Loss of the Y Chromosome: A Review of Molecular Mechanisms, Age Inference, and Implications for Men’s Health
Abstract
1. Introduction
2. Molecular Mechanisms of the Loss of the Y Chromosome
3. Possible Fate of Y Chromosome after It Is Loss
4. Other Factors Associated with the Loss of the Y Chromosome
5. Techniques for Detection of Loss of the Y Chromosome
6. Implications of Loss of the Y Chromosome for Men’s Health
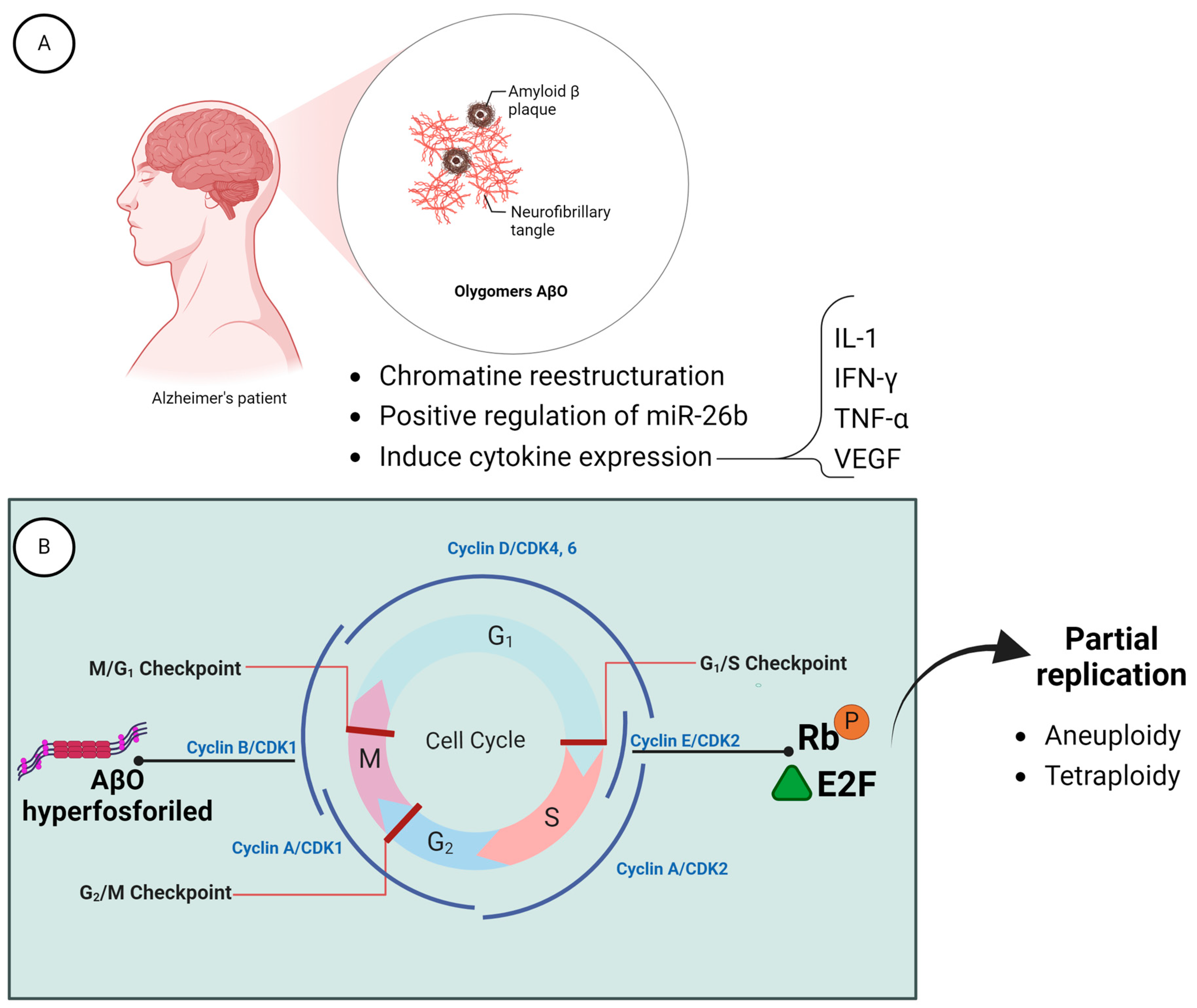
6.1. Loss of the Y Chromosome and Alzheimer Disease
6.2. Loss of the Y Chromosome in Kidney Disease
6.3. Loss of the Y Chromosome in Cardiovascular Disease
6.4. Loss of Y Chromosome and Cancer
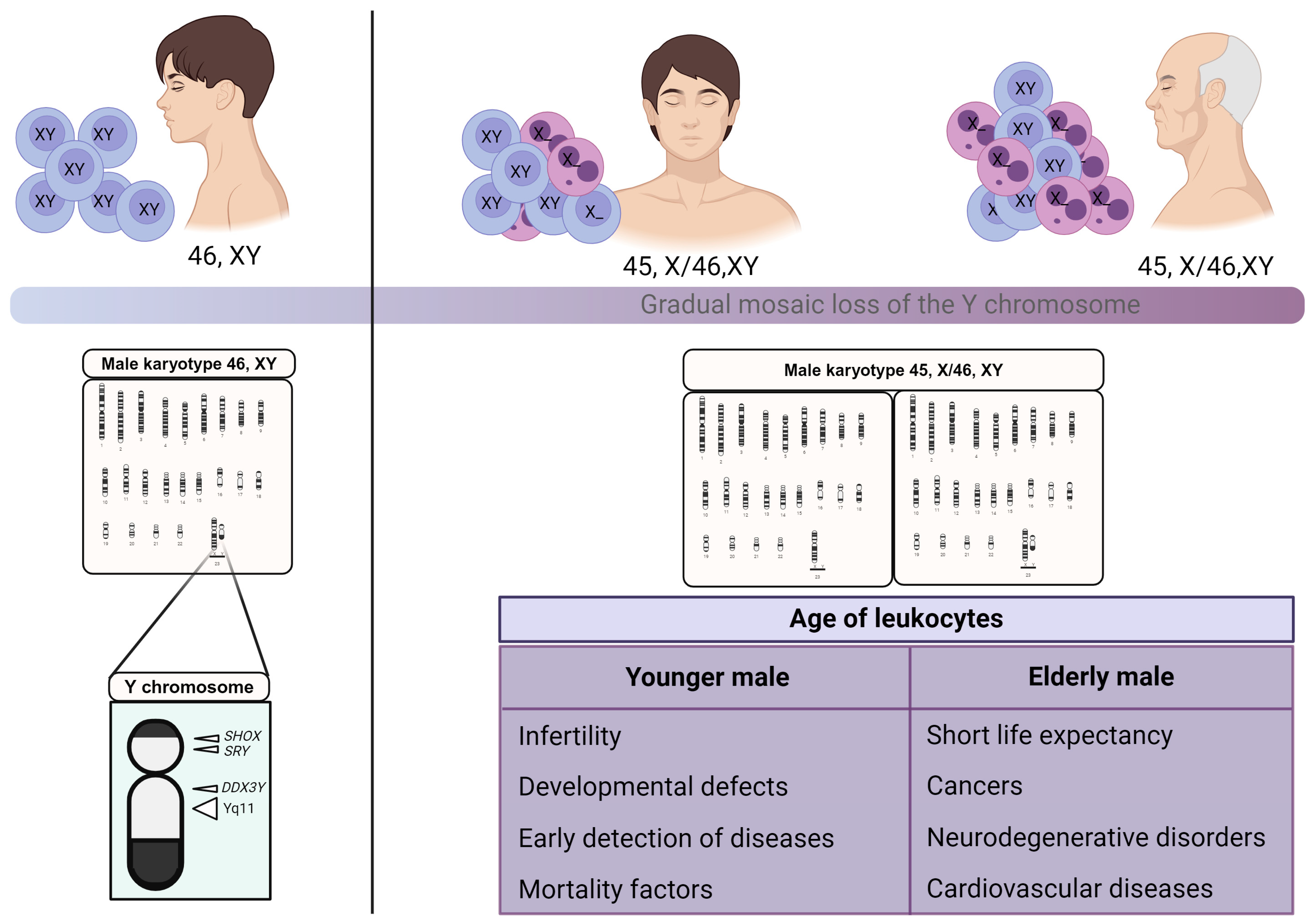
7. Loss of Y Chromosome as a Potential Marker to Infer Men’s Age
8. Conclusions
Funding
Conflicts of Interest
References
- Guo, X.; Dai, X.; Zhou, T.; Wang, H.; Ni, J.; Xue, J.; Wang, X. Mosaic Loss of Human Y Chromosome: What, How and Why. Hum. Genet. 2020, 139, 421–446. [Google Scholar] [CrossRef] [PubMed]
- Rhie, A.; Nurk, S.; Cechova, M.; Hoyt, S.J.; Taylor, D.J.; Altemose, N.; Hook, P.W.; Koren, S.; Rautiainen, M.; Alexandrov, I.A.; et al. The Complete Sequence of a Human Y Chromosome. Nature 2023, 621, 344–354. [Google Scholar] [CrossRef]
- Jobling, M.A.; Tyler-Smith, C. Human Y-Chromosome Variation in the Genome-Sequencing Era. Nat. Rev. Genet. 2017, 18, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Maan, A.A.; Eales, J.; Akbarov, A.; Rowland, J.; Xu, X.; Jobling, M.A.; Charchar, F.J.; Tomaszewski, M. The Y Chromosome: A Blueprint for Men’s Health? Eur. J. Hum. Genet. 2017, 25, 1181–1188. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, L.A.; Rasi, C.; Malmqvist, N.; Davies, H.; Pasupulati, S.; Pakalapati, G.; Sandgren, J.; de Ståhl, T.D.; Zaghlool, A.; Giedraitis, V.; et al. Mosaic Loss of Chromosome Y in Peripheral Blood Is Associated with Shorter Survival and Higher Risk of Cancer. Nat. Genet. 2014, 46, 624–628. [Google Scholar] [CrossRef]
- Dumanski, J.P.; Rasi, C.; Lönn, M.; Davies, H.; Ingelsson, M.; Giedraitis, V.; Lannfelt, L.; Magnusson, P.K.E.; Lindgren, C.M.; Morris, A.P.; et al. Smoking Is Associated with Mosaic Loss of Chromosome Y. Science 2015, 347, 81–83. [Google Scholar] [CrossRef]
- Forsberg, L.A. Loss of Chromosome Y (LOY) in Blood Cells Is Associated with Increased Risk for Disease and Mortality in Aging Men. Hum. Genet. 2017, 136, 657–663. [Google Scholar] [CrossRef]
- Miyado, M.; Fukami, M. Losing Maleness: Somatic Y Chromosome Loss at Every Stage of a Man’s Life. FASEB Bioadv. 2019, 1, 350. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Li, J.; Xue, J.; Fenech, M.; Wang, X. Loss of Y Chromosome: An Emerging next-Generation Biomarker for Disease Prediction and Early Detection? Mutat. Res./Rev. Mutat. Res. 2021, 788, 108389. [Google Scholar] [CrossRef] [PubMed]
- Vakilian, H.; Mirzaei, M.; Sharifi Tabar, M.; Pooyan, P.; Habibi Rezaee, L.; Parker, L.; Haynes, P.A.; Gourabi, H.; Baharvand, H.; Salekdeh, G.H. DDX3Y, a Male-Specific Region of Y Chromosome Gene, May Modulate Neuronal Differentiation. J. Proteome Res. 2015, 14, 3474–3483. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.J.; Genovese, G.; Halvardson, J.; Ulirsch, J.C.; Wright, D.J.; Terao, C.; Davidsson, O.B.; Day, F.R.; Sulem, P.; Jiang, Y.; et al. Genetic Predisposition to Mosaic Y Chromosome Loss in Blood. Nature 2019, 575, 652–657. [Google Scholar] [CrossRef] [PubMed]
- Wilson, P.C.; Verma, A.; Yoshimura, Y.; Muto, Y.; Li, H.; Malvin, N.P.; Dixon, E.E.; Humphreys, B.D. Mosaic Loss of Y Chromosome Is Associated with Aging and Epithelial Injury in Chronic Kidney Disease. Genome Biol. 2024, 25, 36. [Google Scholar] [CrossRef] [PubMed]
- Agahozo, M.C.; Timmermans, M.A.M.; Sleddens, H.F.B.M.; Foekens, R.; Trapman-Jansen, A.M.A.C.; Schröder, C.P.; van Leeuwen-Stok, E.; Martens, J.W.M.; Dinjens, W.N.M.; van Deurzen, C.H.M. Loss of Y-Chromosome during Male Breast Carcinogenesis. Cancers 2020, 12, 631. [Google Scholar] [CrossRef] [PubMed]
- Heydari, R.; Jangravi, Z.; Maleknia, S.; Seresht-Ahmadi, M.; Bahari, Z.; Salekdeh, G.H.; Meyfour, A. Y Chromosome Is Moving out of Sex Determination Shadow. Cell Biosci. 2022, 12, 4. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Min, K.; Kim, M.; Lee, S.K. Sex Chromosomes Are Severely Disrupted in Gastric Cancer Cell Lines. Int. J. Mol. Sci. 2020, 21, 4598. [Google Scholar] [CrossRef] [PubMed]
- Ly, P.; Teitz, L.S.; Kim, D.H.; Shoshani, O.; Skaletsky, H.; Fachinetti, D.; Page, D.C.; Cleveland, D.W. Selective Y Centromere Inactivation Triggers Chromosome Shattering in Micronuclei and Repair by Non-Homologous End Joining. Nat. Cell Biol. 2017, 19, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Fukami, M.; Miyado, M. Mosaic Loss of the Y Chromosome and Men’s Health. Reprod. Med. Biol. 2022, 21, e12445. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, J.; Chen, X.; Czajkowsky, D.M.; Shao, Z. Quantitative Super-Resolution Microscopy Reveals the Relationship between CENP-A Stoichiometry and Centromere Physical Size. Int. J. Mol. Sci. 2023, 24, 15871. [Google Scholar] [CrossRef] [PubMed]
- Thakur, J.; Packiaraj, J.; Henikoff, S. Sequence, Chromatin and Evolution of Satellite DNA. Int. J. Mol. Sci. 2021, 22, 4309. [Google Scholar] [CrossRef]
- Fachinetti, D.; Han, J.S.; McMahon, M.A.; Ly, P.; Abdullah, A.; Wong, A.J.; Cleveland, D.W. DNA Sequence-Specific Binding of CENP-B Enhances the Fidelity of Human Centromere Function. Dev. Cell 2015, 33, 314–327. [Google Scholar] [CrossRef] [PubMed]
- Ohzeki, J.; Nakano, M.; Okada, T.; Masumoto, H. CENP-B Box Is Required for de Novo Centromere Chromatin Assembly on Human Alphoid DNA. J. Cell Biol. 2002, 159, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Giunta, S.; Hervé, S.; White, R.R.; Wilhelm, T.; Dumont, M.; Scelfo, A.; Gamba, R.; Wong, C.K.; Rancati, G.; Smogorzewska, A.; et al. CENP-A Chromatin Prevents Replication Stress at Centromeres to Avoid Structural Aneuploidy. Proc. Natl. Acad. Sci. USA 2021, 118, e2015634118. [Google Scholar] [CrossRef]
- Bodor, D.L.; Mata, J.F.; Sergeev, M.; David, A.F.; Salimian, K.J.; Panchenko, T.; Cleveland, D.W.; Black, B.E.; Shah, J.V.; Jansen, L.E. The Quantitative Architecture of Centromeric Chromatin. eLife 2014, 3, e02137. [Google Scholar] [CrossRef]
- Maehara, K.; Takahashi, K.; Saitoh, S. CENP-A Reduction Induces a P53-Dependent Cellular Senescence Response to Protect Cells from Executing Defective Mitoses. Mol. Cell Biol. 2010, 30, 2090–2104. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.Y.Y.; Margolis, H.G.; Machiela, M.; Zhou, W.; Odden, M.C.; Psaty, B.M.; Robbins, J.; Jones, R.R.; Rotter, J.I.; Chanock, S.J.; et al. Outdoor Air Pollution and Mosaic Loss of Chromosome Y in Older Men from the Cardiovascular Health Study. Environ. Int. 2018, 116, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Ya, J.; Bayraktutan, U. Vascular Ageing: Mechanisms, Risk Factors, and Treatment Strategies. Int. J. Mol. Sci. 2023, 24, 11538. [Google Scholar] [CrossRef] [PubMed]
- Borghini, A.; Ndreu, R.; Canale, P.; Campolo, J.; Marinaro, I.; Mercuri, A.; Turchi, S.; Andreassi, M.G. Telomere Length, Mitochondrial DNA, and Micronucleus Yield in Response to Oxidative Stress in Peripheral Blood Mononuclear Cells. Int. J. Mol. Sci. 2024, 25, 1428. [Google Scholar] [CrossRef]
- Di Bona, M.; Bakhoum, S.F. Micronuclei and Cancer. Cancer Discov. 2024, 14, 214–226. [Google Scholar] [CrossRef]
- Danielsson, M.; Halvardson, J.; Davies, H.; Torabi Moghadam, B.; Mattisson, J.; Rychlicka-Buniowska, E.; Jaszczyński, J.; Heintz, J.; Lannfelt, L.; Giedraitis, V.; et al. Longitudinal Changes in the Frequency of Mosaic Chromosome Y Loss in Peripheral Blood Cells of Aging Men Varies Profoundly between Individuals. Eur. J. Hum. Genet. 2020, 28, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Machiela, M.J.; Freedman, N.D.; Rothman, N.; Malats, N.; Dagnall, C.; Caporaso, N.; Teras, L.T.; Gaudet, M.M.; Gapstur, S.M.; et al. Mosaic Loss of Chromosome Y Is Associated with Common Variation near TCL1A. Nat. Genet. 2016, 48, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Loftfield, E.; Zhou, W.; Graubard, B.I.; Yeager, M.; Chanock, S.J.; Freedman, N.D.; Machiela, M.J. Predictors of Mosaic Chromosome Y Loss and Associations with Mortality in the UK Biobank. Sci. Rep. 2018, 8, 12316. [Google Scholar] [CrossRef]
- Wright, D.J.; Day, F.R.; Kerrison, N.D.; Zink, F.; Cardona, A.; Sulem, P.; Thompson, D.J.; Sigurjonsdottir, S.; Gudbjartsson, D.F.; Helgason, A.; et al. Genetic Variants Associated with Mosaic Y Chromosome Loss Highlight Cell Cycle Genes and Overlap with Cancer Susceptibility. Nat. Genet. 2017, 49, 674–679. [Google Scholar] [CrossRef] [PubMed]
- Siffroi, J.P.; Le Bourhis, C.; Krausz, C.; Barbaux, S.; Quintana-Murci, L.; Kanafani, S.; Rouba, H.; Bujan, L.; Bourrouillou, G.; Seifer, I.; et al. Sex Chromosome Mosaicism in Males Carrying Y Chromosome Long Arm Deletions. Hum. Reprod. 2000, 15, 2559–2562. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, H.; Yang, Y.; Zhang, H.; Wang, R.; Jiang, Y.; Liu, R. High Frequency of Y Chromosome Microdeletions in Male Infertility Patients with 45,X/46,XY Mosaicism. Braz. J. Med. Biol. Res. 2020, 53, e8980. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, E.; Kobori, Y.; Katsumi, M.; Ushijima, K.; Uchiyama, T.; Okada, H.; Miyado, M.; Fukami, M. Copy-number Analysis of Y-linked Loci in Young Men with Non-obstructive Azoospermia: Implications for the Rarity of Early Onset Mosaic Loss of Chromosome Y. Reprod. Med. Biol. 2020, 19, 178–181. [Google Scholar] [CrossRef]
- Rehen, S.K.; McConnell, M.J.; Kaushal, D.; Kingsbury, M.A.; Yang, A.H.; Chun, J. Chromosomal Variation in Neurons of the Developing and Adult Mammalian Nervous System. Proc. Natl. Acad. Sci. USA 2001, 98, 13361–13366. [Google Scholar] [CrossRef]
- Faggioli, F.; Vijg, J.; Montagna, C. Chromosomal Aneuploidy in the Aging Brain. Mech. Ageing Dev. 2011, 132, 429–436. [Google Scholar] [CrossRef]
- Bonda, D.J.; Evans, T.A.; Santocanale, C.; Llosá, J.C.; Viňa, J.; Bajic, V.P.; Castellani, R.J.; Siedlak, S.L.; Perry, G.; Smith, M.A.; et al. Evidence for the Progression through S-Phase in the Ectopic Cell Cycle Re-Entry of Neurons in Alzheimer Disease. Aging 2009, 1, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Guan, P.-P.; Ding, W.-Y.; Wang, P. Molecular Mechanism of Acetylsalicylic Acid in Improving Learning and Memory Impairment in APP/PS1 Transgenic Mice by Inhibiting the Abnormal Cell Cycle Re-Entry of Neurons. Front. Mol. Neurosci. 2022, 15, 1006216. [Google Scholar] [CrossRef]
- Frade, J.M.; López-Sánchez, N. Neuronal Tetraploidy in Alzheimer and Aging. Aging 2017, 9, 2014–2015. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, J.F.; Rennie, K.; Chakravarthy, B. Alzheimer’s Disease and Its Possible Evolutionary Origin: Hypothesis. Cells 2023, 12, 1618. [Google Scholar] [CrossRef] [PubMed]
- Frade, J.M.; Ovejero-Benito, M.C. Neuronal Cell Cycle: The Neuron Itself and Its Circumstances. Cell Cycle 2015, 14, 712–720. [Google Scholar] [CrossRef] [PubMed]
- Reed, E.G.; Keller-Norrell, P.R. Minding the Gap: Exploring Neuroinflammatory and Microglial Sex Differences in Alzheimer’s Disease. Int. J. Mol. Sci. 2023, 24, 17377. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, M.C.; Pearse, R.; Young-Pearse, T.; Mostafavi, S. Mosaic Loss of Chromosome Y in Aged Human Microglia. Genome Res. 2022, 32, 1795–1807. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Su, P.-Y.P.; Leff, J.; Gao, X.; Chen, J.; Guan, A.K.; Kalyanasundaram, G.; Ma, A.; Guan, Z. Distinct Phases of Adult Microglia Proliferation: A Myc-Mediated Early Phase and a Tnfaip3-Mediated Late Phase. Cell Discov. 2022, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Caceres, A.; Jene, A.; Esko, T.; Perez-Jurado, L.A.; Gonzalez, J.R. Extreme Downregulation of Chromosome Y and Alzheimer’s Disease in Men. Neurobiol. Aging 2020, 90, 150.e1–150.e4. [Google Scholar] [CrossRef] [PubMed]
- Dumanski, J.P.; Lambert, J.-C.; Rasi, C.; Giedraitis, V.; Davies, H.; Grenier-Boley, B.; Lindgren, C.M.; Campion, D.; Dufouil, C.; Pasquier, F.; et al. Mosaic Loss of Chromosome Y in Blood Is Associated with Alzheimer Disease. Am. J. Hum. Genet. 2016, 98, 1208–1219. [Google Scholar] [CrossRef]
- García-González, P.; de Rojas, I.; Moreno-Grau, S.; Montrreal, L.; Puerta, R.; Alarcón-Martín, E.; Quintela, I.; Orellana, A.; Andrade, V.; Adami, P.V.M.; et al. Mendelian Randomisation Confirms the Role of Y-Chromosome Loss in Alzheimer’s Disease Aetiopathogenesis in Men. Int. J. Mol. Sci. 2023, 24, 898. [Google Scholar] [CrossRef]
- Riaz, M.; Mattisson, J.; Polekhina, G.; Bakshi, A.; Halvardson, J.; Danielsson, M.; Ameur, A.; McNeil, J.; Forsberg, L.A.; Lacaze, P. A Polygenic Risk Score Predicts Mosaic Loss of Chromosome Y in Circulating Blood Cells. Cell Biosci. 2021, 11, 205. [Google Scholar] [CrossRef]
- Hes, O.; Šíma, R.; Němcová, J.; Hora, M.; Bulimbasic, S.; Kazakov, D.V.; Ürge, T.; Reischig, T.; Dvořák, M.; Michal, M. End-Stage Kidney Disease: Gains of Chromosomes 7 and 17 and Loss of Y Chromosome in Non-Neoplastic Tissue. Virchows Arch. 2008, 453, 313–319. [Google Scholar] [CrossRef]
- Kovacs, G. The Value of Molecular Genetic Analysis in the Diagnosis and Prognosis of Renal Cell Tumours. World J. Urol. 1994, 12, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Büscheck, F.; Fraune, C.; Garmestani, S.; Simon, R.; Kluth, M.; Hube-Magg, C.; Ketterer, K.; Eichelberg, C.; Höflmayer, D.; Jacobsen, F.; et al. Y-Chromosome Loss Is Frequent in Male Renal Tumors. Ann. Transl. Med. 2021, 9, 209. [Google Scholar] [CrossRef] [PubMed]
- Pallauf, M.; Ged, Y.; Singla, N. Molecular Differences in Renal Cell Carcinoma between Males and Females. World J. Urol. 2023, 41, 1727–1739. [Google Scholar] [CrossRef] [PubMed]
- Peired, A.J.; Campi, R.; Angelotti, M.L.; Antonelli, G.; Conte, C.; Lazzeri, E.; Becherucci, F.; Calistri, L.; Serni, S.; Romagnani, P. Sex and Gender Differences in Kidney Cancer: Clinical and Experimental Evidence. Cancers 2021, 13, 4588. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Ruiz, I. Age-Related Loss of Y Chromosome in Leukocytes Linked to Cardiac Fibrosis. Nat. Rev. Cardiol. 2022, 19, 641. [Google Scholar] [CrossRef] [PubMed]
- Mas-Peiro, S.; Abplanalp, W.T.; Rasper, T.; Berkowitsch, A.; Leistner, D.M.; Dimmeler, S.; Zeiher, A.M. Mosaic Loss of Y Chromosome in Monocytes Is Associated with Lower Survival after Transcatheter Aortic Valve Replacement. Eur. Heart J. 2023, 44, 1943–1952. [Google Scholar] [CrossRef]
- Sano, S.; Horitani, K.; Ogawa, H.; Halvardson, J.; Chavkin, N.W.; Wang, Y.; Sano, M.; Mattisson, J.; Hata, A.; Danielsson, M.; et al. Hematopoietic Loss of Y Chromosome Leads to Cardiac Fibrosis and Heart Failure Mortality. Science 2022, 377, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Sano, S.; Walsh, K. Mosaic Loss of Chromosome Y and Cardiovascular Disease. Nat. Rev. Cardiol. 2024, 21, 151–152. [Google Scholar] [CrossRef]
- Xu, X.; Zhou, R.; Duan, Q.; Miao, Y.; Zhang, T.; Wang, M.; Jones, O.D.; Xu, M. Circulating Macrophages as the Mechanistic Link between Mosaic Loss of Y-Chromosome and Cardiac Disease. Cell Biosci. 2023, 13, 135. [Google Scholar] [CrossRef]
- Jackson, S.S.; Marks, M.A.; Katki, H.A.; Cook, M.B.; Hyun, N.; Freedman, N.D.; Kahle, L.L.; Castle, P.E.; Graubard, B.I.; Chaturvedi, A.K. Sex Disparities in the Incidence of 21 Cancer Types: Quantification of the Contribution of Risk Factors. Cancer 2022, 128, 3531–3540. [Google Scholar] [CrossRef]
- Kobayashi, T.; Hachiya, T.; Ikehata, Y.; Horie, S. Genetic Association of Mosaic Loss of Chromosome Y with Prostate Cancer in Men of European and East Asian Ancestries: A Mendelian Randomization Study. Front. Aging 2023, 4, 1176451. [Google Scholar] [CrossRef] [PubMed]
- Łysiak, M.; Smits, A.; Roodakker, K.R.; Sandberg, E.; Dimberg, A.; Mudaisi, M.; Bratthäll, C.; Strandeus, M.; Milos, P.; Hallbeck, M.; et al. Deletions on Chromosome Y and Downregulation of the SRY Gene in Tumor Tissue Are Associated with Worse Survival of Glioblastoma Patients. Cancers 2021, 13, 1619. [Google Scholar] [CrossRef] [PubMed]
- Noveski, P.; Madjunkova, S.; Sukarova Stefanovska, E.; Matevska Geshkovska, N.; Kuzmanovska, M.; Dimovski, A.; Plaseska-Karanfilska, D. Loss of Y Chromosome in Peripheral Blood of Colorectal and Prostate Cancer Patients. PLoS ONE 2016, 11, e0146264. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Pang, J.; Mitsiades, I.; Lane, A.A.; Rheinbay, E. Loss of Chromosome Y in Primary Tumors. Cell 2023, 186, 3125–3136.e11. [Google Scholar] [CrossRef] [PubMed]
- Stańkowska, W.; Sarkisyan, D.; Bruhn-Olszewska, B.; Duzowska, K.; Bieńkowski, M.; Jąkalski, M.; Wójcik-Zalewska, M.; Davies, H.; Drężek-Chyła, K.; Pęksa, R.; et al. Tumor Predisposing Post-Zygotic Chromosomal Alterations in Bladder Cancer—Insights from Histologically Normal Urothelium. Cancers 2024, 16, 961. [Google Scholar] [CrossRef]
- Wang, Y.; Sano, S. Why Y Matters? The Implication of Loss of Y Chromosome in Blood and Cancer. Cancer Sci. 2024, 115, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Hafiz, H.A.; Schafer, J.M.; Chen, X.; Xiao, T.; Gauntner, T.D.; Li, Z.; Theodorescu, D. Y Chromosome Loss in Cancer Drives Growth by Evasion of Adaptive Immunity. Nature 2023, 619, 624–631. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Zhao, Z.; Feng, B.; Yu, W.; Li, J.; Guo, H.; Yang, R. CD8+CD39+ T Cells Mediate Anti-Tumor Cytotoxicity in Bladder Cancer. OncoTargets Ther. 2021, 14, 2149–2161. [Google Scholar] [CrossRef] [PubMed]
- Timperi, E.; Barnaba, V. CD39 Regulation and Functions in T Cells. Int. J. Mol. Sci. 2021, 22, 8068. [Google Scholar] [CrossRef]
- Li, J.; Lan, Z.; Liao, W.; Horner, J.W.; Xu, X.; Liu, J.; Yoshihama, Y.; Jiang, S.; Shim, H.S.; Slotnik, M.; et al. Histone Demethylase KDM5D Upregulation Drives Sex Differences in Colon Cancer. Nature 2023, 619, 632–639. [Google Scholar] [CrossRef]
- Li, N.; Dhar, S.S.; Chen, T.-Y.; Kan, P.-Y.; Wei, Y.; Kim, J.-H.; Chan, C.-H.; Lin, H.-K.; Hung, M.-C.; Lee, M.G. JARID1D Is a Suppressor and Prognostic Marker of Prostate Cancer Invasion and Metastasis. Cancer Res. 2016, 76, 831–843. [Google Scholar] [CrossRef] [PubMed]
- Mattisson, J.; Danielsson, M.; Hammond, M.; Davies, H.; Gallant, C.J.; Nordlund, J.; Raine, A.; Edén, M.; Kilander, L.; Ingelsson, M.; et al. Leukocytes with Chromosome Y Loss Have Reduced Abundance of the Cell Surface Immunoprotein CD99. Sci. Rep. 2021, 11, 15160. [Google Scholar] [CrossRef]
- Kayser, M. Forensic DNA phenotyping: Predicting human appearance from crime scene material for investigative purposes. Forensic Sci. Int. Genet. 2015, 18, 33–48. [Google Scholar] [CrossRef] [PubMed]
- Walsh, S.; Liu, F.; Ballantyne, K.N.; van Oven, M.; Lao, O.; Kayser, M. IrisPlex: A sensitive DNA tool for accurate prediction of blue and brown eye colour in the absence of ancestry information. Forensic Sci. Int. Genet. 2011, 5, 170–180. [Google Scholar] [CrossRef]
- Walsh, S.; Chaitanya, L.; Clarisse, L.; Wirken, L.; Draus-Barini, J.; Kovatsi, L.; Maeda, H.; Ishikawa, T.; Sijen, T.; de Knijff, P.; et al. Developmental validation of the HIrisPlex system: DNA-based eye and hair colour prediction for forensic and anthropological usage. Forensic Sci. Int. Genet. 2014, 9, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Walsh, S.; Liu, F.; Wollstein, A.; Kovatsi, L.; Ralf, A.; Kosiniak-Kamysz, A.; Branicki, W.; Kayser, M. The HIrisPlex system for simultaneous prediction of hair and eye colour from DNA. Forensic Sci. Int. Genet. 2013, 7, 98–115. [Google Scholar] [CrossRef]
- Peng, F.; Zhu, G.; Hysi, P.G.; Eller, R.J.; Chen, Y.; Li, Y.; Hamer, M.A.; Zeng, C.; Hopkins, R.L.; Jacobus, C.L.; et al. Genome-wide association studies identify multiple genetic loci influencing eyebrow color variation in Europeans. J. Investig. Dermatol. 2019, 139, 1601–1605. [Google Scholar] [CrossRef]
- Hernando, B.; Ibañez, M.V.; Deserio-Cuesta, J.A.; Soria-Navarro, R.; Vilar-Sastre, I.; Martinez-Cadenas, C. Genetic determinants of freckle occurrence in the Spanish population: Towards ephelides prediction from human DNA samples. Forensic Sci. Int. Genet. 2018, 33, 38–47. [Google Scholar] [CrossRef]
- Pośpiech, E.; Karłowska-Pik, J.; Marcińska, M.; Abidi, S.; Andersen, J.D.; Van Den Berge, M.; Carracedo, Á.; Eduardoff, M.; Freire-Aradas, A.; Morling, N.; et al. Evaluation of the predictive capacity of DNA variants associated with straight hair in Europeans. Forensic Sci. Int. Genet. 2015, 19, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Marcińska, M.; Pośpiech, E.; Abidi, S.; Andersen, J.D.; Van Den Berge, M.; Carracedo, Á.; Eduardoff, M.; Marczakiewicz-Lustig, A.; Morling, N.; Sijen, T.; et al. Evaluation of DNA variants associated with androgenetic alopecia and their potential to predict male pattern baldness. PLoS ONE 2015, 10, e0127852. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhong, K.; Jing, X.; Uitterlinden, A.G.; Hendriks, A.E.J.; Drop, S.L.S.; Kayser, M. Update on the predictability of tall stature from DNA markers in Europeans. Forensic Sci. Int. Genet. 2019, 42, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Ambroa-Conde, A.; Casares de Cal, M.A.; Gómez-Tato, A.; Robinson, O.; Mosquera-Miguel, A.; de la Puente, M.; Ruiz-Ramírez, J.; Phillips, C.; Lareu, M.V.; Freire-Aradas, A. Inference of tobacco and alcohol consumption habits from DNA methylation analysis of blood. Forensic Sci. Int. Genet. 2024, 28, 103022. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.; Egan, S.; Hughes, S.; Chapman, B. The Sexome—A proof of concept study into microbial transfer between heterosexual couples after sexual intercourse. Forensic Sci. Int. 2023, 348, 111711. [Google Scholar] [CrossRef] [PubMed]
- Kayser, M.; Branicki, W.; Parson, W.; Phillips, C. Recent advances in Forensic DNA Phenotyping of appearance, ancestry and age. Forensic Sci. Int. Genet. 2023, 65, 102870. [Google Scholar] [CrossRef] [PubMed]
- Koch, C.M.; Wagner, W. Epigenetic-aging-signature to determine age in different tissues. Aging 2011, 3, 1018–1027. [Google Scholar] [CrossRef] [PubMed]
- Bocklandt, S.; Lin, W.; Sehl, M.E.; Sanchez, F.J.; Sinsheimer, J.S.; Horvath, S.; Vilain, E. Epigenetic predictor of age. PLoS ONE 2011, 6, e14821. [Google Scholar] [CrossRef]
- Jung, S.E.; Shin, K.J.; Lee, H.-Y. DNA methylation-based age prediction from various tissues and body fluids. BMB Rep. 2017, 50, 546–553. [Google Scholar] [CrossRef]
- Cho, S.; Jung, S.-E.; Hong, S.R.; Lee, E.H.; Lee, J.H.; Lee, S.D.; Lee, H.Y. Independent validation of DNA-based approaches for age prediction in blood. Forensic Sci. Int. Genet. 2017, 29, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Freire-Aradas, A.; Phillips, C.; Girón-Santamaría, L.; Miguel, A.M.; Gómez-Tato, A.; de Cal, M.C.; Alvarez-Dios, J.A.; Lareu, M.V. Tracking age-correlated DNA methylation markers in the young. Forensic Sci. Int. Genet. 2018, 36, 50–59. [Google Scholar] [CrossRef]
- McEwen, L.M.; O’Donnell, K.J.; McGill, M.G.; Edgar, R.D.; Jones, M.J.; MacIsaac, J.L.; Lin, D.T.S.; Ramadori, K.; Morin, A.; Gladish, N.; et al. The PedBE clock accurately estimates DNA methylation age in pediatric buccal cells. Proc. Natl. Acad. Sci. USA 2020, 117, 23329–23335. [Google Scholar] [CrossRef] [PubMed]
- Freire-Aradas, A.; Girón-Santamaría, L.; Mosquera-Miguel, A.; Ambroa-Conde, A.; Phillips, C.; de Cal, M.C.; Gómez-Tato, A.; Álvarez-Dios, J.; Pospiech, E.; Aliferi, A.; et al. A common epigenetic clock from childhood to old age. Forensic Sci. Int. Genet. 2022, 60, 102743. [Google Scholar] [CrossRef] [PubMed]
- Koop, B.E.; Reckert, A.; Becker, J.; Han, Y.; Wagner, W.; Ritz-Timme, S. Epigenetic clocks may come out of rhythm-implications for the estimation of chronological age in forensic casework. Int J. Leg. Med. 2020, 134, 2215–2228. [Google Scholar] [CrossRef]
- Spolnicka, M.; Pośpiech, E.; Pepłońska, B.; Zbieć-Piekarska, R.; Makowska, Ż.; Pięta, A.; Karłowska-Pik, J.; Ziemkiewicz, B.; Wężyk, M.; Gasperowicz, P.; et al. DNA methylation in ELOVL2 and C1orf132 correctly predicted chronological age of individuals from three disease groups. Int J. Leg. Med. 2018, 132, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Spolnicka, M.; Zbieć-Piekarska, R.; Karp, M.; Machnicki, M.M.; Własiuk, P.; Makowska, Ż.; Pięta, A.; Gambin, T.; Gasperowicz, P.; Branicki, W.; et al. DNA methylation signature in blood does not predict calendar age in patients with chronic lymphocytic leukemia but may alert to the presence of disease. Forensic Sci. Int. Genet. 2018, 34, e15–e17. [Google Scholar] [CrossRef] [PubMed]
- Spolnicka, M.; Pośpiech, E.; Adamczyk, J.G.; Freire-Aradas, A.; Pepłońska, B.; Zbieć-Piekarska, R.; Makowska, Ż.; Pięta, A.; Lareu, M.V.; Phillips, C.; et al. Modified aging of elite athletes revealed by analysis of epigenetic age markers. Aging 2018, 10, 241–252. [Google Scholar] [CrossRef]
- Zbiec-Piekarska, R.; Spólnicka, M.; Kupiec, T.; Makowska, Ż.; Spas, A.; Parys-Proszek, A.; Kucharczyk, K.; Płoski, R.; Branicki, W. Examination of DNA methylation status of the ELOVL2 marker may be useful for human age prediction in forensic science. Forensic Sci. Int. Genet. 2015, 14, 161–167. [Google Scholar] [CrossRef]
- Mayer, F.; Becker, J.; Reinauer, C.; Böhme, P.; Eickhoff, S.B.; Koop, B.; Gündüz, T.; Blum, J.; Wagner, W.; Ritz-Timme, S. Altered DNA methylation at age-associated CpG sites in children with growth disorders: Impact on age estimation? Int J. Leg. Med. 2022, 136, 987–996. [Google Scholar] [CrossRef]
- Piniewska-Rog, D.; Heidegger, A.; Pośpiech, E.; Xavier, C.; Pisarek, A.; Jarosz, A.; Woźniak, A.; Wojtas, M.; Phillips, C.; Kayser, M.; et al. Impact of excessive alcohol abuse on age prediction using the VISAGE enhanced tool for epigenetic age estimation in blood. Int J. Leg. Med. 2021, 135, 2209–2219. [Google Scholar] [CrossRef] [PubMed]
- Foox, J.; Richards, J.; Smith, A.; Johnson, B.; Thompson, C. Epigenetic forensics for suspect identification and age prediction. Forensic Genom. 2021, 1, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Choung, C.M.; Jung, J.Y.; Lee, H.Y.; Lim, S.-K. A validation study of DNA methylation-based age prediction using semen in forensic casework samples. Leg. Med. 2018, 31, 74–77. [Google Scholar] [CrossRef] [PubMed]
- Aliferi, A.; Ballard, D.; Gallidabino, M.D.; Thurtle, H.; Barron, L.; Court, D.S. DNA methylation-based age prediction using massively parallel sequencing data and multiple machine learning models. Forensic Sci. Int. Genet. 2018, 37, 215–226. [Google Scholar] [CrossRef]
- Naue, J.; Hoefsloot, H.C.; Kloosterman, A.D.; Verschure, P.J. Forensic DNA methylation profiling from minimal traces: How low can we go? Forensic Sci. Int. Genet. 2018, 33, 17–23. [Google Scholar] [CrossRef]
- Aliferi, A.; Sundaram, S.; Ballard, D.; Freire-Aradas, A.; Phillips, C.; Lareu, M.V.; Court, D.S. Combining current knowledge on DNA methylation-based age estimation towards the development of a superior forensic DNA intelligence tool. Forensic Sci. Int. Genet. 2022, 57, 102637. [Google Scholar] [CrossRef]
- Smith, R.W.; Monroe, C.; Bolnick, D.A. Detection of Cytosine methylation in ancient DNA from five native american populations using bisulfite sequencing. PLoS ONE 2015, 10, e0125344. [Google Scholar] [CrossRef] [PubMed]
- Correia Dias, H.; Cordeiro, C.; Real, F.C.; Cunha, E.; Manco, L. Age estimation based on DNA methylation using blood samples from deceased individuals. J. Forensic Sci. 2020, 65, 465–470. [Google Scholar] [CrossRef]
- Freire-Aradas, A.; Pośpiech, E.; Aliferi, A.; Girón-Santamaría, L.; Mosquera-Miguel, A.; Pisarek, A.; Ambroa-Conde, A.; Phillips, C.; de Cal, M.A.C.; Gómez-Tato, A.; et al. A comparison of forensic age prediction models using data from four DNA methylation technologies. Front. Genet. 2020, 11, 932. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Feng, L.; Chen, J.; Wang, L.; Li, P.; Ji, A.; Zeng, C.; Liu, F.; Li, C. Validation of methylation-based forensic age estimation in timeseries bloodstains on FTA cards and gauze at room temperature conditions. Forensic Sci. Int. Genet. 2019, 40, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Peng, F.; Li, S.; Jiang, L.; Sun, H.; Ji, A.; Zeng, C.; Li, C.; Liu, F. Systematic feature selection improves accuracy of methylation based forensic age estimation in Han Chinese males. Forensic Sci. Int. Genet. 2018, 35, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Pisarek, A.; Pośpiech, E.; Heidegger, A.; Xavier, C.; Papież, A.; Piniewska-Róg, D.; Kalamara, V.; Potabattula, R.; Bochenek, M.; Sikora-Polaczek, M.; et al. Epigenetic age prediction in semen—Marker selection and model development. Aging 2021, 13, 19145–19164. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.R.; Shin, K.-J.; Jung, S.-E.; Lee, E.H.; Lee, H.Y. Platform-independent models for age prediction using DNA methylation data. Forensic Sci. Int. Genet. 2019, 38, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Fleckhaus, J.; Schneider, P.M. Novel multiplex strategy for DNA methylation based age prediction from small amounts of DNA via Pyrosequencing. Forensic Sci. Int. Genet. 2020, 44, 102189. [Google Scholar] [CrossRef] [PubMed]
- Manco, L.; Dias, H.C. DNA methylation analysis of ELOVL2 gene using droplet digital PCR for age estimation purposes. Forensic Sci. Int. Genet. 2022, 333, 111206. [Google Scholar] [CrossRef]
- Takahashi, Y.; Asari, M.; Isozaki, S.; Hoshina, C.; Okuda, K.; Mori, K.; Namba, R.; Ochiai, W.; Shimizu, K. Age prediction by methylation analysis of small amounts of DNA using locked nucleic acids. J. Forensic Sci. 2023, 68, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Lee, J.M.; Naue, J.; Fleckhaus, J.; Freire-Aradas, A.; Neubauer, J.; Pośpiech, E.; McCord, B.; Kalamara, V.; Gauthier, Q.; et al. A collaborative exercise on DNA methylation-based age prediction and body fluid typing. Forensic Sci. Int. Genet. 2022, 57, 102656. [Google Scholar] [CrossRef] [PubMed]
- Montesanto, A.; D’aquila, P.; Lagani, V.; Paparazzo, E.; Geracitano, S.; Formentini, L.; Giacconi, R.; Cardelli, M.; Provinciali, M.; Bellizzi, D.; et al. A new robust epigenetic model for forensic age prediction. J. Forensic Sci. 2020, 65, 1424–1431. [Google Scholar] [CrossRef]
- Zbiec-Piekarska, R.; Spólnicka, M.; Kupiec, T.; Parys-Proszek, A.; Makowska, Ż.; Pałeczka, A.; Kucharczyk, K.; Płoski, R.; Branicki, W. Development of a forensically useful age prediction method based on DNA methylation analysis. Forensic Sci. Int. Genet. 2015, 17, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.I.; Hughes, G.; Sadda, S.; Klotzle, B.; Bibikova, M.; Fan, J.B.; Gao, Y.; et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 2013, 49, 359–367. [Google Scholar] [CrossRef]
- Naue, J.; Sänger, T.; Hoefsloot, H.C.; Lutz-Bonengel, S.; Kloosterman, A.D.; Verschure, P.J. Proof of concept study of age-dependent DNA methylation markers across different tissues by massive parallel sequencing. Forensic Sci. Int. Genet. 2018, 36, 152–159. [Google Scholar] [CrossRef]
- Slieker, R.C.; Relton, C.L.; Gaunt, T.R.; Slagboom, P.E.; Heijmans, B.T. Age-related DNA methylation changes are tissue-specific with ELOVL2 promoter methylation as exception. Epigenet. Chrom. 2018, 11, 25. [Google Scholar]
- Dias, H.C.; Cordeiro, C.; Pereira, J.; Pinto, C.; Real, F.C.; Cunha, E.; Manco, L. DNA methylation age estimation in blood samples of living and deceased individuals using a multiplex SNaPshot assay. Forensic Sci. Int. 2020, 311, 110267. [Google Scholar] [CrossRef]
- Heyn, H.; Moran, S.; Hernando-Herraez, I.; Sayols, S.; Gomez, A.; Sandoval, J.; Monk, D.; Hata, K.; Marques-Bonet, T.; Wang, L.; et al. DNA methylation contributes to natural human variation. Genome Res. 2013, 23, 1363–1372. [Google Scholar] [CrossRef]
- Naue, J.; Hoefsloot, H.C.; Mook, O.R.; Rijlaarsdam-Hoekstra, L.; van der Zwalm, M.C.; Henneman, P.; Kloosterman, A.D.; Verschure, P.J. Chronological age prediction based on DNA methylation: Massive parallel sequencing and random forest regression. Forensic Sci. Int. Genet. 2017, 31, 19–28. [Google Scholar] [CrossRef]
- Hamano, Y.; Manabe, S.; Morimoto, C.; Fujimoto, S.; Tamaki, K. Forensic age prediction for saliva samples using methylationsensitive high resolution melting: Exploratory application for cigarette butts. Sci. Rep. 2017, 7, 10444. [Google Scholar] [CrossRef]
- Xin, Y.; Dong, K.; Cao, F.; Tian, Y.; Sun, J.; Peng, M.; Liu, W.; Shi, P. Studies of hTERT DNA methylation assays on the human age prediction. Int J. Leg. Med. 2019, 133, 1333–1339. [Google Scholar] [CrossRef]
- Mawlood, S.K.; Dennany, L.; Watson, N.; Dempster, J.; Pickard, B.S. Quantification of global mitochondrial DNA methylation levels and inverse correlation with age at two CpG sites. Aging 2016, 8, 636–641. [Google Scholar] [CrossRef] [PubMed]
- Lau, P.Y.; Fung, W.K. Evaluation of marker selection methods and statistical models for chronological age prediction based on DNA methylation. Leg. Med. 2020, 47, 101744. [Google Scholar] [CrossRef]
- Vidaki, A.; González, D.M.; Jiménez, B.P.; Kayser, M. Male-specific age estimation based on Y-chromosomal DNA methylation. Aging 2021, 13, 6442–6458. [Google Scholar] [CrossRef] [PubMed]
- Hamano, Y.; Manabe, S.; Morimoto, C.; Fujimoto, S.; Ozeki, M.; Tamaki, K. Forensic age prediction for dead or living samples by use of methylation-sensitive high resolution melting. Leg. Med. 2016, 21, 5–10. [Google Scholar] [CrossRef]
- Hong, S.R.; Jung, S.-E.; Lee, E.H.; Shin, K.-J.; Yang, W.I.; Lee, H.Y. DNA methylation-based age prediction from saliva: High age predictability by combination of 7 CpG markers. Forensic Sci. Int. Genet. 2017, 29, 118–125. [Google Scholar] [CrossRef]
- Eipel, M.; Mayer, F.; Arent, T.; Ferreira, M.R.; Birkhofer, C.; Gerstenmaier, U.; Costa, I.G.; Ritz-Timme, S.; Wagner, W. Epigenetic age predictions based on buccal swabs are more precise in combination with cell type-specific DNA methylation signatures. Aging 2016, 8, 1034–1048. [Google Scholar] [CrossRef]
- Koop, B.E.; Mayer, F.; Gündüz, T.; Blum, J.; Becker, J.; Schaffrath, J.; Wagner, W.; Han, Y.; Boehme, P.; Ritz-Timme, S. Postmortem age estimation via DNA methylation analysis in buccal swabs from corpses in different stages of decomposition—A “proof of principle” study. Int J. Leg. Med 2021, 135, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Ambroa-Conde, A.; Girón-Santamaría, L.; Mosquera-Miguel, A.; Phillips, C.; de Cal, M.C.; Gómez-Tato, A.; Álvarez-Dios, J.; de la Puente, M.; Ruiz-Ramírez, J.; Lareu, M.; et al. Epigenetic age estimation in saliva and in buccal cells. Forensic Sci. Int. Genet. 2022, 61, 102770. [Google Scholar] [CrossRef] [PubMed]
- Márquez-Ruiz, A.B.; González-Herrera, L.; Luna, J.D.; Valenzuela, A. DNA methylation levels and telomere length in human teeth: Usefulness for age estimation. Int. J. Leg. Med. 2020, 134, 451–459. [Google Scholar] [CrossRef]
- Giuliani, C.; Cilli, E.; Bacalini, M.G.; Pirazzini, C.; Sazzini, M.; Gruppioni, G.; Franceschi, C.; Garagnani, P.; Luiselli, D. Inferring chronological age from DNA methylation patterns of human teeth. Am. J. Phys. Anthropol. 2016, 159, 585–595. [Google Scholar] [CrossRef]
- Forsberg, L.A.; Gisselsson, D.; Dumanski, J.P. Mosaicism in Health and Disease—Clones Picking up Speed. Nat. Rev. Genet. 2017, 18, 128–142. [Google Scholar] [CrossRef]
- Guttenbach, M.; Koschorz, B.; Bernthaler, U.; Grimm, T.; Schmid, M. Sex Chromosome Loss and Aging: In Situ Hybridization Studies on Human Interphase Nuclei. Am. J. Hum. Genet. 1995, 57, 1143–1150. [Google Scholar] [PubMed]
- Kang, Y.G.; Suh, E.; Lee, J.; Kim, D.W.; Cho, K.H.; Bae, C.-Y. Biological Age as a Health Index for Mortality and Major Age-Related Disease Incidence in Koreans: National Health Insurance Service–Health Screening 11-Year Follow-up Study. Clin. Interv. Aging 2018, 13, 429–436. [Google Scholar] [CrossRef]
- Jylhävä, J.; Pedersen, N.L.; Hägg, S. Biological Age Predictors. EBioMedicine 2017, 21, 29–36. [Google Scholar] [CrossRef]
- Song, M.; Jiang, L.; Wang, X.; Zhou, W.; Wang, N.; Hou, Y.; Song, F. Loss of Y Chromosome in Leukocytes Can Be Regarded as a Male-Specific Age Predictor for Age Group Estimation in Forensic Genetics. Mol. Genet. Genom. 2023, 298, 1073–1085. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gutiérrez-Hurtado, I.A.; Sánchez-Méndez, A.D.; Becerra-Loaiza, D.S.; Rangel-Villalobos, H.; Torres-Carrillo, N.; Gallegos-Arreola, M.P.; Aguilar-Velázquez, J.A. Loss of the Y Chromosome: A Review of Molecular Mechanisms, Age Inference, and Implications for Men’s Health. Int. J. Mol. Sci. 2024, 25, 4230. https://doi.org/10.3390/ijms25084230
Gutiérrez-Hurtado IA, Sánchez-Méndez AD, Becerra-Loaiza DS, Rangel-Villalobos H, Torres-Carrillo N, Gallegos-Arreola MP, Aguilar-Velázquez JA. Loss of the Y Chromosome: A Review of Molecular Mechanisms, Age Inference, and Implications for Men’s Health. International Journal of Molecular Sciences. 2024; 25(8):4230. https://doi.org/10.3390/ijms25084230
Chicago/Turabian StyleGutiérrez-Hurtado, Itzae Adonai, Astrid Desireé Sánchez-Méndez, Denisse Stephania Becerra-Loaiza, Héctor Rangel-Villalobos, Norma Torres-Carrillo, Martha Patricia Gallegos-Arreola, and José Alonso Aguilar-Velázquez. 2024. "Loss of the Y Chromosome: A Review of Molecular Mechanisms, Age Inference, and Implications for Men’s Health" International Journal of Molecular Sciences 25, no. 8: 4230. https://doi.org/10.3390/ijms25084230
APA StyleGutiérrez-Hurtado, I. A., Sánchez-Méndez, A. D., Becerra-Loaiza, D. S., Rangel-Villalobos, H., Torres-Carrillo, N., Gallegos-Arreola, M. P., & Aguilar-Velázquez, J. A. (2024). Loss of the Y Chromosome: A Review of Molecular Mechanisms, Age Inference, and Implications for Men’s Health. International Journal of Molecular Sciences, 25(8), 4230. https://doi.org/10.3390/ijms25084230









