, 10, 2181. [Google Scholar] [CrossRef]
- Cui, Y.C.; Li, M.J.; Yin, X.M.; Song, S.F.; Xu, G.Y.; Wang, M.L.; Li, C.Y.; Peng, C.; Xia, X.J. OsDSSR1, a novel small peptide, enhances drought tolerance in transgenic rice. Plant Sci. 2018, 270, 85–96. [Google Scholar] [CrossRef]
- Jing, X.-Q.; Shi, P.-T.; Zhang, R.; Zhou, M.-R.; Shalmani, A.; Wang, G.-F.; Liu, W.-T.; Li, W.-Q.; Chen, K.-M. Rice kinase OsMRLK63 contributes to drought tolerance by regulating reactive oxygen species production. Plant Physiol. 2023, 194, 2679–2696. [Google Scholar] [CrossRef]
- Xu, K.X.; Tian, D.D.; Wang, T.J.; Zhang, A.J.; Elsadek, M.A.Y.; Liu, W.H.; Chen, L.P.; Guo, Y.F. Small secreted peptides (SSPs) in tomato and their potential roles in drought stress response. Mol. Hortic. 2023, 1, 17. [Google Scholar] [CrossRef]
- Tian, D.D.; Xie, Q.; Deng, Z.C.; Xue, J.; Li, W.; Zhang, Z.L.; Dai, Y.F.; Zheng, B.; Lu, T.G.; De Smet, I.; et al. Small secreted peptides encoded on the wheat (Triticum aestivum L.) genome and their potential roles in stress responses. Front. Plant Sci. 2022, 18, 1000297. [Google Scholar] [CrossRef]
- Christmann, A.; Grill, E. Peptide signal alerts plants to drought. Nature 2018, 7700, 178–179. [Google Scholar] [CrossRef]
- Takahashi, F.; Suzuki, T.; Osakabe, Y.; Betsuyaku, S.; Kondo, Y.; Dohmae, N.; Fukuda, H.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 2018, 7700, 235–238. [Google Scholar] [CrossRef]
- Farago, D.; Zsigmond, L.; Benyo, D.; Alcazar, R.; Rigo, G.; Ayaydin, F.; Rabilu, S.A.; Hunyadi-Gulyas, E.; Szabados, L. Small paraquat resistance proteins modulate paraquat and ABA responses and confer drought tolerance to overexpressing Arabidopsis plants. Plant Cell Environ. 2022, 7, 1985–2003. [Google Scholar] [CrossRef]
- Nagar, P.; Kumar, A.; Jain, M.; Kumari, S.; Mustafiz, A. Genome-wide analysis and transcript profiling of PSKR gene family members in Oryza sativa. PLoS ONE 2020, 7, e0236349. [Google Scholar] [CrossRef]
- Reichardt, S.; Piepho, H.P.; Stintzi, A.; Schaller, A. Peptide signaling for drought-induced tomato flower drop. Science 2020, 6485, 1482–1485. [Google Scholar] [CrossRef]
- Hao, Z.D.; Wu, H.; Zheng, R.H.; Li, R.; Zhu, Z.L.; Chen, Y.; Lu, Y.; Cheng, T.L.; Shi, J.S.; Chen, J.H. The plant peptide hormone phytosulfokine promotes somatic embryogenesis by maintaining redox homeostasis in Cunninghamia lanceolata. Plant J. 2023, 113, 716–733. [Google Scholar] [CrossRef]
- Zhang, J.; Yuan, J.; Li, Z.; Fu, C.; Xu, M.; Yang, J.; Jiang, X.; Zhou, B.; Ye, X.; Xu, C. Exploring and exploiting plant cyclic peptides for drug discovery and development. Med. Res. Rev. 2021, 6, 3096–3117. [Google Scholar] [CrossRef]
- Nakaminami, K.; Okamoto, M.; Higuchi-Takeuchi, M.; Yoshizumi, T.; Yamaguchi, Y.; Fukao, Y.; Shimizu, M.; Ohashi, C.; Tanaka, M.; Matsui, M.; et al. AtPep3 is a hormone-like peptide that plays a role in the salinity stress tolerance of plants. Proc. Natl. Acad. Sci. USA 2018, 22, 5810–5815. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, Z.H.; Wu, D.S.; Yu, F. RALF-FERONIA Signaling: Linking Plant Immune Response with Cell Growth. Plant Commun. 2020, 4, 100084. [Google Scholar] [CrossRef]
- Zhao, C.Z.; Zayed, O.; Yu, Z.P.; Jiang, W.; Zhu, P.P.; Hsu, C.C.; Zhang, L.R.; Tao, W.A.; Lozano-Duran, R.; Zhu, J.K. Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 51, 13123–13128. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Yu, Z.P.; Zhang, D.; Huang, J.G.; Wu, C.G.; Yang, G.D.; Yan, K.; Zhang, S.Z.; Zheng, C.C. CYSTM, a Novel Non-Secreted Cysteine-Rich Peptide Family, Involved in Environmental Stresses in Arabidopsis thaliana. Plant Cell Physiol. 2018, 2, 423–438. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Yu, Z.P.; Zhang, S.Z.; Wu, C.A.; Yang, G.D.; Yan, K.; Zheng, C.C.; Huang, J.G. CYSTM3 negatively regulates salt stress tolerance in Arabidopsis. Plant Mol. Biol. 2019, 99, 395–406. [Google Scholar] [CrossRef]
- Guo, Z.H.; Cai, L.J.; Liu, C.X.; Chen, Z.Q.; Guan, S.W.; Ma, W.D.; Pan, G.J. Low-temperature stress affects reactive oxygen species, osmotic adjustment substances, and antioxidants in rice (Oryza sativa L.) at the reproductive stage. Sci. Rep. 2022, 1, 13. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.A.; Fariduddin, Q.; Yusuf, M. Low-temperature stress: Is. phytohormones application a remedy? Environ. Sci. Pollut. Res. 2017, 27, 21574–21590. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.J.; Zhang, L.P.; Song, S.R.; Wang, L.; Xu, W.P.; Zhang, C.X.; Wang, S.P.; Liu, H.F.; Ma, C. vvi-miPEP172b and vvi-miPEP3635b increase cold tolerance of grapevine by regulating the corresponding MIRNA genes. Plant Sci. 2022, 10, 111450. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhang, G.; Wu, M. CLE Peptide Signaling and Crosstalk with Phytohormones and Environmental Stimuli. Front. Plant Sci. 2015, 1211, 167813. [Google Scholar] [CrossRef]
- Meng, X.X.; Li, W.F.; Shen, R.F.; Lan, P. Ectopic expression of IMA small peptide genes confers tolerance to cadmium stress in Arabidopsis through activating the iron deficiency response. J. Hazard. Mater. 2022, 14, 126913. [Google Scholar] [CrossRef]
- Lu, L.; Chen, X.; Chen, J.; Zhang, Z.; Zhang, Z.; Sun, Y.; Wang, Y.; Xie, S.; Ma, Y.; Song, Y.; et al. MicroRNA-encoded regulatory peptides modulate cadmium tolerance and accumulation in rice. Plant Cell Environ. 2024. [Google Scholar] [CrossRef]
- Vie, A.K.; Najafi, J.; Winge, P.; Cattan, E.; Wrzaczek, M.; Kangasjarvi, J.; Miller, G.; Brembu, T.; Bones, A.M. The IDA-LIKE peptides IDL6 and IDL7 are negative modulators of stress responses in Arabidopsis thaliana. J. Exp. Bot. 2017, 13, 3557–3571. [Google Scholar] [CrossRef]
- Segonzac, C.; Monaghan, J. Modulation of plant innate immune signaling by small peptides. Curr. Opin. Plant Biol. 2019, 51, 22–28. [Google Scholar] [CrossRef]
- Tang, J.; Wu, D.S.; Li, X.X.; Wang, L.F.; Xu, L.; Zhang, Y.; Xu, F.; Liu, H.B.; Xie, Q.J.; Dai, S.J.; et al. Plant immunity suppression via PHR1-RALF-FERONIA shapes the root microbiome to alleviate phosphate starvation. EMBO J. 2022, 6, e109102. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Zhang, N.; Zhang, L.N.; He, Y.X.; Cai, C.; Zhou, J.G.; Li, J.; Meng, X.Z. Perception of the pathogen-induced peptide RGF7 by the receptor-like kinases RGI4 and RGI5 triggers innate immunity in Arabidopsis thaliana. New Phytol. 2021, 3, 1110–1125. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Liu, S.Y.; Zou, J.Z.; Zhao, J.J.; Zhu, F.F.; Chai, L.X.; Wang, Y.; Han, C.G.; Wang, X.B. A small peptide inhibits siRNA amplification in plants by mediating autophagic degradation of SGS3/RDR6 bodies. EMBO J. 2021, 15, e108050. [Google Scholar] [CrossRef] [PubMed]
- Combest, M.M.; Moroz, N.; Tanaka, K.; Rogan, C.J.; Anderson, J.C.; Thura, L.; Rakotondrafara, A.M.; Goyer, A. StPIP1, a PAMP-induced peptide in potato, elicits plant defenses and is associated with disease symptom severity in a compatible interaction with Potato virus Y. J. Exp. Bot. 2021, 12, 4472–4488. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, S.; Kawasaki, A.; Makino, Y. Characterization of Oligopeptides in Solanum lycopersicum Xylem Exudates. Life 2022, 4, 592. [Google Scholar] [CrossRef] [PubMed]
- Koenig, A.M.; Hoffmann-Benning, S. The interplay of phloem-mobile signals in plant development and stress response. Biosci. Rep. 2020, 20, BSR20193329. [Google Scholar] [CrossRef]
- Chapman, K.; Ivanovici, A.; Taleski, M.; Sturrock, C.J.; Ng, J.L.P.; Mohd-Radzman, N.A.; Frugier, F.; Bennett, M.J.; Mathesius, U.; Djordjevic, M.A. CEP receptor signalling controls root system architecture in Arabidopsis and Medicago. New Phytol. 2020, 6, 1809–1821. [Google Scholar] [CrossRef] [PubMed]
- Chapman, K.; Taleski, M.; Ogilvie, H.A.; Imin, N.; Djordjevic, M.A. CEP-CEPR1 signalling inhibits the sucrose-dependent enhancement of lateral root growth. J. Exp. Bot. 2019, 15, 3955–3967. [Google Scholar] [CrossRef]
- Xie, Y.H.; Zhang, F.J.; Sun, P.; Li, Z.Y.; Zheng, P.F.; Gu, K.D.; Hao, Y.J.; Zhang, Z.L.; You, C.X. Apple receptor-like kinase FERONIA regulates salt tolerance and ABA sensitivity in Malus domestica. J. Plant Physiol. 2022, 270, 1536161. [Google Scholar] [CrossRef]
- Zhao, C.Z.; Jiang, W.; Zayed, O.; Liu, X.; Tang, K.; Nie, W.F.; Li, Y.L.; Xie, S.J.; Li, Y.; Long, T.D.; et al. The LRXs-RALFs-FER module controls plant growth and salt stress responses by modulating multiple plant hormones. Natl. Sci. Rev. 2021, 1, nwaa149. [Google Scholar] [CrossRef] [PubMed]
- Lucena, C.; Waters, B.M.; Romera, F.J.; García, M.J.; Morales, M.; Alcántara, E.; Pérez-Vicente, R. Ethylene could influence ferric reductase, iron transporter, and H-ATPase gene expression by affecting FER or FER-like) gene activity. J. Exp. Bot. 2006, 15, 4145–4154. [Google Scholar] [CrossRef]
- Yang, R.R.; Wang, Z.C.; Zhao, L.; Liu, J.; Meng, J.; Luan, Y.S. Secreted Peptide SpPIP1 Modulates Disease Resistance and Salt Tolerance in Tomato. J. Agric. Food Chem. 2023, 32, 12264–12279. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.H.; Hu, X.; Ye, H.Z.; Wang, Y.; Yang, Q.; Liang, X.D.; Wang, Z.L.; Zhou, Y.F.; Wen, M.M.; Yuan, X.Y.; et al. Cell-specific clock-controlled gene expression program regulates rhythmic fiber cell growth in cotton. Genome Biol. 2023, 1, 49. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.; Zhu, S.S.; Joos, L.; Roberts, I.; Nikonorova, N.; Vu, L.D.; Stes, E.; Cho, H.; Larrieu, A.; Xuan, W.; et al. The CEP5 Peptide Promotes Abiotic Stress Tolerance, As Revealed by Quantitative Proteomics, and Attenuates the AUX/IAA Equilibrium in Arabidopsis. Mol. Cell. Proteom. 2020, 8, 1248–1262. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.Y.; Hu, X.C.; Ren, M.F.; Ma, F.; Fu, J.; Cui, H.C. Stem-cell-expressed DEVIL-like small peptides maintain root growth under abiotic stress via abscisic acid signaling. Plant Physiol. 2023, 194, 2372–2386. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.A.; Wang, C.Y.; Ji, Z.; Lu, J.Y.; Zhang, L.; Li, C.L.; Huang, J.G.; Yang, G.D.; Yan, K.; Zhang, S.Z.; et al. Regulation of drought tolerance in Arabidopsis involves the PLATZ4-mediated transcriptional repression of plasma membrane aquaporin PIP2;8. Plant J. 2023, 115, 434–451. [Google Scholar] [CrossRef] [PubMed]
- Chapman, K.; Taleski, M.; Frank, M.; Djordjevic, M.A. C-TERMINALLY ENCODED PEPTIDE (CEP) and cytokinin hormone signaling intersect to promote shallow lateral root angles. J. Exp. Bot. 2023, 75, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Song, H.W.; Wang, X.F.; Hu, W.C.; Yang, X.H.; Diao, E.J.; Shen, T.; Qiang, Q. A cold-induced phytosulfokine peptide is related to the improvement of loquat fruit chilling tolerance. Food Chem. 2017, 232, 434–442. [Google Scholar] [CrossRef]
- Li, Z.S.; Jin, J.R.; Wang, Y.; Long, W.T.; Ding, Y.H.; Hu, H.Y.; Wei, L.Y. ExamPle: Explainable deep learning framework for the prediction of plant small secreted peptides. Bioinformatics 2023, 3, btad108. [Google Scholar] [CrossRef]
- Teufel, F.; Armenteros, J.J.A.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 7, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- Song, X.F.; Guo, P.; Ren, S.C.; Xu, T.T.; Liu, C.M. Antagonistic Peptide Technology for Functional Dissection of CLV3/ESR Genes in Arabidopsis. Plant Physiol. 2013, 3, 1076–1085. [Google Scholar] [CrossRef] [PubMed]
- Bruck, S.; Pfannstiel, J.; Ingram, G.; Stintzi, A.; Schaller, A. Analysis of peptide hormone maturation and processing specificity using isotope-labeled peptides. In Methods in Molecular Biology; Springer: Clifton, NJ, USA, 2023; pp. 323–335. [Google Scholar]
- Ma, D.C.; Endo, S.; Betsuyaku, E.; Fujiwara, T.; Betsuyaku, S.; Fukuda, H. Root-specific CLE3 expression is required for WRKY33 activation in Arabidopsis shoots. Plant Mol. Biol. 2022, 3, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, S.; Shinohara, H.; Mori, T.; Matsubayashi, Y.; Kawaguchi, M. Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat. Commun. 2013, 4, 2191. [Google Scholar] [CrossRef] [PubMed]

| Types | Small Peptides | Function | References |
|---|---|---|---|
| Non-secretory small peptides | early nodulin | root nodule development, intracellular localization of proteins, etc. | [5,6,7,8,9] |
| ROT4 | regulates plant immunity, modulates cell proliferation, etc. | [11] | |
| Post-translationally modified peptides | PSK | abiotic stress, cell division, root development, etc. | [2] |
| TDIF | tracheary element differentiation inhibitory factor, root development, etc. | [12] | |
| CLE | abiotic stress, root development, leaf senescence, etc. | [13] | |
| CLV3 | differentiation of the apical meristem, etc. | [14] | |
| CIF | formation of the Casparian strip, etc. | [15] | |
| CEP | abiotic stress, root development, nodulation, etc. | [16] | |
| miPEP | abiotic stress, fruit ripening, flower development, photosynthesis, nodulation, etc. | [17] | |
| RGF | maintaining the activity of the root apical meristem, abiotic stress, etc. | [18] | |
| IDA | floral organ abscission, etc. | [19] | |
| PIP | abiotic stress, etc. | [20] | |
| PSY | abiotic stress, root development, etc. | [21] | |
| Cysteine-rich peptides | RALFs | root development, cell wall remodeling, immune response, abiotic stress, etc. | [22] |
| LURE | promoting pollen tube germination, etc. | [23] | |
| TAX | regulating plant-specific metabolism, etc. | [24] | |
| EPF | regulating epidermal cell patterns, etc. | [23] | |
| LAT52 | pollen hydration process, pollen tube growth, etc. | [25] | |
| SCOOPs | root development, immune response, etc. | [26,27] | |
| CYSTM | abiotic stress, etc. | [28] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, G.; Zhang, Y.; Chen, Z.; Xue, X.; Fan, H. Research Progress of Small Plant Peptides on the Regulation of Plant Growth, Development, and Abiotic Stress. Int. J. Mol. Sci. 2024, 25, 4114. https://doi.org/10.3390/ijms25074114
Ren G, Zhang Y, Chen Z, Xue X, Fan H. Research Progress of Small Plant Peptides on the Regulation of Plant Growth, Development, and Abiotic Stress. International Journal of Molecular Sciences. 2024; 25(7):4114. https://doi.org/10.3390/ijms25074114
Chicago/Turabian StyleRen, Guocheng, Yanling Zhang, Zengting Chen, Xin Xue, and Hai Fan. 2024. "Research Progress of Small Plant Peptides on the Regulation of Plant Growth, Development, and Abiotic Stress" International Journal of Molecular Sciences 25, no. 7: 4114. https://doi.org/10.3390/ijms25074114
APA StyleRen, G., Zhang, Y., Chen, Z., Xue, X., & Fan, H. (2024). Research Progress of Small Plant Peptides on the Regulation of Plant Growth, Development, and Abiotic Stress. International Journal of Molecular Sciences, 25(7), 4114. https://doi.org/10.3390/ijms25074114





