Energy Transfer and Radical-Pair Dynamics in Photosystem I with Different Red Chlorophyll a Pigments
Abstract
1. Introduction
2. Results
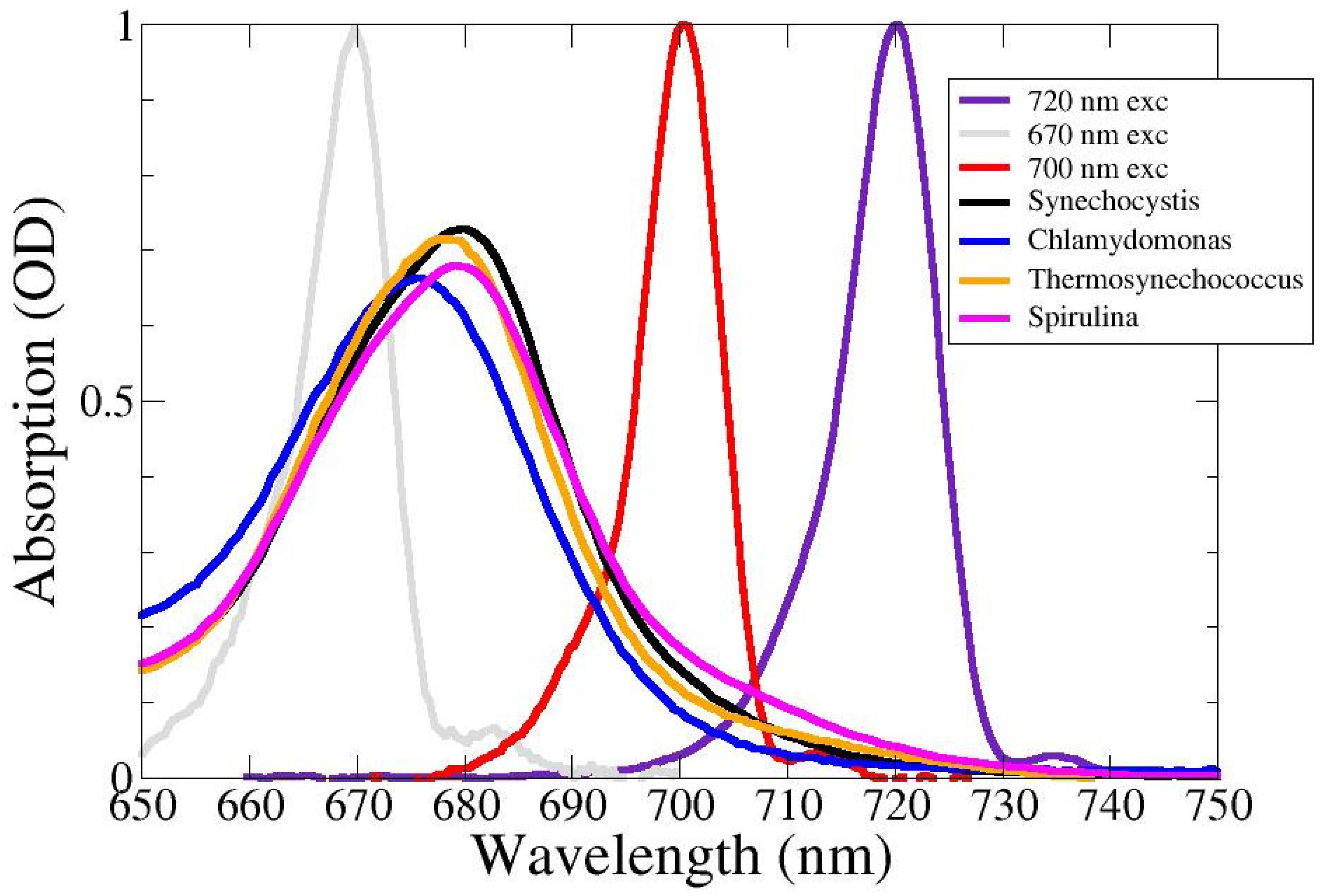
2.1. Absorption Spectra
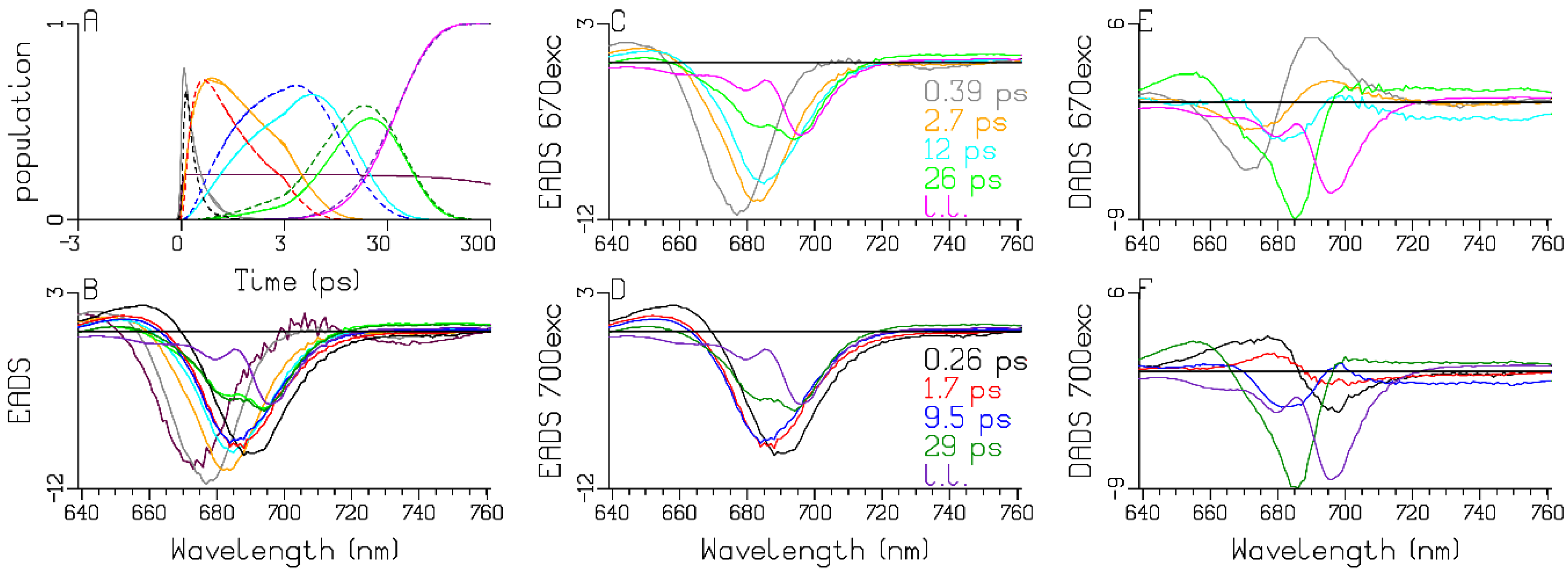
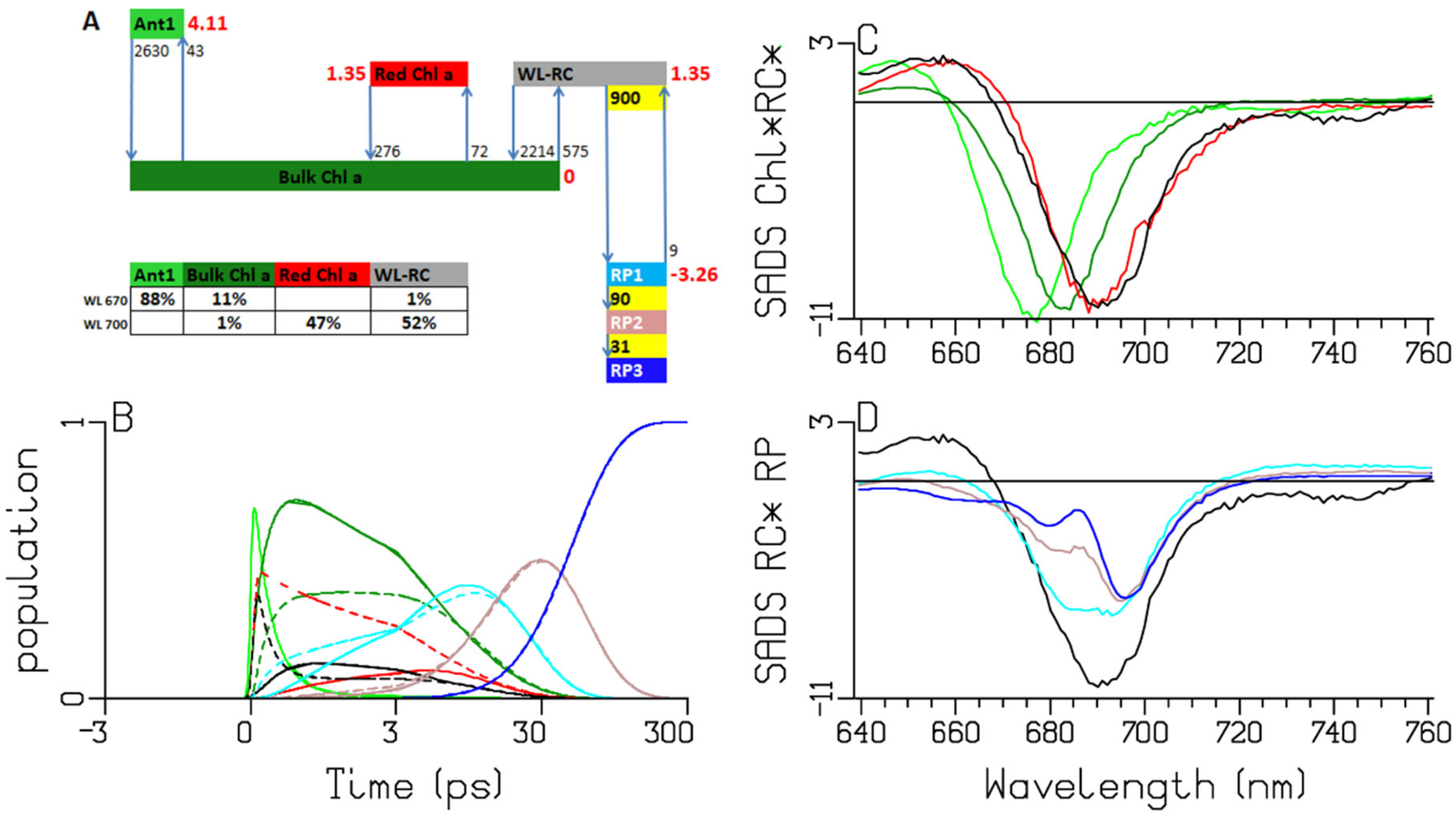
2.2. Chlamydomonas reinhardtii
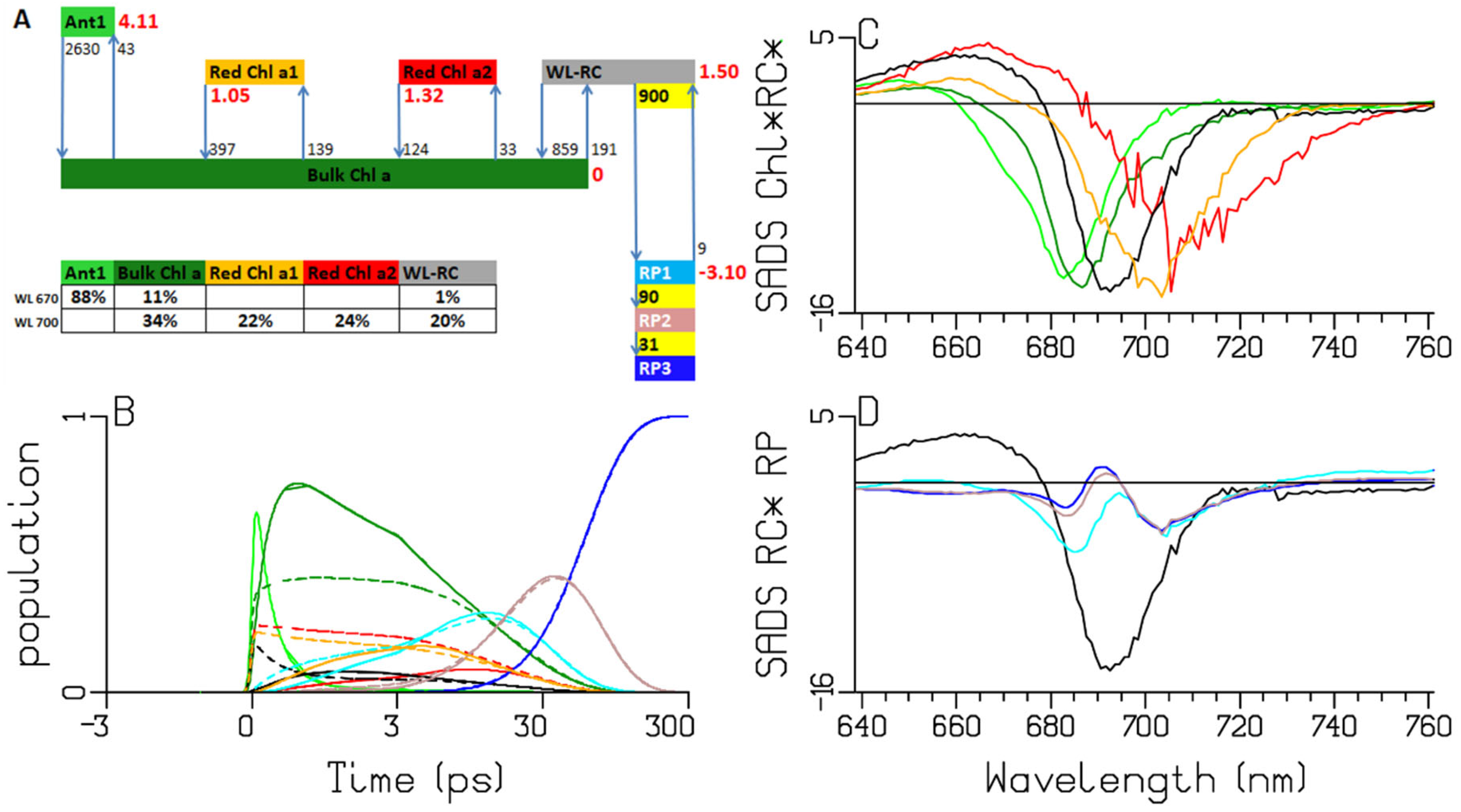
2.3. Synechocystis PCC6803
2.4. Thermosynechococcus elongatus
2.5. Spirulina Platensis
3. Discussion
3.1. Sub-ps Dynamics
3.2. Thermodynamic Considerations
3.3. Comparison of the RC and RP SADS
4. Materials and Methods
4.1. Transient-Absorption Experiments
4.2. Time-Resolved Fluorescence Spectra
4.3. Global and Target Analysis of Time-Resolved Spectra
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Model for the Observations
References
- Jordan, P.; Fromme, P.; Witt, H.T.; Klukas, O.; Saenger, W.; Krauss, N. Three-dimensional structure of cyanobacterial photosystem I at 2.5 angstrom resolution. Nature 2001, 411, 909–917. [Google Scholar] [CrossRef]
- Blankenship, R.E. Molecular Mechanisms of Photosynthesis, 3rd ed.; Wiley: Chichester, UK, 2021. [Google Scholar]
- Melkozernov, A.N.; Barber, J.; Blankenship, R.E. Light Harvesting in Photosystem I Supercomplexes. Biochemistry 2006, 45, 331–345. [Google Scholar] [CrossRef] [PubMed]
- Savikhin, S. Ultrafast Optical Spectroscopy of Photosystem I. In Photosystem I: The Light-Driven Plastocyanin: Ferredoxin Oxidoreductase; Golbeck, J.H., Ed.; Springer: Dordrecht, The Netherlands, 2006; pp. 155–175. [Google Scholar]
- Croce, R.; van Amerongen, H. Light-harvesting in photosystem I. Photosynth. Res. 2013, 116, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Holzwarth, A.R. Ultrafast Primary Reactions in the Photosystems of Oxygen-Evolving Organisms. In Ultrashort Laser Pulses in Biology and Medicine; Braun, M., Gilch, P., Zinth, W., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 141–164. [Google Scholar]
- Govindjee; Shevela, D.; Björn, L.O. Evolution of the Z-scheme of photosynthesis: A perspective. Photosynth. Res. 2017, 133, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Cherepanov, D.A.; Shelaev, I.V.; Gostev, F.E.; Petrova, A.; Aybush, A.V.; Nadtochenko, V.A.; Xu, W.; Golbeck, J.H.; Semenov, A.Y. Primary charge separation within the structurally symmetric tetrameric Chl2APAPBChl2B chlorophyll exciplex in photosystem I. J. Photochem. Photobiol. B Biol. 2021, 217, 112154. [Google Scholar] [CrossRef] [PubMed]
- van Grondelle, R.; Dekker, J.P.; Gillbro, T.; Sundström, V. Energy-Transfer and Trapping in Photosynthesis. Biochim. Biophys. Acta 1994, 1187, 1–65. [Google Scholar] [CrossRef]
- Gobets, B.; Valkunas, L.; van Grondelle, R. Bridging the Gap between Structural and Lattice Models: A Parameterization of Energy Transfer and Trapping in Photosystem I. Biophys. J. 2003, 85, 3872–3882. [Google Scholar] [CrossRef][Green Version]
- Gorka, M.; Charles, P.; Kalendra, V.; Baldansuren, A.; Lakshmi, K.V.; Golbeck, J.H. A dimeric chlorophyll electron acceptor differentiates type I from type II photosynthetic reaction centers. iScience 2021, 24, 102719. [Google Scholar] [CrossRef]
- Gobets, B.; van Stokkum, I.H.M.; Rogner, M.; Kruip, J.; Schlodder, E.; Karapetyan, N.V.; Dekker, J.P.; van Grondelle, R. Time-resolved fluorescence emission measurements of photosystem I particles of various cyanobacteria: A unified compartmental model. Biophys. J. 2001, 81, 407–424. [Google Scholar] [CrossRef]
- Müller, M.G.; Niklas, J.; Lubitz, W.; Holzwarth, A.R. Ultrafast transient absorption studies on Photosystem I reaction centers from Chlamydomonas reinhardtii. 1. A new interpretation of the energy trapping and early electron transfer steps in Photosystem I. Biophys. J. 2003, 85, 3899–3922. [Google Scholar] [CrossRef]
- Holzwarth, A.R.; Müller, M.G.; Niklas, J.; Lubitz, W. Ultrafast Transient Absorption Studies on Photosystem I Reaction Centers from Chlamydomonas reinhardtii. 2: Mutations near the P700 Reaction Center Chlorophylls Provide New Insight into the Nature of the Primary Electron Donor. Biophys. J. 2006, 90, 552–565. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.G.; Slavov, C.; Luthra, R.; Redding, K.E.; Holzwarth, A.R. Independent initiation of primary electron transfer in the two branches of the photosystem I reaction center. Proc. Natl. Acad. Sci. USA 2010, 107, 4123–4128. [Google Scholar] [CrossRef]
- Snellenburg, J.J.; Dekker, J.P.; van Grondelle, R.; van Stokkum, I.H.M. Functional Compartmental Modeling of the Photosystems in the Thylakoid Membrane at 77 K. J. Phys. Chem. B 2013, 117, 11363–11371. [Google Scholar] [CrossRef] [PubMed]
- van Stokkum, I.H.M.; Larsen, D.S.; van Grondelle, R. Global and target analysis of time-resolved spectra. Biochim. Biophys. Acta 2004, 1657, 82–104. [Google Scholar] [CrossRef]
- van Stokkum, I.H.M.; Müller, M.G.; Weißenborn, J.; Weigand, S.; Snellenburg, J.J.; Holzwarth, A.R. Energy transfer and trapping in photosystem I with and without chlorophyll-f. iScience 2023, 26, 107650. [Google Scholar] [CrossRef]
- Guergova-Kuras, M.; Boudreaux, B.; Joliot, A.; Joliot, P.; Redding, K. Evidence for two active branches for electron transfer in photosystem I. Proc. Natl. Acad. Sci. USA 2001, 98, 4437–4442. [Google Scholar] [CrossRef]
- Li, Y.; van der Est, A.; Lucas, M.G.; Ramesh, V.M.; Gu, F.; Petrenko, A.; Lin, S.; Webber, A.N.; Rappaport, F.; Redding, K. Directing electron transfer within Photosystem I by breaking H-bonds in the cofactor branches. Proc. Natl. Acad. Sci. USA 2006, 103, 2144–2149. [Google Scholar] [CrossRef] [PubMed]
- Badshah, S.L.; Sun, J.; Mula, S.; Gorka, M.; Baker, P.; Luthra, R.; Lin, S.; van der Est, A.; Golbeck, J.H.; Redding, K.E. Mutations in algal and cyanobacterial Photosystem I that independently affect the yield of initial charge separation in the two electron transfer cofactor branches. Biochim. Biophys. Acta (BBA) Bioenerg. 2018, 1859, 42–55. [Google Scholar] [CrossRef]
- Snellenburg, J.J.; Wlodarczyk, L.M.; Dekker, J.P.; van Grondelle, R.; van Stokkum, I.H.M. A model for the 77K excited state dynamics in Chlamydomonas reinhardtii in state 1 and state 2. Biochim. Biophys. Acta (BBA) Bioenerg. 2017, 1858, 64–72. [Google Scholar] [CrossRef]
- Schoffman, H.; Keren, N. Function of the IsiA pigment–protein complex in vivo. Photosynth. Res. 2019, 141, 343–353. [Google Scholar] [CrossRef]
- Chen, C.; Gong, N.; Li, Z.; Sun, C.; Men, Z. Concentration Effect on Quenching of Chlorophyll a Fluorescence by All-Trans-β-Carotene in Photosynthesis. Molecules 2017, 22, 1585. [Google Scholar] [CrossRef]
- Akhtar, P.; Caspy, I.; Nowakowski, P.J.; Malavath, T.; Nelson, N.; Tan, H.-S.; Lambrev, P.H. Two-Dimensional Electronic Spectroscopy of a Minimal Photosystem I Complex Reveals the Rate of Primary Charge Separation. J. Am. Chem. Soc. 2021, 143, 14601–14612. [Google Scholar] [CrossRef] [PubMed]
- Cherepanov, D.A.; Shelaev, I.V.; Gostev, F.E.; Aybush, A.V.; Mamedov, M.D.; Shen, G.; Nadtochenko, V.A.; Bryant, D.A.; Semenov, A.Y.; Golbeck, J.H. Evidence that chlorophyll f functions solely as an antenna pigment in far-red-light photosystem I from Fischerella thermalis PCC 7521. Biochim. Biophys. Acta (BBA) Bioenerg. 2020, 1861, 148184. [Google Scholar] [CrossRef] [PubMed]
- Shelaev, I.V.; Gostev, F.E.; Mamedov, M.D.; Sarkisov, O.M.; Nadtochenko, V.A.; Shuvalov, V.A.; Semenov, A.Y. Femtosecond primary charge separation in Synechocystis sp. PCC 6803 photosystem I. Biochim. Biophys. Acta (BBA) Bioenerg. 2010, 1797, 1410–1420. [Google Scholar] [CrossRef]
- Cherepanov, D.A.; Shelaev, I.V.; Gostev, F.E.; Mamedov, M.D.; Petrova, A.A.; Aybush, A.V.; Shuvalov, V.A.; Semenov, A.Y.; Nadtochenko, V.A. Mechanism of adiabatic primary electron transfer in photosystem I: Femtosecond spectroscopy upon excitation of reaction center in the far-red edge of the QY band. Biochim. Biophys. Acta (BBA) Bioenerg. 2017, 1858, 895–905. [Google Scholar] [CrossRef]
- Cherepanov, D.A.; Brady, N.G.; Shelaev, I.V.; Nguyen, J.; Gostev, F.E.; Mamedov, M.D.; Nadtochenko, V.A.; Bruce, B.D. PSI-SMALP, a Detergent-free Cyanobacterial Photosystem I, Reveals Faster Femtosecond Photochemistry. Biophys. J. 2020, 118, 337–351. [Google Scholar] [CrossRef]
- Cherepanov, D.A.; Shelaev, I.V.; Gostev, F.E.; Nadtochenko, V.A.; Xu, W.; Golbeck, J.H.; Semenov, A.Y. Symmetry breaking in photosystem I: Ultrafast optical studies of variants near the accessory chlorophylls in the A- and B-branches of electron transfer cofactors. Photochem. Photobiol. Sci. 2021, 20, 1209–1227. [Google Scholar] [CrossRef] [PubMed]
- Kramer, T.; Noack, M.; Reimers, J.R.; Reinefeld, A.; Rodríguez, M.; Yin, S. Energy flow in the Photosystem I supercomplex: Comparison of approximative theories with DM-HEOM. Chem. Phys. 2018, 515, 262–271. [Google Scholar] [CrossRef]
- Byrdin, M.; Jordan, P.; Krauss, N.; Fromme, P.; Stehlik, D.; Schlodder, E. Light Harvesting in Photosystem I: Modeling Based on the 2.5-Å Structure of Photosystem I from Synechococcus elongatus. Biophys. J. 2002, 83, 433–457. [Google Scholar] [CrossRef]
- Yang, M.; Damjanović, A.; Vaswani, H.M.; Fleming, G.R. Energy Transfer in Photosystem I of Cyanobacteria Synechococcus elongatus: Model Study with Structure-Based Semi-Empirical Hamiltonian and Experimental Spectral Density. Biophys. J. 2003, 85, 140–158. [Google Scholar] [CrossRef]
- Şener, M.; Strümpfer, J.; Hsin, J.; Chandler, D.; Scheuring, S.; Hunter, C.N.; Schulten, K. Förster Energy Transfer Theory as Reflected in the Structures of Photosynthetic Light-Harvesting Systems. ChemPhysChem 2011, 12, 518–531. [Google Scholar] [CrossRef] [PubMed]
- Sener, M.K.; Lu, D.; Ritz, T.; Park, S.; Fromme, P.; Schulten, K. Robustness and Optimality of Light Harvesting in Cyanobacterial Photosystem I. J. Phys. Chem. B 2002, 106, 7948–7960. [Google Scholar] [CrossRef]
- Damjanović, A.; Vaswani, H.M.; Fromme, P.; Fleming, G.R. Chlorophyll Excitations in Photosystem I of Synechococcus elongatus. J. Phys. Chem. B 2002, 106, 10251–10262. [Google Scholar] [CrossRef]
- Kramer, T.; Rodríguez, M.; Zelinskyy, Y. Modeling of Transient Absorption Spectra in Exciton–Charge-Transfer Systems. J. Phys. Chem. B 2017, 121, 463–470. [Google Scholar] [CrossRef] [PubMed]
- van Stokkum, I.H.M.; Akhtar, P.; Biswas, A.; Lambrev, P.H. Energy transfer from phycobilisomes to photosystem I at 77 K. Front. Plant Sci. 2023, 14, 1293813. [Google Scholar] [CrossRef] [PubMed]
- Witt, H.; Schlodder, E.; Teutloff, C.; Niklas, J.; Bordignon, E.; Carbonera, D.; Kohler, S.; Labahn, A.; Lubitz, W. Hydrogen Bonding to P700: Site-Directed Mutagenesis of Threonine A739 of Photosystem I in Chlamydomonas reinhardtii. Biochemistry 2002, 41, 8557–8569. [Google Scholar] [CrossRef] [PubMed]
- Kruip, J.; Boekema, E.J.; Bald, D.; Boonstra, A.F.; Rögner, M. Isolation and structural characterization of monomeric and trimeric photosystem I complexes (P700.FA/FB and P700.FX) from the cyanobacterium Synechocystis PCC 6803. J. Biol. Chem. 1993, 268, 23353–23360. [Google Scholar] [CrossRef] [PubMed]
- Fromme, P.; Witt, H.T. Improved isolation and crystallization of photosystem I for structural analysis. Biochim. Biophys. Acta (BBA) Bioenerg. 1998, 1365, 175–184. [Google Scholar] [CrossRef]
- Karapetyan, N.V.; Holzwarth, A.R.; Rögner, M. The photosystem I trimer of cyanobacteria: Molecular organization, excitation dynamics and physiological significance. FEBS Lett. 1999, 460, 395–400. [Google Scholar] [CrossRef]
- Kruip, J.; Karapetyan, N.V.; Terekhova, I.V.; Rögner, M. In vitro oligomerization of a membrane protein complex. J. Biol. Chem. 1999, 274, 18181–18188. [Google Scholar] [CrossRef]
- Müller, M.G.; Hucke, M.; Reus, M.; Holzwarth, A.R. Annihilation processes in the isolated D1-D2-cyt-b559 reaction center complex of photosystem II. An intensity-dependence study of femtosecond transient absorption. J. Phys. Chem. 1996, 100, 9537–9544. [Google Scholar] [CrossRef]
- van Stokkum, I.H.M.; Kloz, M.; Polli, D.; Viola, D.; Weißenborn, J.; Peerbooms, E.; Cerullo, G.; Kennis, J.T.M. Vibronic dynamics resolved by global and target analysis of ultrafast transient absorption spectra. J. Chem. Phys. 2021, 155, 114113. [Google Scholar] [CrossRef]
- Kovalenko, S.A.; Dobryakov, A.L.; Ruthmann, J.; Ernsting, N.P. Femtosecond spectroscopy of condensed phases with chirped supercontinuum probing. Phys. Rev. A 1999, 59, 2369–2384. [Google Scholar] [CrossRef]
- van Stokkum, I.H.M.; Weißenborn, J.; Weigand, S.; Snellenburg, J.J. Pyglotaran: A lego-like Python framework for global and target analysis of time resolved spectra. Photochem. Photobiol. Sci. 2023, 22, 2413–2431. [Google Scholar] [CrossRef]
- Gobets, B.; van Grondelle, R. Energy transfer and trapping in photosystem I. Biochim. Biophys. Acta-Bioenerg. 2001, 1507, 80–99. [Google Scholar] [CrossRef]
- Godfrey, K. Compartmental Models and Their Application; Academic Press: London, UK, 1983. [Google Scholar]
- Golub, G.H.; LeVeque, R.J. Extensions and Uses of the variable projection algorithm for solving nonlinear least squares problems. In Proceedings of the 1979 Army Numerical Analysis and Computers Conference ARO Report 79-3; U.S. Army Research Office, Research Triangle Park: Durham, NC, USA, 1979; pp. 1–12. [Google Scholar]
- Nagle, J.F. Solving complex photocycle kinetics—Theory and direct method. Biophys. J. 1991, 59, 476–487. [Google Scholar] [CrossRef][Green Version]
- Henry, E.R.; Hofrichter, J. Singular Value Decomposition—Application to analysis of experimental-data. Methods Enzymol. 1992, 210, 129–192. [Google Scholar]


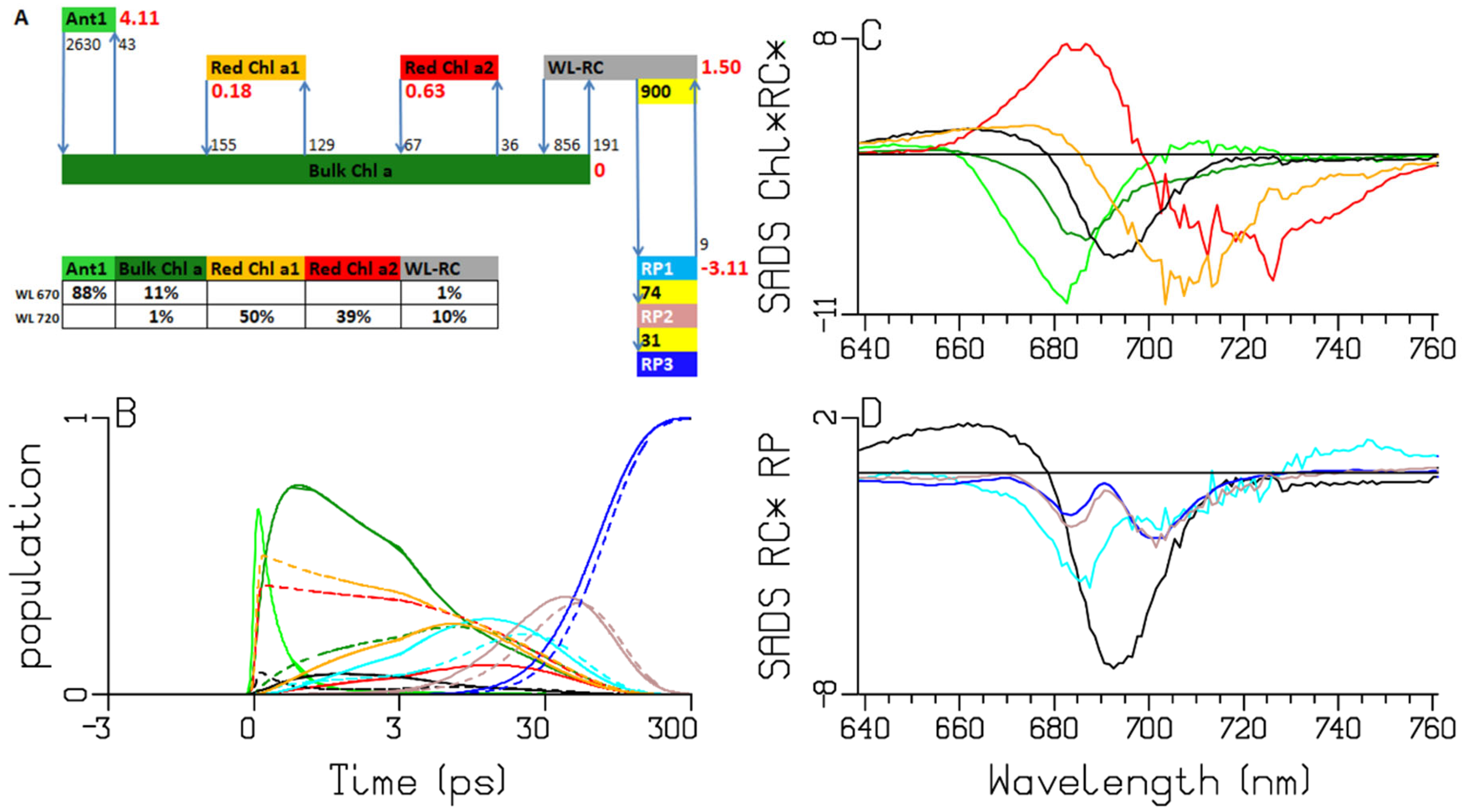

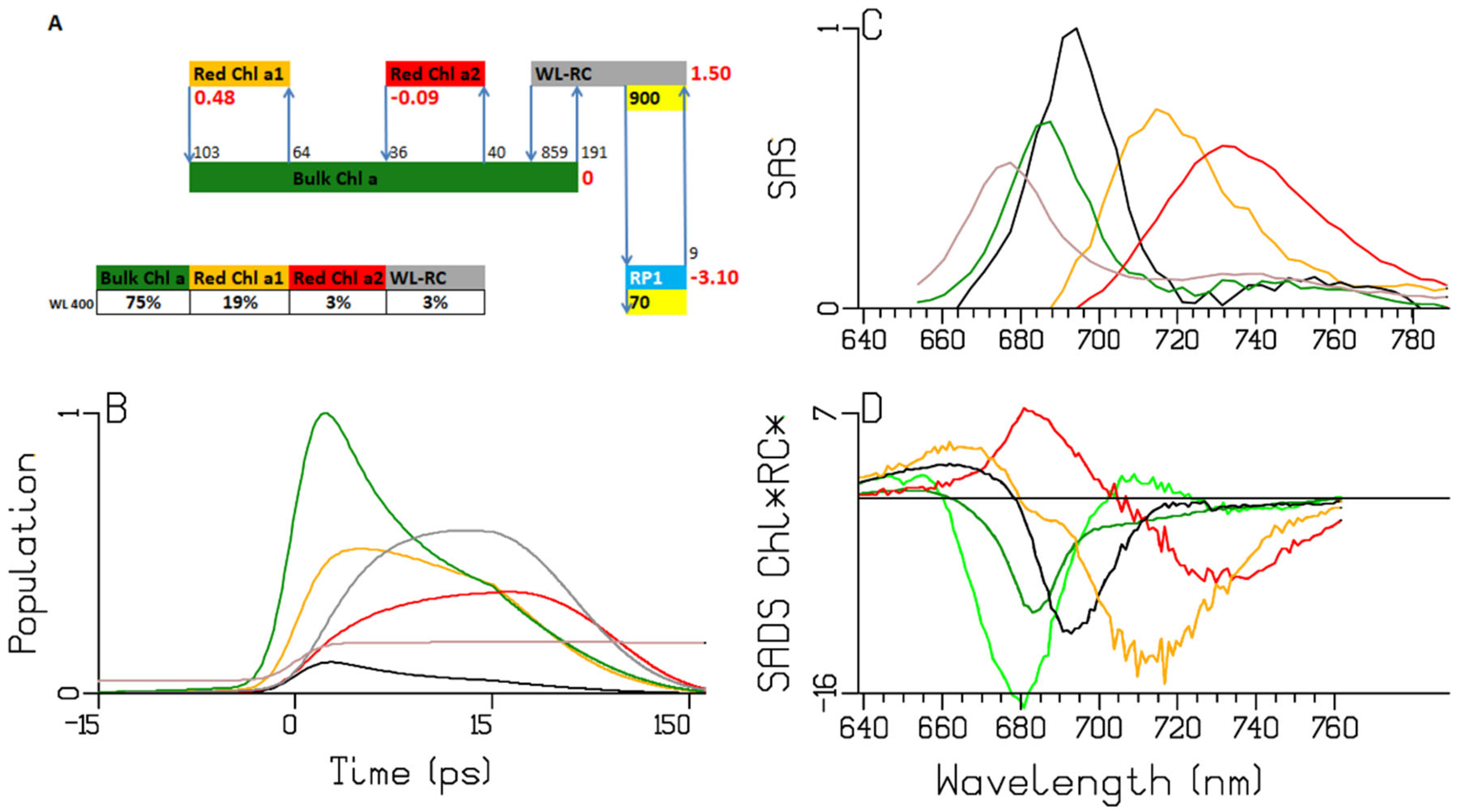

| A | species\τ (ps) | 0.28 | 0.38 | 2.65 | 7.92 | 13.3 | 32.3 | long lived | input |
| Ant1 | 0.015 | 0.850 | 0.007 | 0.005 | 0.004 | 0 | 0 | 0.88 | |
| Bulk Chl a | −0.335 | −0.475 | 0.361 | 0.316 | 0.241 | 0 | 0 | 0.11 | |
| Red Chl a | 0.007 | 0.014 | −0.258 | 0.150 | 0.086 | 0 | 0 | 0 | |
| WL-RC | 0.418 | −0.589 | 0.075 | 0.055 | 0.051 | 0 | 0 | 0.01 | |
| RP1 | −0.108 | 0.208 | −0.242 | −1.826 | 1.968 | 0 | 0 | 0 | |
| RP2 | 0.003 | −0.007 | 0.063 | 1.724 | −3.984 | 2.201 | 0 | 0 | |
| RP3 | 0.000 | 0.000 | −0.005 | −0.423 | 1.637 | −2.203 | 0.994 | 0 | |
| sum | 0 | 0 | 0 | 0 | 0.004 | −0.002 | 0.994 | 1 | |
| B | species\τ (ps) | 0.28 | 0.38 | 2.65 | 7.92 | 13.3 | 32.3 | long lived | input |
| Ant1 | 0.016 | −0.022 | −0.005 | 0.006 | 0.004 | 0 | 0 | 0 | |
| Bulk Chl a | −0.355 | 0.012 | −0.273 | 0.371 | 0.255 | 0 | 0 | 0.01 | |
| Red Chl a | 0.008 | 0.000 | 0.195 | 0.177 | 0.091 | 0 | 0 | 0.47 | |
| WL-RC | 0.443 | 0.015 | −0.057 | 0.065 | 0.054 | 0 | 0 | 0.52 | |
| RP1 | −0.114 | −0.005 | 0.184 | −2.142 | 2.079 | 0 | 0 | 0 | |
| RP2 | 0.003 | 0.000 | −0.048 | 2.024 | −4.207 | 2.228 | 0 | 0 | |
| RP3 | 0.000 | 0.000 | 0.004 | −0.497 | 1.729 | −2.230 | 0.994 | 0 | |
| sum | 0 | 0 | 0 | 0 | 0.005 | −0.002 | 0.994 | 1 |
| A | Chlamydomonas reinhardtii | Ant1 | Bulk Chl a | Red Chl a | WL-RC | |
| λmax (nm) | 675 | 682 | 689 | 690 | ||
| (1/cm) | 14,805 | 14,654 | 14,519 | 14,483 | ||
| (kB.T) | 72.7 | 72.0 | 71.3 | 71.1 | ||
| ΔH (kB.T) | 0.7 | −0.7 | −0.8 | |||
| ΔG (kB.T) | 4.1 | 1.3 | 1.3 | |||
| ΔH − ΔG = TΔS (kB.T) | −3.4 | −2.0 | −2.2 | |||
| “N” | 2.7 | 78 | 10.4 | 8.7 | ||
| B | Synechocystis PCC6803 | Ant1 | Bulk Chl a | Red Chl a1 | Red Chl a2 | WL-RC |
| λmax (nm) | 683 | 685 | 701 | 709 | 693 | |
| (1/cm) | 14,635 | 14,594 | 14,260 | 14,096 | 14,438 | |
| (kB.T) | 71.9 | 71.7 | 70.0 | 69.2 | 70.9 | |
| ΔH (kB.T) | 0.2 | −1.6 | −2.4 | −0.8 | ||
| ΔG (kB.T) | 4.1 | 1.1 | 1.3 | 1.5 | ||
| ΔH – ΔG = TΔS (kB.T) | −3.9 | −2.7 | −3.8 | −2.3 | ||
| “N” | 1.6 | 78 | 5.3 | 1.8 | 8.1 | |
| C | Thermosynechococcus elongatus | Ant1 | Bulk Chl a | Red Chl a1 | Red Chl a2 | WL-RC |
| λmax (nm) | 681 | 686 | 707 | 723 | 693 | |
| (1/cm) | 14,682 | 14,581 | 14,152 | 13,824 | 14,438 | |
| (kB.T) | 72.1 | 71.6 | 69.5 | 67.9 | 70.9 | |
| ΔH (kB.T) | 0.5 | −2.1 | −3.7 | −0.7 | ||
| ΔG (kB.T) | 4.1 | 0.2 | 0.6 | 1.5 | ||
| ΔH – ΔG = TΔS (kB.T) | −3.6 | −2.3 | −4.3 | −2.2 | ||
| “N” | 2.1 | 78 | 7.9 | 1.0 | 8.6 | |
| D | Spirulina platensis | Ant1 | Bulk Chl a | Red Chl a1 | Red Chl a2 | WL-RC |
| λmax (nm) | 679 | 684 | 713 | 732 | 693 | |
| (1/cm) | 14,722 | 14,630 | 14,031 | 13,670 | 14,438 | |
| (kB.T) | 72.3 | 71.8 | 68.9 | 67.1 | 70.9 | |
| ΔH (kB.T) | 0.5 | −2.9 | −4.7 | −0.9 | ||
| ΔG (kB.T) | 4.1 | 0.1 | −0.4 | 1.5 | ||
| ΔH – ΔG = TΔS (kB.T) | −3.7 | −3.1 | −4.3 | −2.4 | ||
| “N” | 2.0 | 78 | 3.6 | 1.0 | 6.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Stokkum, I.H.M.; Müller, M.G.; Holzwarth, A.R. Energy Transfer and Radical-Pair Dynamics in Photosystem I with Different Red Chlorophyll a Pigments. Int. J. Mol. Sci. 2024, 25, 4125. https://doi.org/10.3390/ijms25074125
van Stokkum IHM, Müller MG, Holzwarth AR. Energy Transfer and Radical-Pair Dynamics in Photosystem I with Different Red Chlorophyll a Pigments. International Journal of Molecular Sciences. 2024; 25(7):4125. https://doi.org/10.3390/ijms25074125
Chicago/Turabian Stylevan Stokkum, Ivo H. M., Marc G. Müller, and Alfred R. Holzwarth. 2024. "Energy Transfer and Radical-Pair Dynamics in Photosystem I with Different Red Chlorophyll a Pigments" International Journal of Molecular Sciences 25, no. 7: 4125. https://doi.org/10.3390/ijms25074125
APA Stylevan Stokkum, I. H. M., Müller, M. G., & Holzwarth, A. R. (2024). Energy Transfer and Radical-Pair Dynamics in Photosystem I with Different Red Chlorophyll a Pigments. International Journal of Molecular Sciences, 25(7), 4125. https://doi.org/10.3390/ijms25074125





