Effect of Low-Level Tragus Stimulation on Cardiac Metabolism in Heart Failure with Preserved Ejection Fraction: A Transcriptomics-Based Analysis
Abstract
1. Introduction
2. Results
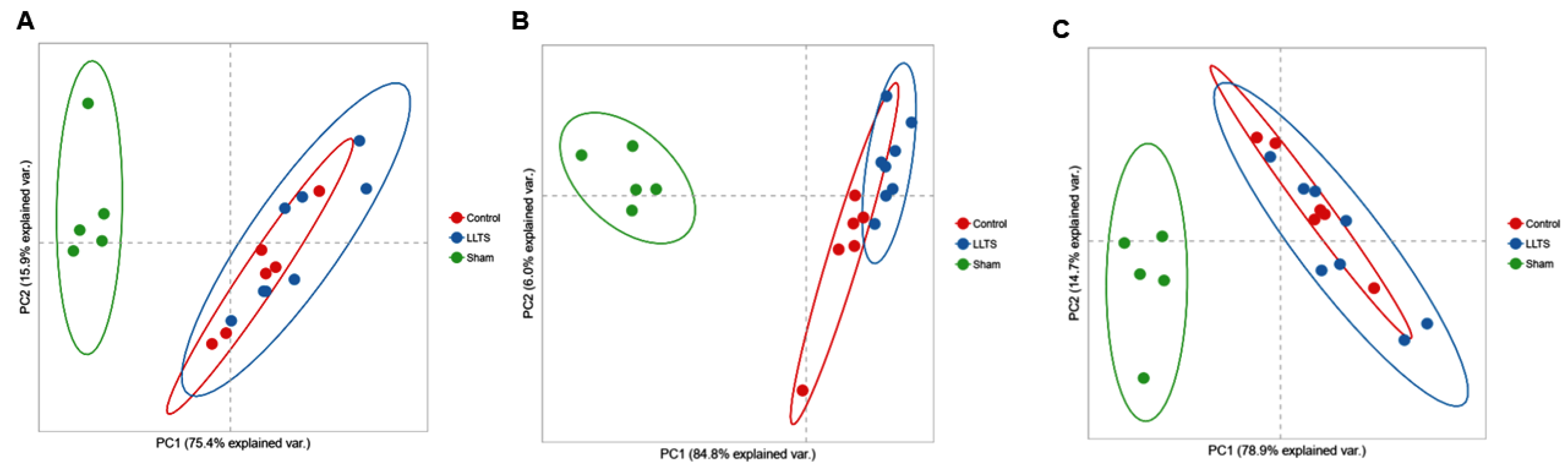
2.1. Differentially Expressed Genes
2.1.1. Effect on Cardiac Phenotype
2.1.2. Control vs. Sham
2.1.3. Active vs. Sham
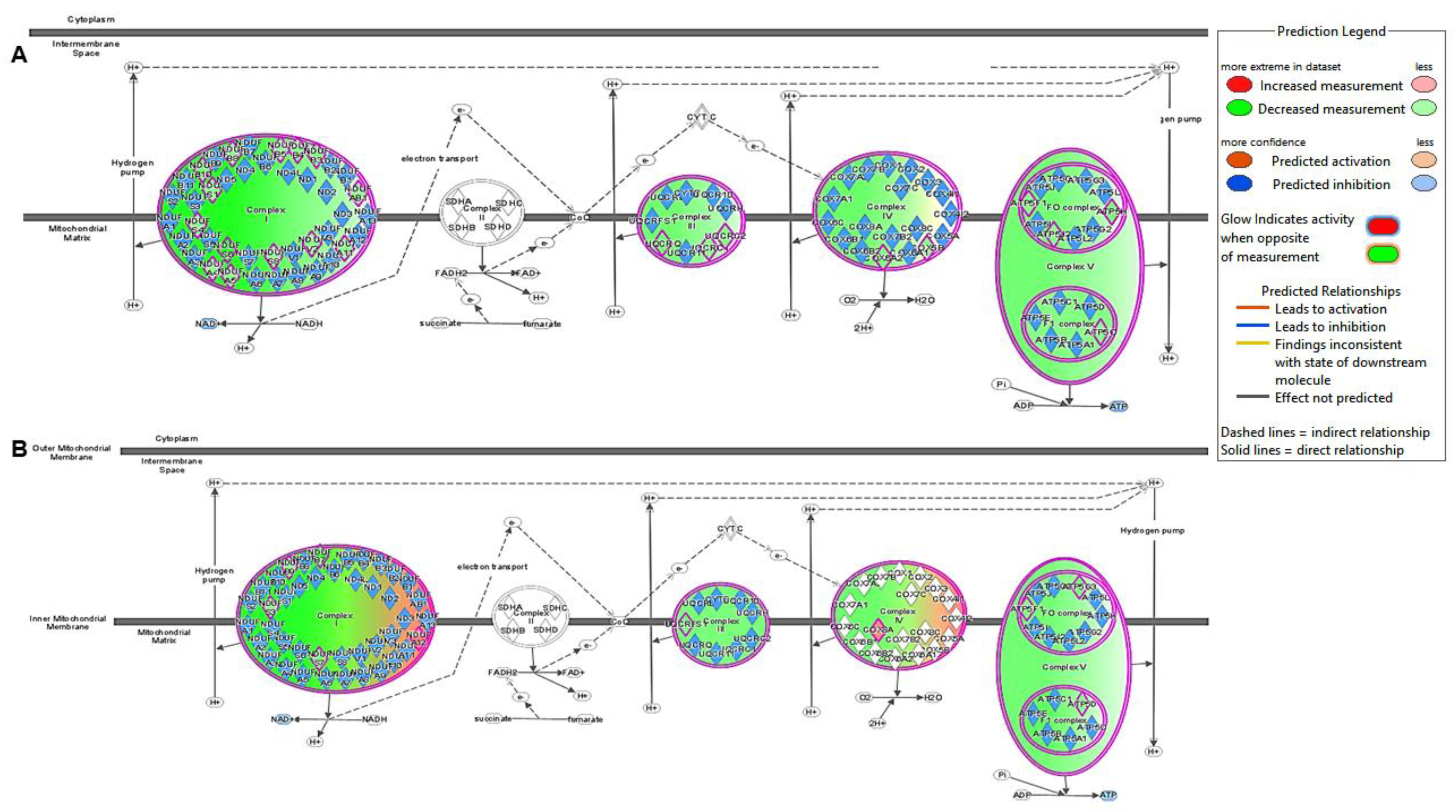
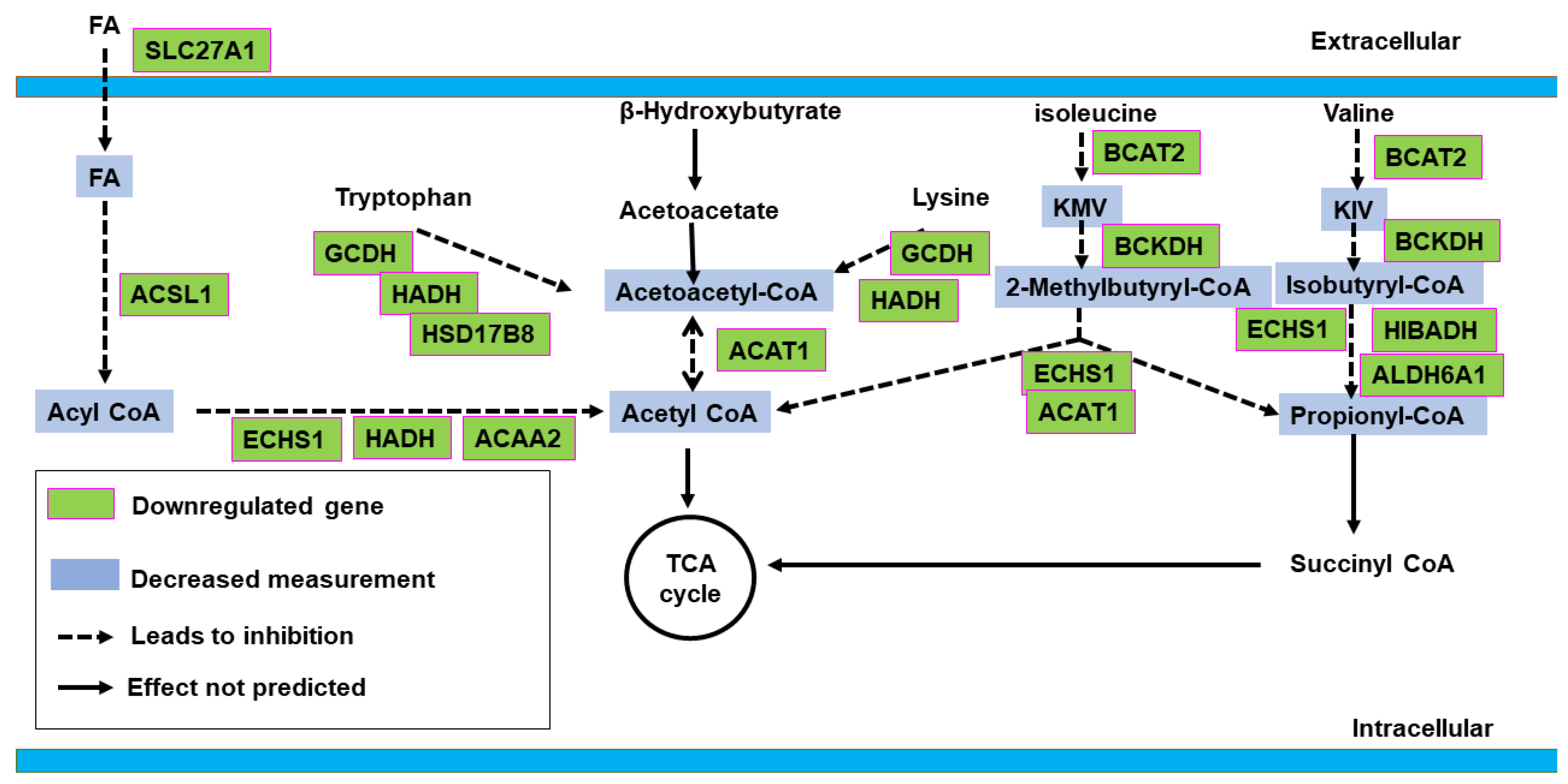
2.2. Canonical Pathway Analysis
2.2.1. Control vs. Sham
2.2.2. Active vs. Sham
2.3. Upstream Regulator Analysis
2.3.1. Control vs. Sham
2.3.2. Active vs. Sham
3. Discussion
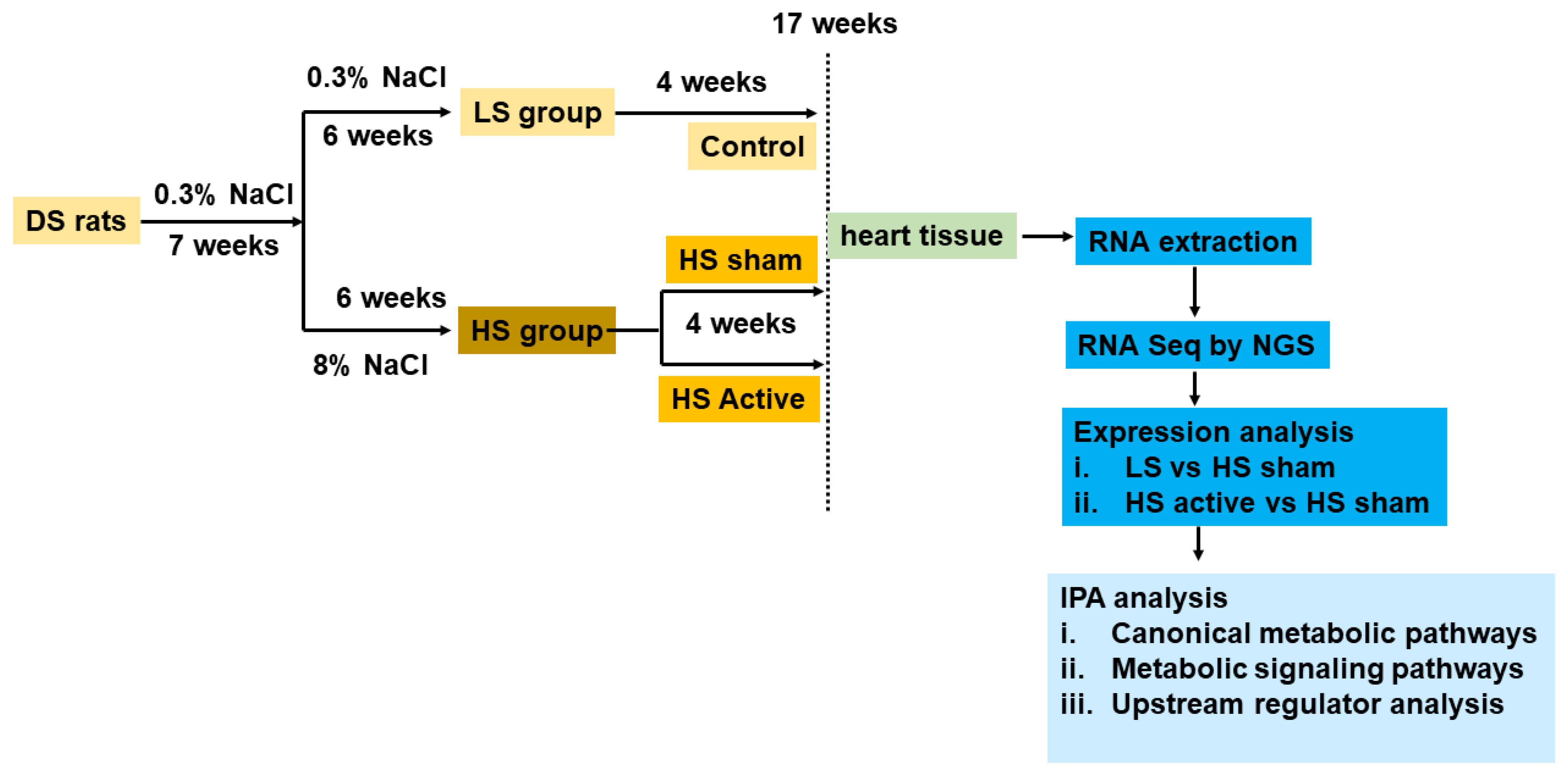
4. Materials and Methods
4.1. Study Protocol
4.2. Next-Generation RNA Sequencing
4.3. Ingenuity Pathway Analysis
4.4. Statistical Analysis
4.5. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Salah, H.M.; Minhas, A.M.K.; Khan, M.S.; Pandey, A.; Michos, E.D.; Mentz, R.J.; Fudim, M. Causes of hospitalization in the USA between 2005 and 2018. Eur. Heart J. Open 2021, 1, oeab001. [Google Scholar] [CrossRef] [PubMed]
- Redfield, M.M.; Borlaug, B.A. Heart Failure with Preserved Ejection Fraction: A Review. JAMA 2023, 329, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Stavrakis, S.; Elkholey, K.; Morris, L.; Niewiadomska, M.; Asad, Z.U.A.; Humphrey, M.B. Neuromodulation of Inflammation to Treat Heart Failure with Preserved Ejection Fraction: A Pilot Randomized Clinical Trial. J. Am. Heart Assoc. 2022, 11, e023582. [Google Scholar] [CrossRef] [PubMed]
- Sha, Y.; Scherlag, B.J.; Yu, L.; Sheng, X.; Jackman, W.M.; Lazzara, R.; Po, S.S. Low-level right vagal stimulation: Anticholinergic and antiadrenergic effects. J. Cardiovasc. Electrophysiol. 2011, 22, 1147–1153. [Google Scholar] [CrossRef] [PubMed]
- Elkholey, K.; Niewiadomska, M.; Morris, L.; Whyte, S.; Houser, J.; Humphrey, M.B.; Stavrakis, S. Transcutaneous Vagus Nerve Stimulation Ameliorates the Phenotype of Heart Failure with Preserved Ejection Fraction through Its Anti-Inflammatory Effects. Circ. Heart Fail. 2022, 15, e009288. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Filiberti, A.; Humphrey, M.B.; Fleming, C.D.; Scherlag, B.J.; Po, S.S.; Stavrakis, S. Low-level transcutaneous vagus nerve stimulation attenuates cardiac remodelling in a rat model of heart failure with preserved ejection fraction. Exp. Physiol. 2019, 104, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Yoshii, A.; Tian, R. Remodeling of cardiac metabolism in heart failure with preserved ejection fraction. Curr. Opin. Physiol. 2022, 27, 100559. [Google Scholar] [CrossRef]
- Lopaschuk, G.D.; Karwi, Q.G.; Tian, R.; Wende, A.R.; Abel, E.D. Cardiac Energy Metabolism in Heart Failure. Circ. Res. 2021, 128, 1487–1513. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, B.V.; Rawls, K.D.; Kolling, G.L.; Vinnakota, K.C.; Wallqvist, A.; Papin, J.A. Identifying functional metabolic shifts in heart failure with the integration of omics data and a heart-specific, genome-scale model. Cell Rep. 2021, 34, 108836. [Google Scholar] [CrossRef] [PubMed]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Chakraborty, P.; Po, S.S.; Scherlag, B.J.; Dasari, T.W. The neurometabolic axis: A novel therapeutic target in heart failure. Life Sci. 2023, 333, 122122. [Google Scholar] [CrossRef]
- Kumar, A.A.; Kelly, D.P.; Chirinos, J.A. Mitochondrial Dysfunction in Heart Failure with Preserved Ejection Fraction. Circulation 2019, 139, 1435–1450. [Google Scholar] [CrossRef] [PubMed]
- Luptak, I.; Sverdlov, A.L.; Panagia, M.; Qin, F.; Pimentel, D.R.; Croteau, D.; Siwik, D.A.; Ingwall, J.S.; Bachschmid, M.M.; Balschi, J.A.; et al. Decreased ATP production and myocardial contractile reserve in metabolic heart disease. J. Mol. Cell Cardiol. 2018, 116, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Nuntaphum, W.; Pongkan, W.; Wongjaikam, S.; Thummasorn, S.; Tanajak, P.; Khamseekaew, J.; Intachai, K.; Chattipakorn, S.C.; Chattipakorn, N.; Shinlapawittayatorn, K. Vagus nerve stimulation exerts cardioprotection against myocardial ischemia/reperfusion injury predominantly through its efferent vagal fibers. Basic Res Cardiol. 2018, 113, 22. [Google Scholar] [CrossRef] [PubMed]
- Xue, R.Q.; Sun, L.; Yu, X.J.; Li, D.L.; Zang, W.J. Vagal nerve stimulation improves mitochondrial dynamics via an M(3) receptor/CaMKKβ/AMPK pathway in isoproterenol-induced myocardial ischaemia. J. Cell Mol. Med. 2017, 21, 58–71. [Google Scholar] [CrossRef]
- Hahn, V.S.; Petucci, C.; Kim, M.S.; Bedi, K.C., Jr.; Wang, H.; Mishra, S.; Koleini, N.; Yoo, E.J.; Margulies, K.B.; Arany, Z.; et al. Myocardial Metabolomics of Human Heart Failure with Preserved Ejection Fraction. Circulation 2023, 147, 1147–1161. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Jiang, L.; Li, T. Aberrant branched-chain amino acid catabolism in cardiovascular diseases. Front Cardiovasc. Med. 2022, 9, 965899. [Google Scholar] [CrossRef]
- Uddin, G.M.; Zhang, L.; Shah, S.; Fukushima, A.; Wagg, C.S.; Gopal, K.; Al Batran, R.; Pherwani, S.; Ho, K.L.; Boisvenue, J.; et al. Impaired branched chain amino acid oxidation contributes to cardiac insulin resistance in heart failure. Cardiovasc. Diabetol. 2019, 18, 86. [Google Scholar] [CrossRef] [PubMed]
- Wolfson, R.L.; Chantranupong, L.; Saxton, R.A.; Shen, K.; Scaria, S.M.; Cantor, J.R.; Sabatini, D.M. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 2016, 351, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Montaigne, D.; Butruille, L.; Staels, B. PPAR control of metabolism and cardiovascular functions. Nat. Rev. Cardiol. 2021, 18, 809–823. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Gao, K.; Liu, C.; Li, T.; Ding, Y.; Ma, J. Pathological mechanism of heart failure with preserved ejection fraction in rats based on iTRAQ technology. PeerJ 2023, 11, e15280. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, S.; Sadoshima, J. The role of sirtuins in cardiac disease. Am. J. Physiol. Heart Circ. Physiol. 2015, 309, H1375–H1389. [Google Scholar] [CrossRef]
- Oka, S.; Alcendor, R.; Zhai, P.; Park, J.Y.; Shao, D.; Cho, J.; Yamamoto, T.; Tian, B.; Sadoshima, J. PPARα-Sirt1 complex mediates cardiac hypertrophy and failure through suppression of the ERR transcriptional pathway. Cell Metab. 2011, 14, 598–611. [Google Scholar] [CrossRef] [PubMed]
- Sundaresan, N.R.; Pillai, V.B.; Wolfgeher, D.; Samant, S.; Vasudevan, P.; Parekh, V.; Raghuraman, H.; Cunningham, J.M.; Gupta, M.; Gupta, M.P. The deacetylase SIRT1 promotes membrane localization and activation of Akt and PDK1 during tumorigenesis and cardiac hypertrophy. Sci. Signal 2011, 4, ra46. [Google Scholar] [CrossRef] [PubMed]
- Scherbakov, N.; Bauer, M.; Sandek, A.; Szabó, T.; Töpper, A.; Jankowska, E.A.; Springer, J.; von Haehling, S.; Anker, S.D.; Lainscak, M.; et al. Insulin resistance in heart failure: Differences between patients with reduced and preserved left ventricular ejection fraction. Eur. J. Heart Fail. 2015, 17, 1015–1021. [Google Scholar] [CrossRef] [PubMed]
- Ashrafian, H.; Frenneaux, M.P.; Opie, L.H. Metabolic Mechanisms in Heart Failure. Circulation 2007, 116, 434–448. [Google Scholar] [CrossRef] [PubMed]
- Saw, E.L.; Pearson, J.T.; Schwenke, D.O.; Munasinghe, P.E.; Tsuchimochi, H.; Rawal, S.; Coffey, S.; Davis, P.; Bunton, R.; Van Hout, I.; et al. Activation of the cardiac non-neuronal cholinergic system prevents the development of diabetes-associated cardiovascular complications. Cardiovasc. Diabetol. 2021, 20, 50. [Google Scholar] [CrossRef] [PubMed]
- Ruthenburg, A.J.; Allis, C.D.; Wysocka, J. Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol. Cell 2007, 25, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Pu, W.T. Cardiomyocyte Maturation. Circ. Res. 2020, 126, 1086–1106. [Google Scholar] [CrossRef] [PubMed]
- Deogharia, M.; Agrawal, A.; Shi, M.; Jain, A.K.; McHugh, K.J.; Altamirano, F.; Marian, A.J.; Gurha, P. Histone demethylase KDM5 regulates cardiomyocyte maturation by promoting fatty acid oxidation, oxidative phosphorylation, and myofibrillar organization. bioRxiv 2023. [Google Scholar] [CrossRef] [PubMed]
- Bullard, J.H.; Purdom, E.; Hansen, K.D.; Dudoit, S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinform. 2010, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Lindsay, H.; Robinson, M.D. Robustly detecting differential expression in RNA sequencing data using observation weights. Nucleic. Acids Res. 2014, 42, e91. [Google Scholar] [CrossRef] [PubMed]
- Kambis, T.N.; Shahshahan, H.R.; Mishra, P.K. Metabolites and Genes behind Cardiac Metabolic Remodeling in Mice with Type 1 Diabetes Mellitus. Int. J. Mol. Sci. 2022, 23, 1392. [Google Scholar] [CrossRef] [PubMed]
- Alimadadi, A.; Aryal, S.; Manandhar, I.; Joe, B.; Cheng, X. Identification of Upstream Transcriptional Regulators of Ischemic Cardiomyopathy Using Cardiac RNA-Seq Meta-Analysis. Int. J. Mol. Sci. 2020, 21, 3472. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chakraborty, P.; Niewiadomska, M.; Farhat, K.; Morris, L.; Whyte, S.; Humphries, K.M.; Stavrakis, S. Effect of Low-Level Tragus Stimulation on Cardiac Metabolism in Heart Failure with Preserved Ejection Fraction: A Transcriptomics-Based Analysis. Int. J. Mol. Sci. 2024, 25, 4312. https://doi.org/10.3390/ijms25084312
Chakraborty P, Niewiadomska M, Farhat K, Morris L, Whyte S, Humphries KM, Stavrakis S. Effect of Low-Level Tragus Stimulation on Cardiac Metabolism in Heart Failure with Preserved Ejection Fraction: A Transcriptomics-Based Analysis. International Journal of Molecular Sciences. 2024; 25(8):4312. https://doi.org/10.3390/ijms25084312
Chicago/Turabian StyleChakraborty, Praloy, Monika Niewiadomska, Kassem Farhat, Lynsie Morris, Seabrook Whyte, Kenneth M. Humphries, and Stavros Stavrakis. 2024. "Effect of Low-Level Tragus Stimulation on Cardiac Metabolism in Heart Failure with Preserved Ejection Fraction: A Transcriptomics-Based Analysis" International Journal of Molecular Sciences 25, no. 8: 4312. https://doi.org/10.3390/ijms25084312
APA StyleChakraborty, P., Niewiadomska, M., Farhat, K., Morris, L., Whyte, S., Humphries, K. M., & Stavrakis, S. (2024). Effect of Low-Level Tragus Stimulation on Cardiac Metabolism in Heart Failure with Preserved Ejection Fraction: A Transcriptomics-Based Analysis. International Journal of Molecular Sciences, 25(8), 4312. https://doi.org/10.3390/ijms25084312





