The Characterization of G-Quadruplexes in Tobacco Genome and Their Function under Abiotic Stress
Abstract
1. Introduction
2. Results
2.1. General Situation of G-Quadruplex Distribution in Tobacco Genome
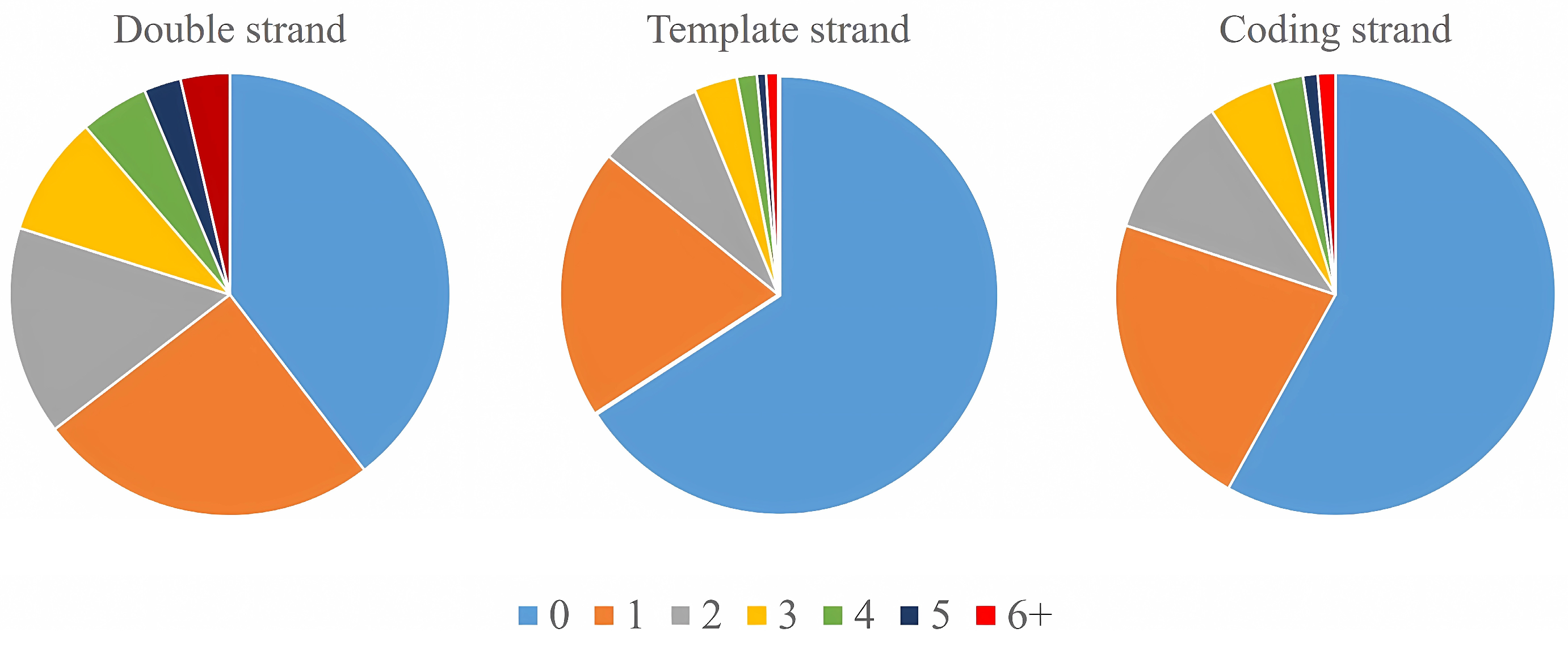
2.2. The Relationship between Tobacco G-Quadruplexes and Genome Characteristics
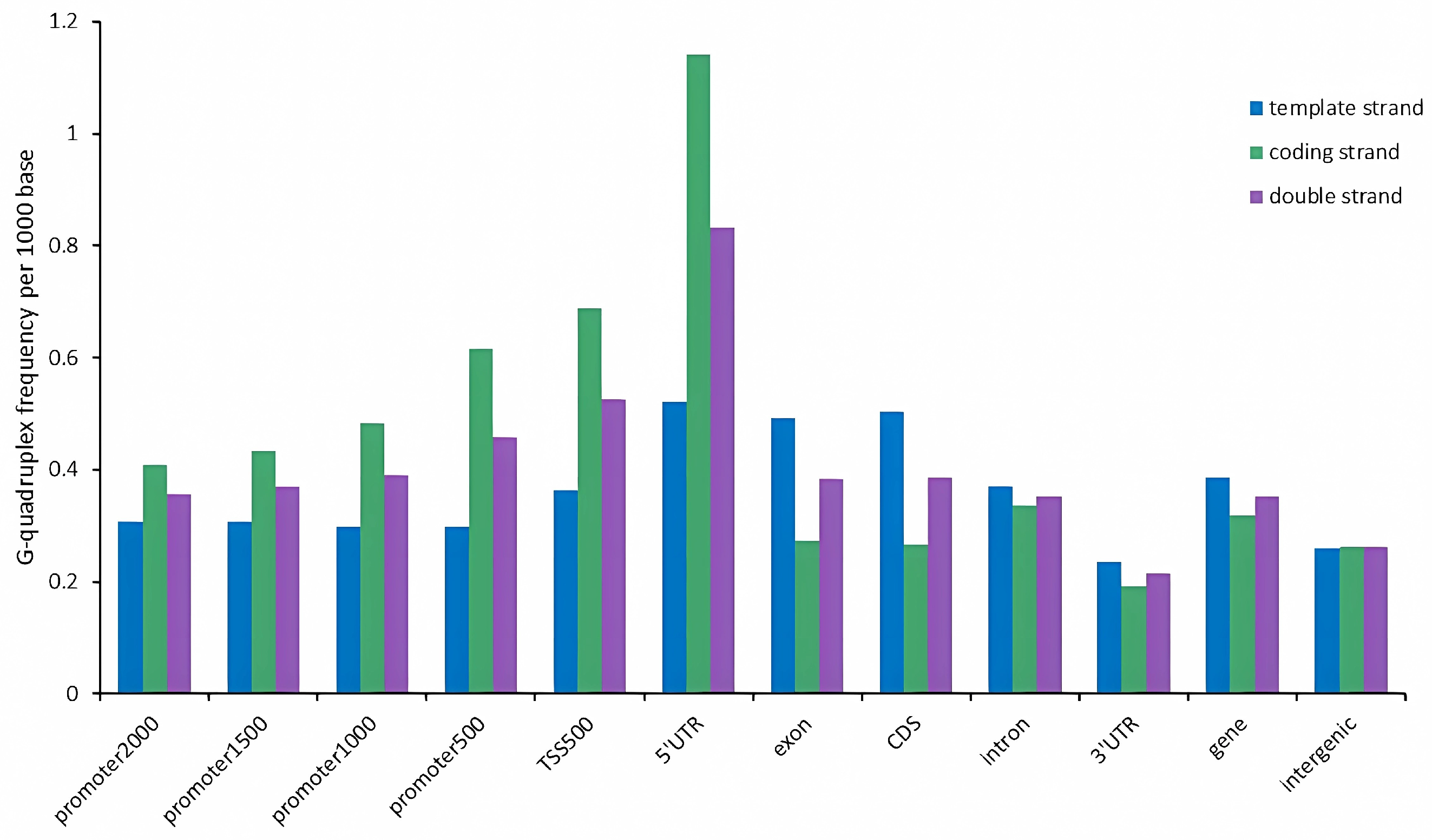
2.3. G-Quadruplex of Feature Regions in Tobacco Genome
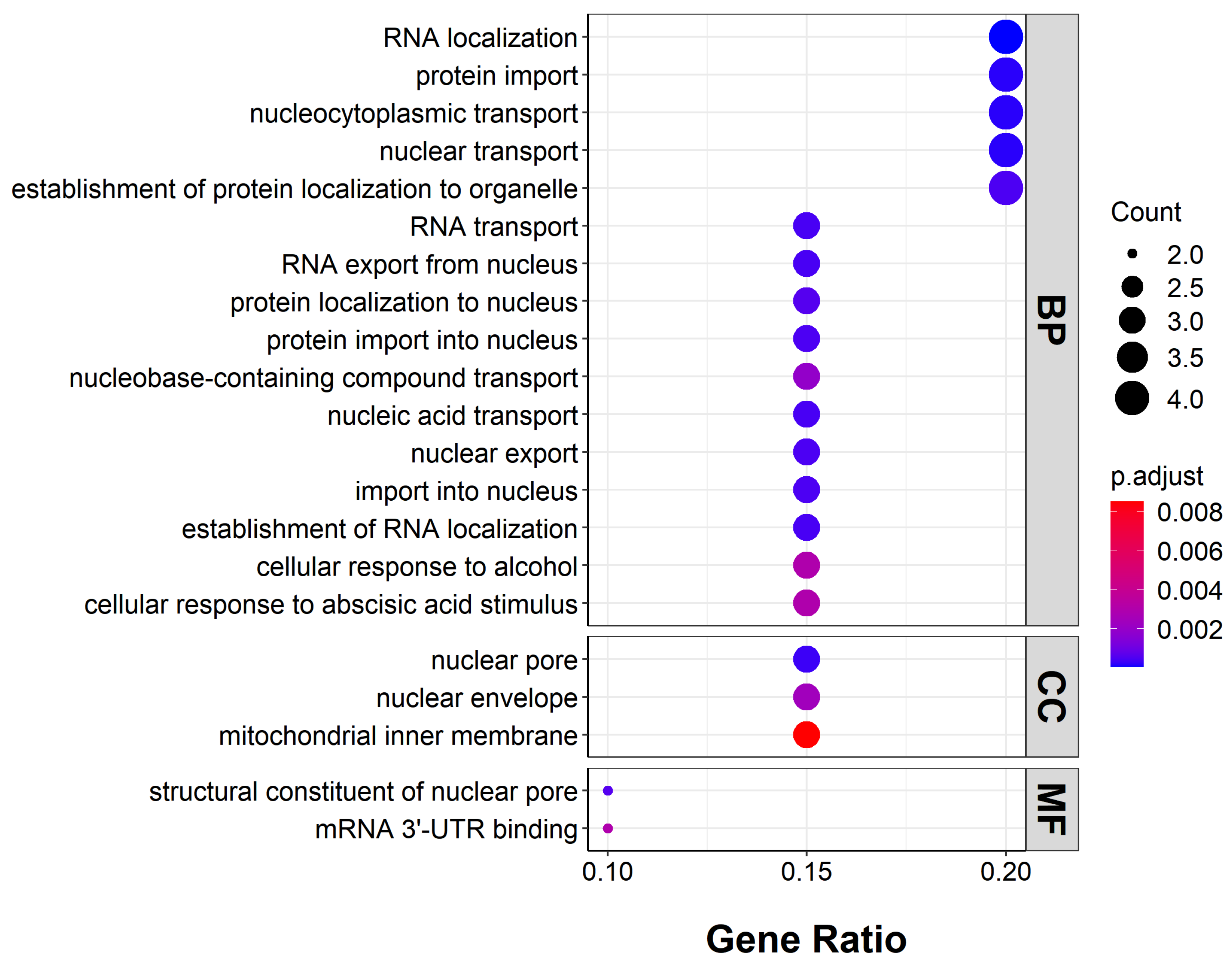
2.4. Functions of Genes with Highly Enriched G-Quadruplexes in Promoters
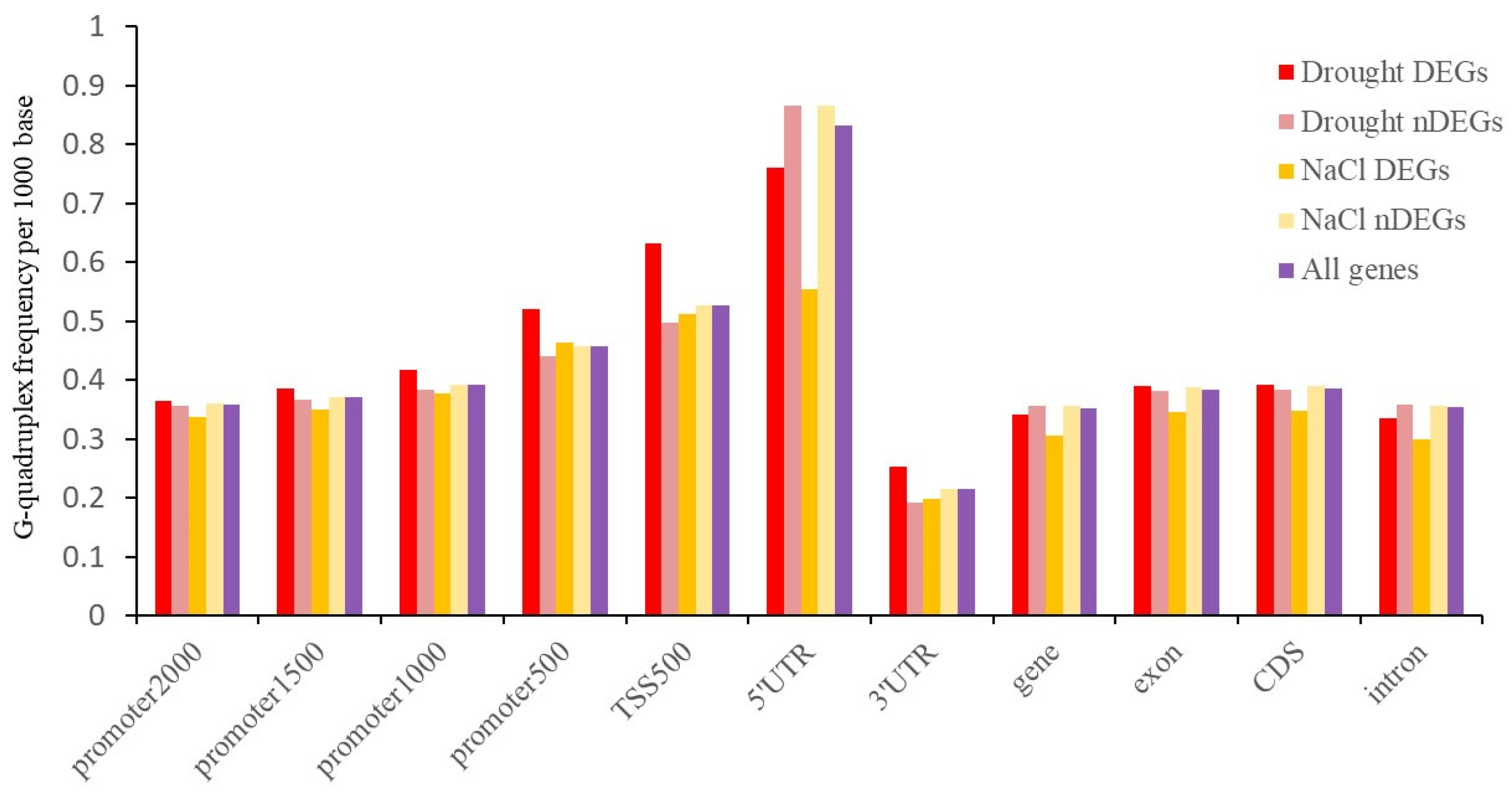
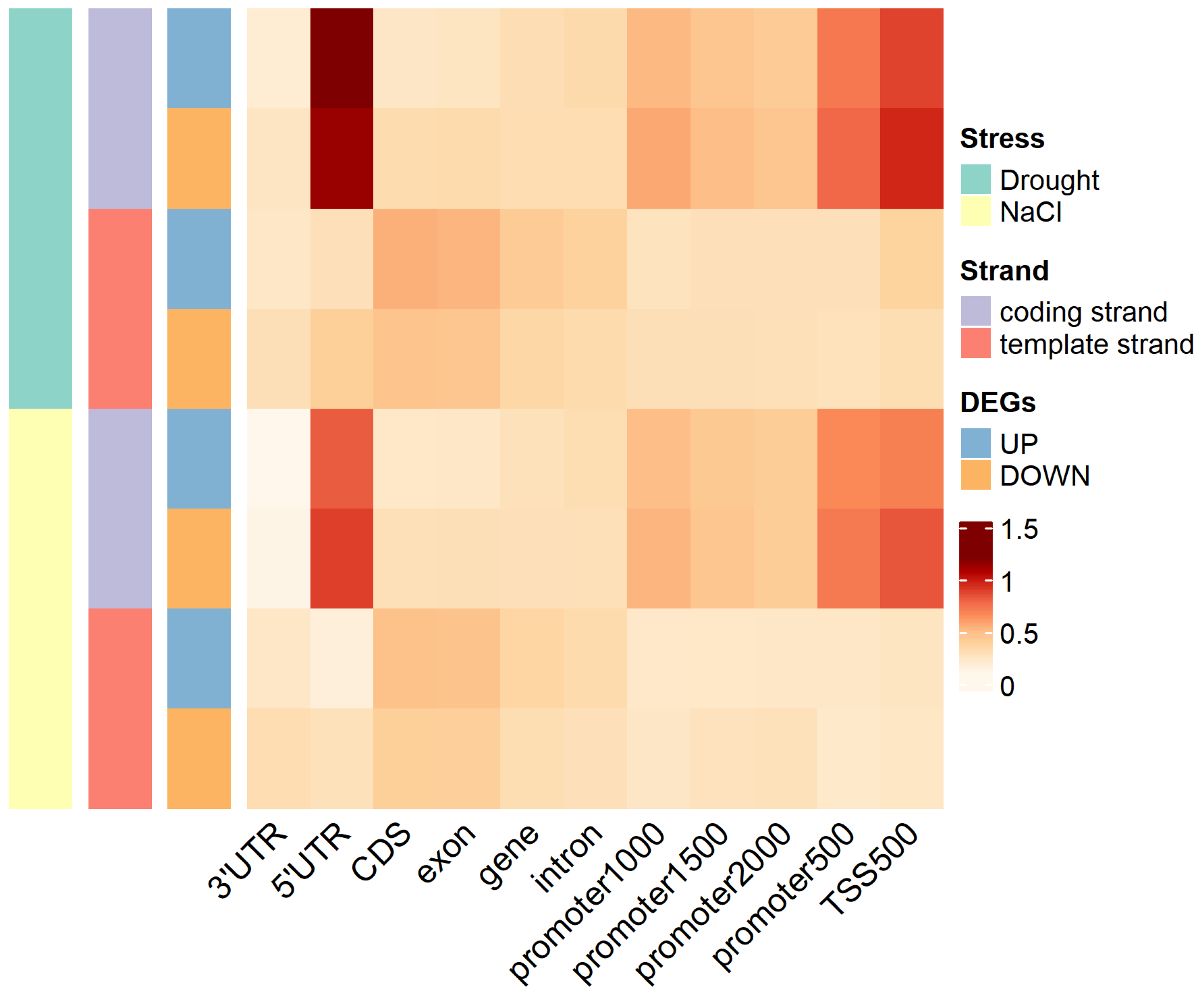
2.5. G-Quadruplex of DEGs under Abiotic Stress
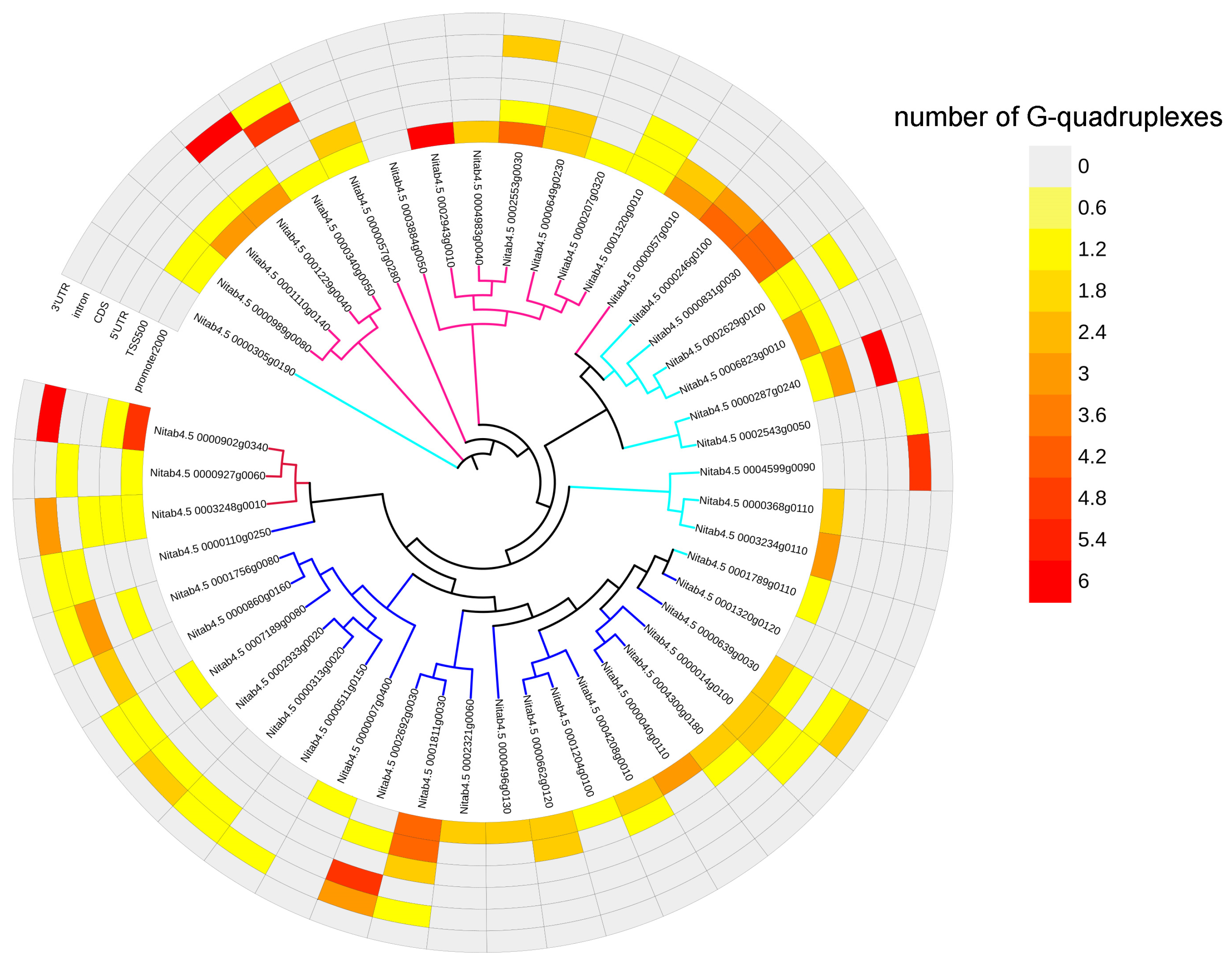
2.6. G-Quadruplexes in Transcription Factor Gene Family
3. Discussion
3.1. Tobacco Organelle Genome Has Higher G-Quadruplex Density than Nuclear Genome
3.2. G-Quadruplex Was Enriched in the Coding Strand of Upstream Regulatory Region
3.3. SSR Provided Conditions for G-Quadruplex Formation
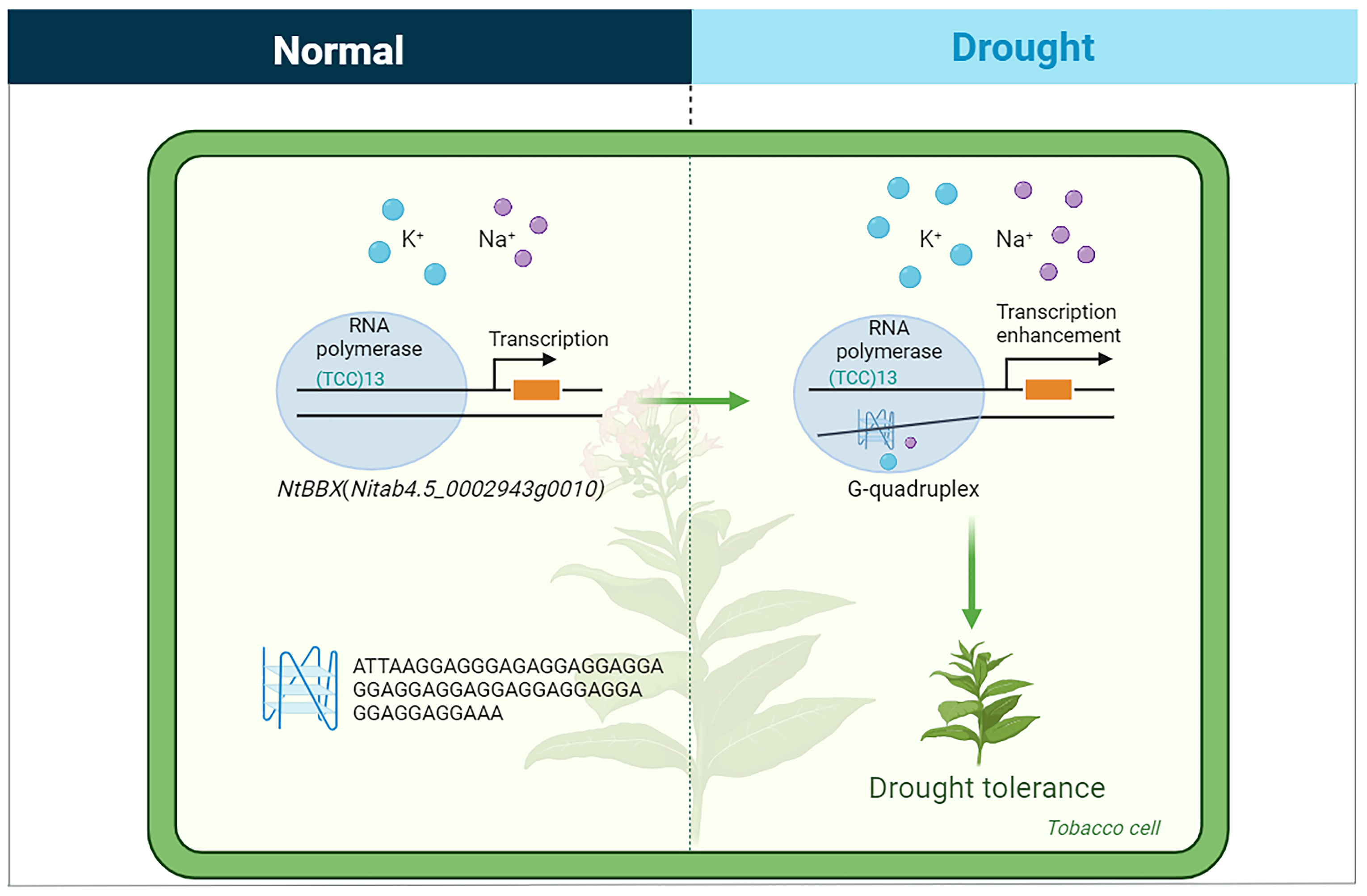
3.4. G-Quadruplex May Be Involved in the Response of Abiotic Stress in Tobacco
4. Materials and Methods
4.1. In Silico Identification and Characterization of G-Quadruplexes in Tobacco Genome
4.2. Number Level of G-Quadruplexes in Promoter and Functional Enrichment Analysis
4.3. G-Quadruplex Analysis of DEGs under Abiotic Stresses
4.4. G-Quadruplex Analysis of Transcription Factor Genes Responding to Drought Stress
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef]
- Moon, J.; Han, J.H.; Kim, D.Y.; Jung, M.J.; Kim, S.K. Effects of deficient of the Hoogsteen base-pairs on the G-quadruplex stabilization and binding mode of a cationic porphyrin. Biochem. Biophys. Rep. 2015, 2, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, D.; Mirihana Arachchilage, G.; Basu, S. Metal Cations in G-Quadruplex Folding and Stability. Front. Chem. 2016, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.; Mouawad, L. Comprehensive analysis of intramolecular G-quadruplex structures: Furthering the understanding of their formalism. Nucleic Acids Res. 2024. [Google Scholar] [CrossRef] [PubMed]
- Yadav, V.; Hemansi; Kim, N.; Tuteja, N.; Yadav, P. G Quadruplex in Plants: A Ubiquitous Regulatory Element and Its Biological Relevance. Front. Plant Sci. 2017, 8, 269762. [Google Scholar] [CrossRef] [PubMed]
- Sparks, M.A.; Singh, S.P.; Burgers, P.M.; Galletto, R. Complementary roles of Pif1 helicase and single stranded DNA binding proteins in stimulating DNA replication through G-quadruplexes. Nucleic Acids Res. 2019, 47, 8595–8605. [Google Scholar] [CrossRef] [PubMed]
- Kopec, P.M.; Karlowski, W.M. Sequence Dynamics of Pre-mRNA G-Quadruplexes in Plants. Front. Plant Sci. 2019, 10, 812. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.Q.; Zhang, M.L.; Song, C.P. A comprehensive evaluation of a typical plant telomeric G-quadruplex (G4) DNA reveals the dynamics of G4 formation, rearrangement, and unfolding. J. Biol. Chem. 2020, 295, 5461–5469. [Google Scholar] [CrossRef]
- Mullen, M.A.; Olson, K.J.; Dallaire, P.; Major, F.; Assmann, S.M.; Bevilacqua, P.C. RNA G-Quadruplexes in the model plant species Arabidopsis thaliana: Prevalence and possible functional roles. Nucleic Acids Res. 2010, 38, 8149–8163. [Google Scholar] [CrossRef]
- Tokan, V.; Puterova, J.; Lexa, M.; Kejnovsky, E. Quadruplex DNA in long terminal repeats in maize LTR retrotransposons inhibits the expression of a reporter gene in yeast. BMC Genom. 2018, 19, 184. [Google Scholar] [CrossRef]
- Vinyard, W.A.; Fleming, A.M.; Ma, J.; Burrows, C.J. Characterization of G-Quadruplexes in Chlamydomonas reinhardtii and the Effects of Polyamine and Magnesium Cations on Structure and Stability. Biochemistry 2018, 57, 6551–6561. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Tao, S.; Zhang, P.; Sperti, F.R.; Liu, G.; Cheng, X.; Zhang, T.; Yu, H.; Wang, X.E.; Chen, C.; et al. Epigenomic features of DNA G-quadruplexes and their roles in regulating rice gene transcription. Plant Physiol. 2022, 188, 1632–1648. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Kishchenko, O.; Stepanenko, A.; Chen, G.; Wang, W.; Zhou, J.; Pan, C.; Borisjuk, N. The Dynamics of NO3− and NH4+ Uptake in Duckweed Are Coordinated with the Expression of Major Nitrogen Assimilation Genes. Plants 2021, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Volna, A.; Bartas, M.; Karlicky, V.; Nezval, J.; Kundratova, K.; Pecinka, P.; Spunda, V.; Cerven, J. G-Quadruplex in Gene Encoding Large Subunit of Plant RNA Polymerase II: A Billion-Year-Old Story. Int. J. Mol. Sci. 2021, 22, 7381. [Google Scholar] [CrossRef] [PubMed]
- Dobrovolna, M.; Bohalova, N.; Peska, V.; Wang, J.; Luo, Y.; Bartas, M.; Volna, A.; Mergny, J.L.; Brazda, V. The Newly Sequenced Genome of Pisum sativum Is Replete with Potential G-Quadruplex-Forming Sequences-Implications for Evolution and Biological Regulation. Int. J. Mol. Sci. 2022, 23, 8482. [Google Scholar] [CrossRef] [PubMed]
- Cagirici, H.B.; Budak, H.; Sen, T.Z. Genome-wide discovery of G-quadruplexes in barley. Sci. Rep. 2021, 11, 7876. [Google Scholar] [CrossRef] [PubMed]
- Kwok, C.K.; Ding, Y.; Shahid, S.; Assmann, S.M.; Bevilacqua, P.C. A stable RNA G-quadruplex within the 5′-UTR of Arabidopsis thaliana ATR mRNA inhibits translation. Biochem. J. 2015, 467, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Yi, C.; Zhang, Z.; Su, H.; Liu, C.; Huang, Y.; Li, W.; Hu, X.; Liu, C.; Birchler, J.A.; et al. Non-B-form DNA tends to form in centromeric regions and has undergone changes in polyploid oat subgenomes. Proc. Natl. Acad. Sci. USA 2023, 120, e2211683120. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Aggarwal, J.; Thakkar, B. Genome-wide discovery of G-quadruplex forming sequences and their functional relevance in plants. Sci. Rep. 2016, 6, 28211. [Google Scholar] [CrossRef]
- Doluca, O. G-Quadruplex enrichment analysis reveals their role as intronic regulatory elements in plants. Turk. J. Bot. 2019, 43, 151–166. [Google Scholar] [CrossRef]
- Yang, X.; Yu, H.; Duncan, S.; Zhang, Y.; Cheema, J.; Liu, H.; Benjamin Miller, J.; Zhang, J.; Kwok, C.K.; Zhang, H.; et al. RNA G-quadruplex structure contributes to cold adaptation in plants. Nat. Commun. 2022, 13, 6224. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Yang, M.; Wu, Y.; Jin, S.; Hou, J.; Mao, Y.; Liu, W.; Shen, Y.; Wu, L. Flower Bud Transcriptome Analysis of Sapium sebiferum (Linn.) Roxb. and Primary Investigation of Drought Induced Flowering: Pathway Construction and G-Quadruplex Prediction Based on Transcriptome. PLoS ONE 2015, 10, e0118479. [Google Scholar] [CrossRef]
- Andorf, C.M.; Kopylov, M.; Dobbs, D.; Koch, K.E.; Stroupe, M.E.; Lawrence, C.J.; Bass, H.W. G-Quadruplex (G4) Motifs in the Maize (Zea mays L.) Genome Are Enriched at Specific Locations in Thousands of Genes Coupled to Energy Status, Hypoxia, Low Sugar, and Nutrient Deprivation. J. Genet. Genom. 2014, 41, 627–647. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Cho, H.S.; Nam, H.; Jo, H.; Yoon, J.; Park, C.; Dang, T.V.T.; Kim, E.; Jeong, J.; Park, S.; et al. Translational control of phloem development by RNA G-quadruplex-JULGI determines plant sink strength. Nat. Plants 2018, 4, 376–390. [Google Scholar] [CrossRef]
- Yang, X.; Cheema, J.; Zhang, Y.; Deng, H.; Duncan, S.; Umar, M.I.; Zhao, J.; Liu, Q.; Cao, X.; Kwok, C.K.; et al. RNA G-quadruplex structures exist and function in vivo in plants. Genome Biol. 2020, 21, 226. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.; Li, G.; Ding, Z.; Li, W.; Zhu, P.; Lei, W.; Shangguan, D. Potential G-quadruplexes within the Promoter Nuclease Hypersensitive Sites of the Heat-Responsive Genes in Rice. Chembiochem 2022, 23, e202200405. [Google Scholar] [CrossRef]
- Sanclemente, M.A.; Ma, F.; Liu, P.; Della Porta, A.; Singh, J.; Wu, S.; Colquhoun, T.; Johnson, T.; Guan, J.C.; Koch, K.E. Sugar modulation of anaerobic-response networks in maize root tips. Plant Physiol. 2021, 185, 295–317. [Google Scholar] [CrossRef]
- Pečinka, P.; Bohálová, N.; Volná, A.; Kundrátová, K.; Brázda, V.; Bartas, M. Analysis of G-Quadruplex-Forming Sequences in Drought Stress-Responsive Genes, and Synthesis Genes of Phenolic Compounds in Arabidopsis thaliana. Life 2023, 13, 199. [Google Scholar] [CrossRef]
- Huang, R.; Feng, Y.; Gao, Z.; Ahmed, A.; Zhang, W. The Epigenomic Features and Potential Functions of PEG- and PDS-Favorable DNA G-Quadruplexes in Rice. Int. J. Mol. Sci. 2024, 25, 634. [Google Scholar] [CrossRef]
- Cagirici, H.B.; Sen, T.Z. Genome-Wide Discovery of G-Quadruplexes in Wheat: Distribution and Putative Functional Roles. G3 Genes|Genomes|Genet. 2020, 10, 2021–2032. [Google Scholar] [CrossRef]
- Falabella, M.; Kolesar, J.E.; Wallace, C.; de Jesus, D.; Sun, L.; Taguchi, Y.V.; Wang, C.; Wang, T.; Xiang, I.M.; Alder, J.K.; et al. G-quadruplex dynamics contribute to regulation of mitochondrial gene expression. Sci. Rep. 2019, 9, 5605. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhao, N.; Li, S.; Grover, C.E.; Nie, H.; Wendel, J.F.; Hua, J. Plant Mitochondrial Genome Evolution and Cytoplasmic Male Sterility. Crit. Rev. Plant Sci. 2017, 36, 55–69. [Google Scholar] [CrossRef]
- Daniell, H.; Jin, S.; Zhu, X.G.; Gitzendanner, M.A.; Soltis, D.E.; Soltis, P.S. Green giant-a tiny chloroplast genome with mighty power to produce high-value proteins: History and phylogeny. Plant Biotechnol. J. 2021, 19, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Forner, J.; Kleinschmidt, D.; Meyer, E.H.; Fischer, A.; Morbitzer, R.; Lahaye, T.; Schottler, M.A.; Bock, R. Targeted introduction of heritable point mutations into the plant mitochondrial genome. Nat. Plants 2022, 8, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Martin Avila, E.; Gisby, M.F.; Day, A. Seamless editing of the chloroplast genome in plants. BMC Plant Biol. 2016, 16, 168. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.; Miller, K.M.; Forment, J.V.; Bradshaw, C.R.; Nikan, M.; Britton, S.; Oelschlaegel, T.; Xhemalce, B.; Balasubramanian, S.; Jackson, S.P. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012, 8, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Hansel-Hertsch, R.; Beraldi, D.; Lensing, S.V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M.; et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016, 48, 1267–1272. [Google Scholar] [CrossRef]
- Hansel-Hertsch, R.; Simeone, A.; Shea, A.; Hui, W.W.I.; Zyner, K.G.; Marsico, G.; Rueda, O.M.; Bruna, A.; Martin, A.; Zhang, X.; et al. Landscape of G-quadruplex DNA structural regions in breast cancer. Nat. Genet. 2020, 52, 878–883. [Google Scholar] [CrossRef] [PubMed]
- Lago, S.; Nadai, M.; Cernilogar, F.M.; Kazerani, M.; Dominiguez Moreno, H.; Schotta, G.; Richter, S.N. Promoter G-quadruplexes and transcription factors cooperate to shape the cell type-specific transcriptome. Nat. Commun. 2021, 12, 3885. [Google Scholar] [CrossRef]
- Zhang, Y.; Fan, C.; Chen, Y.; Wang, R.R.; Zhang, X.; Han, F.; Hu, Z. Genome evolution during bread wheat formation unveiled by the distribution dynamics of SSR sequences on chromosomes using FISH. BMC Genom. 2021, 22, 55. [Google Scholar] [CrossRef]
- Kaur, S.; Panesar, P.S.; Bera, M.B.; Kaur, V. Simple sequence repeat markers in genetic divergence and marker-assisted selection of rice cultivars: A review. Crit. Rev. Food Sci. Nutr. 2015, 55, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Song, K.; Li, B.; Zhang, X.; Wang, D.; Dong, S.; Yang, L. CRISPR/Cas9 Editing Sites Identification and Multi-Elements Association Analysis in Camellia sinensis. Int. J. Mol. Sci. 2023, 24, 15317. [Google Scholar] [CrossRef]
- Hoang, X.L.T.; Nhi, D.N.H.; Thu, N.B.A.; Thao, N.P.; Tran, L.-S.P. Transcription Factors and Their Roles in Signal Transduction in Plants under Abiotic Stresses. Curr. Genom. 2017, 18, 483–497. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Mo, Z.; Fan, Y.; Li, K.; Yang, M.; Li, D.; Ke, Y.; Zhang, Q.; Wang, F.; Fan, Y.; et al. Genome-wide identification and expression analysis of the bZIP transcription factor family genes in response to abiotic stress in Nicotiana tabacum L. BMC Genom. 2022, 23, 318. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Sharoni, A.M.; Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013, 4, 248. [Google Scholar] [CrossRef]
- Song, K.; Li, B.; Wu, H.; Sha, Y.; Qin, L.; Chen, X.; Liu, Y.; Tang, H.; Yang, L. The Function of BBX Gene Family under Multiple Stresses in Nicotiana tabacum. Genes 2022, 13, 1841. [Google Scholar] [CrossRef]
- Bai, G.; Yang, D.H.; Cao, P.; Yao, H.; Zhang, Y.; Chen, X.; Xiao, B.; Li, F.; Wang, Z.Y.; Yang, J.; et al. Genome-Wide Identification, Gene Structure and Expression Analysis of the MADS-Box Gene Family Indicate Their Function in the Development of Tobacco (Nicotiana tabacum L.). Int. J. Mol. Sci. 2019, 20, 5043. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic stress responses in plants. Nat. Rev. Genet. 2021, 23, 104–119. [Google Scholar] [CrossRef]
- Fernandez-Pozo, N.; Menda, N.; Edwards, J.D.; Saha, S.; Tecle, I.Y.; Strickler, S.R.; Bombarely, A.; Fisher-York, T.; Pujar, A.; Foerster, H.; et al. The Sol Genomics Network (SGN)—From genotype to phenotype to breeding. Nucleic Acids Res. 2015, 43, D1036–D1041. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- Bedrat, A.; Lacroix, L.; Mergny, J.L. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016, 44, 1746–1759. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Thiel, T.; Munch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernandez-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Katz, K.; Shutov, O.; Lapoint, R.; Kimelman, M.; Brister, J.R.; O‘Sullivan, C. The Sequence Read Archive: A decade more of explosive growth. Nucleic Acids Res. 2022, 50, D387–D390. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
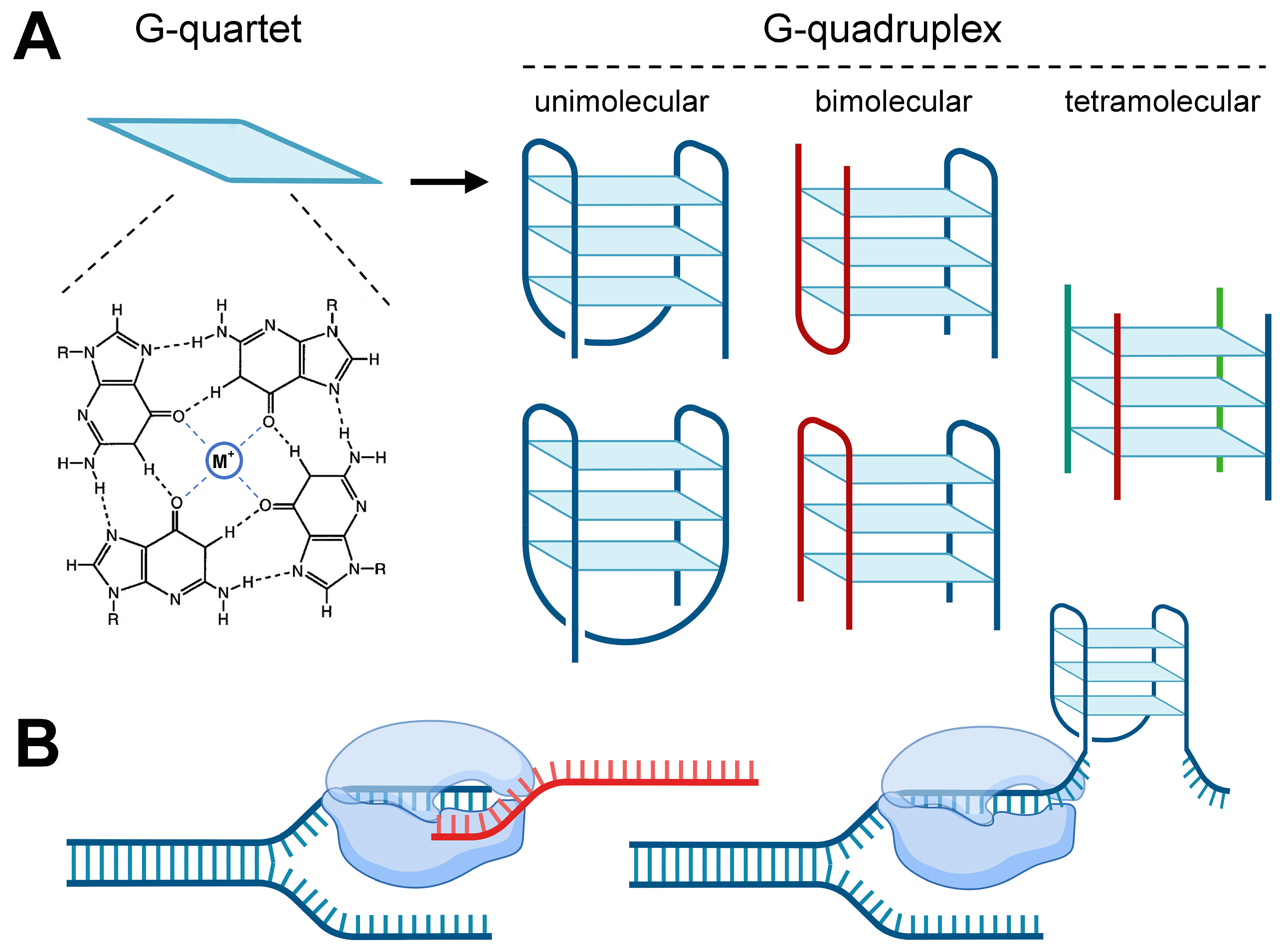
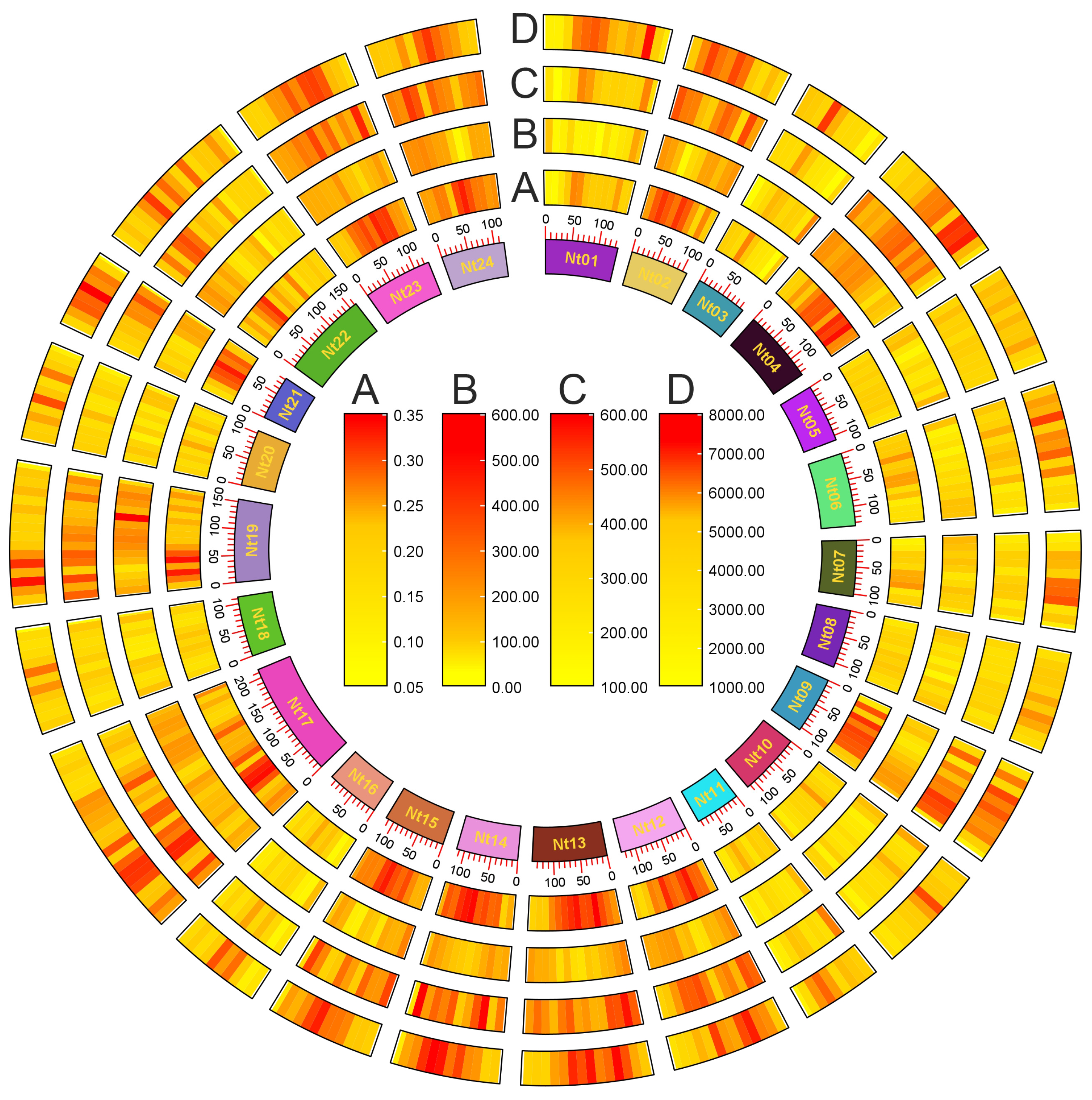
| Chromosome | Length (bp) | GC Content (%) | G-Quadruplex Number | G-Quadruplex Density (per kbp) |
|---|---|---|---|---|
| Nt01 | 135,559,120 | 39.5 | 69,547 | 0.5 |
| Nt02 | 109,624,155 | 38.7 | 60,928 | 0.6 |
| Nt03 | 97,104,660 | 39.1 | 43,104 | 0.4 |
| Nt04 | 136,037,944 | 38.8 | 75,885 | 0.6 |
| Nt05 | 109,337,480 | 39.3 | 53,838 | 0.5 |
| Nt06 | 136,518,381 | 39.3 | 70,455 | 0.5 |
| Nt07 | 105,049,242 | 39.2 | 51,317 | 0.5 |
| Nt08 | 108,393,918 | 39.4 | 52,757 | 0.5 |
| Nt09 | 106,147,314 | 38.8 | 57,485 | 0.5 |
| Nt10 | 116,194,611 | 39.1 | 55,884 | 0.5 |
| Nt11 | 84,914,900 | 39.2 | 42,881 | 0.5 |
| Nt12 | 127,111,110 | 38.8 | 71,171 | 0.6 |
| Nt13 | 139,740,185 | 38.8 | 82,542 | 0.6 |
| Nt14 | 115,579,444 | 38.8 | 69,329 | 0.6 |
| Nt15 | 115,424,069 | 39 | 66,956 | 0.6 |
| Nt16 | 99,759,613 | 39.2 | 45,931 | 0.5 |
| Nt17 | 215,930,317 | 38.8 | 116,804 | 0.5 |
| Nt18 | 113,077,399 | 39 | 51,195 | 0.5 |
| Nt19 | 155,028,365 | 38.6 | 81,044 | 0.5 |
| Nt20 | 105,109,812 | 39.2 | 52,074 | 0.5 |
| Nt21 | 82,751,733 | 39 | 47,122 | 0.6 |
| Nt22 | 163,185,734 | 39 | 85,914 | 0.5 |
| Nt23 | 128,150,892 | 38.7 | 71,924 | 0.6 |
| Nt24 | 118,540,604 | 38.7 | 66,729 | 0.6 |
| mitochondrion | 430,597 | 45 | 705 | 1.6 |
| chloroplast | 155,943 | 37.8 | 148 | 0.9 |
| Sequence | Number | Positive Strand | Negative Strand | Length (bp) | ABS_Score |
|---|---|---|---|---|---|
| GGGGGTGTGTACAGACTCCGGAGGGG | 1302 | 635 | 667 | 26 | 1.423077 |
| GGGGGCCTCGGGTGTGTTTCGGATG | 605 | 294 | 311 | 25 | 1.200000 |
| GGGGTGTGTACAGACTCCGGAGGGG | 587 | 306 | 281 | 25 | 1.320000 |
| CGGGGGGTTGACTTTTTGATATCGGGGT | 599 | 304 | 295 | 28 | 1.357143 |
| CTGGGGGTGTACAGACTCCGGAGGGGCT | 575 | 283 | 292 | 28 | 1.214286 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, K.; Li, B.; Li, H.; Zhang, R.; Zhang, X.; Luan, R.; Liu, Y.; Yang, L. The Characterization of G-Quadruplexes in Tobacco Genome and Their Function under Abiotic Stress. Int. J. Mol. Sci. 2024, 25, 4331. https://doi.org/10.3390/ijms25084331
Song K, Li B, Li H, Zhang R, Zhang X, Luan R, Liu Y, Yang L. The Characterization of G-Quadruplexes in Tobacco Genome and Their Function under Abiotic Stress. International Journal of Molecular Sciences. 2024; 25(8):4331. https://doi.org/10.3390/ijms25084331
Chicago/Turabian StyleSong, Kangkang, Bin Li, Haozhen Li, Rui Zhang, Xiaohua Zhang, Ruiwei Luan, Ying Liu, and Long Yang. 2024. "The Characterization of G-Quadruplexes in Tobacco Genome and Their Function under Abiotic Stress" International Journal of Molecular Sciences 25, no. 8: 4331. https://doi.org/10.3390/ijms25084331
APA StyleSong, K., Li, B., Li, H., Zhang, R., Zhang, X., Luan, R., Liu, Y., & Yang, L. (2024). The Characterization of G-Quadruplexes in Tobacco Genome and Their Function under Abiotic Stress. International Journal of Molecular Sciences, 25(8), 4331. https://doi.org/10.3390/ijms25084331





