Molecular Mechanism of Resistance to Alternaria alternata Apple Pathotype in Apple by Alternative Splicing of Transcription Factor MdMYB6-like
Abstract
1. Introduction
2. Results
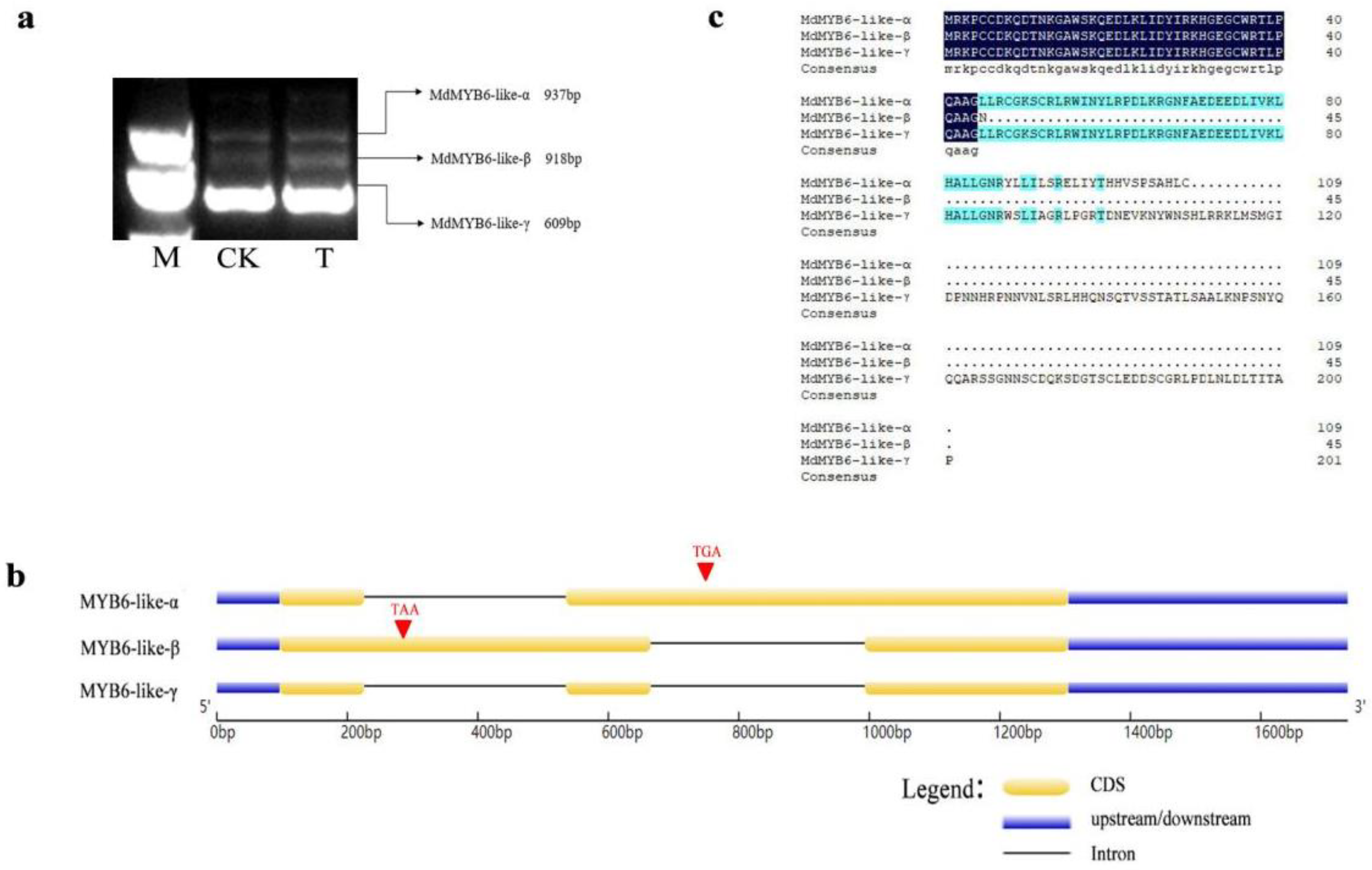
2.1. Three Alternative Spliced Variants of MdMYB6-like in Apple
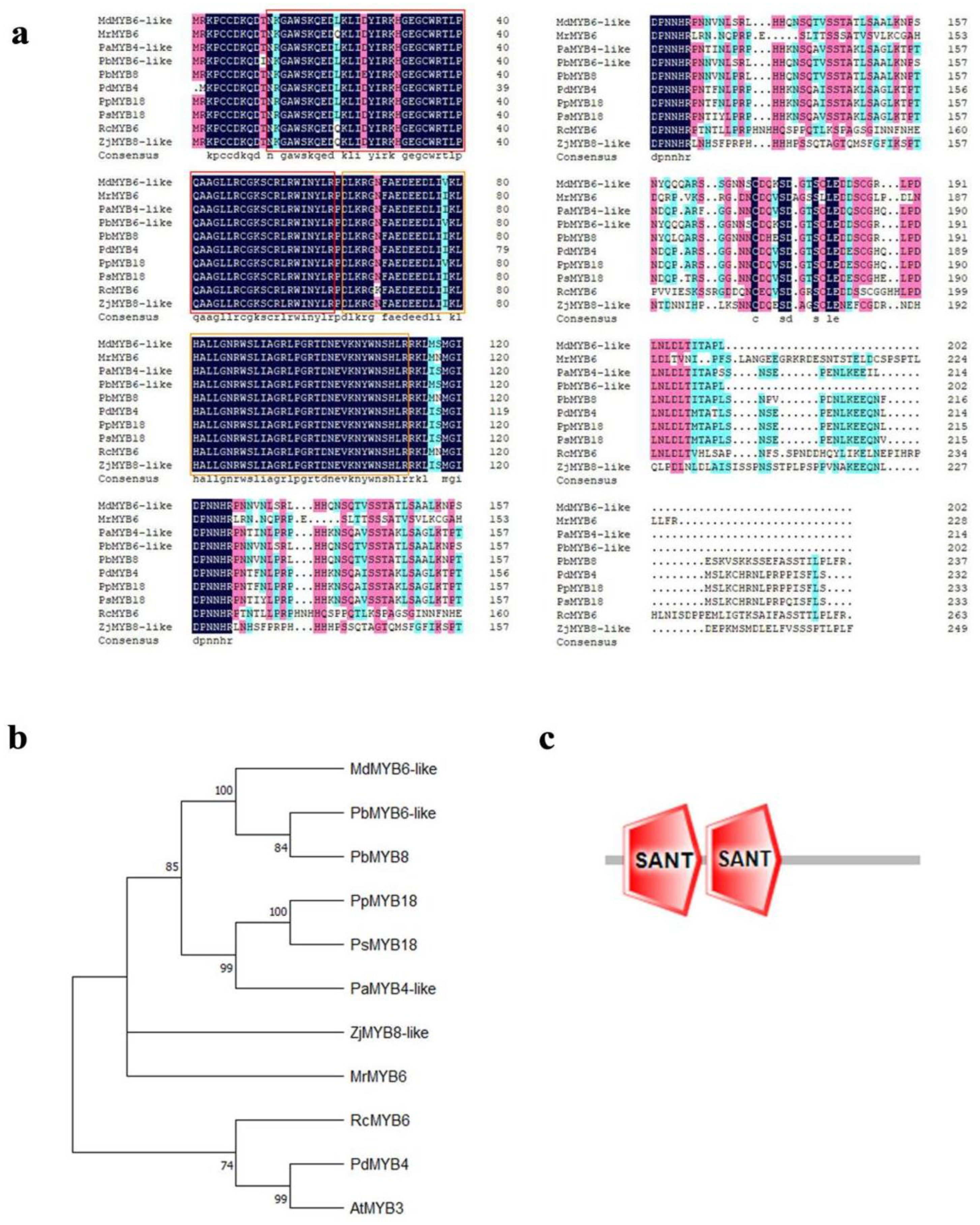
2.2. The Evolutionary Analysis of MdMYB6-like in Apple
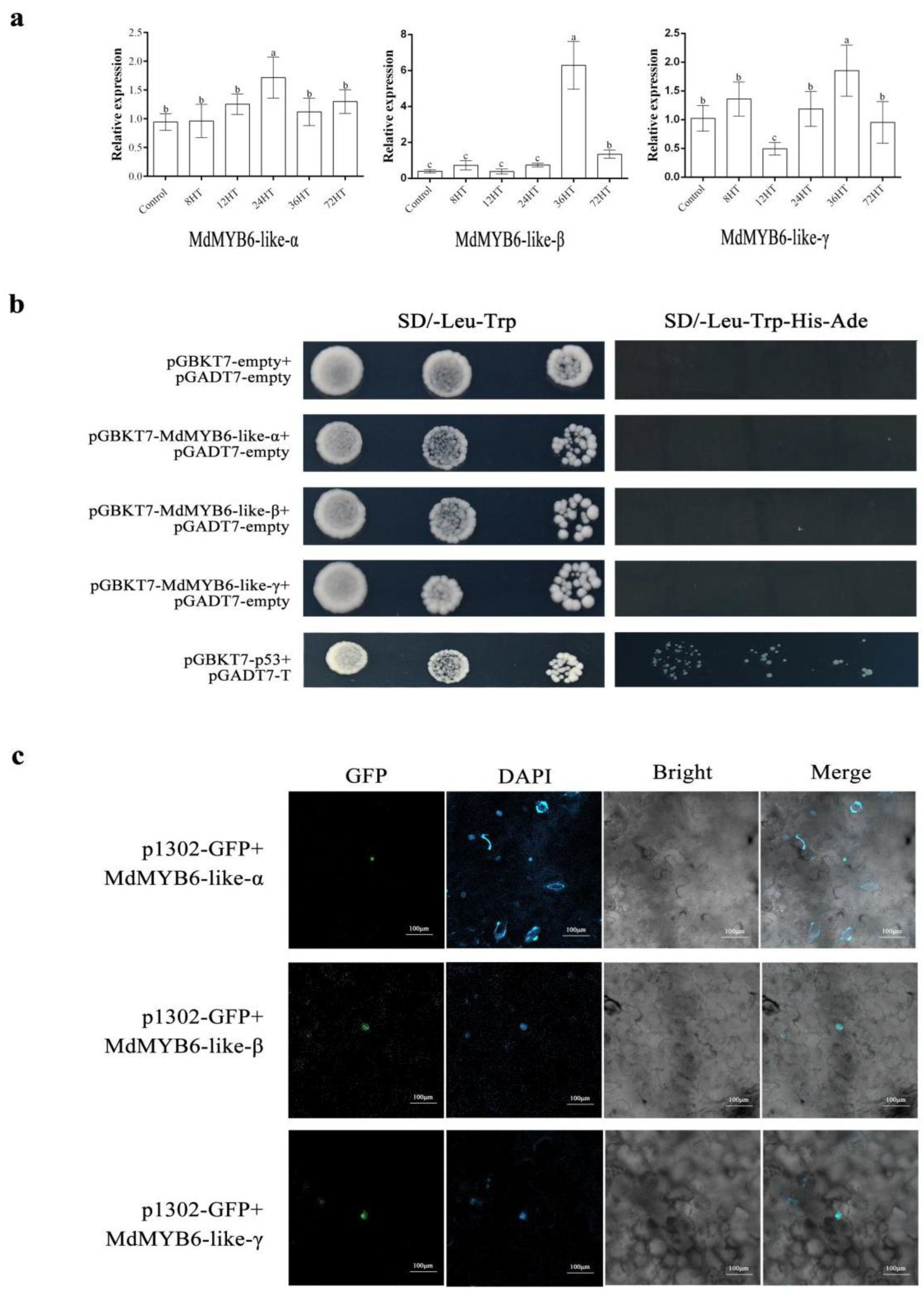
2.3. The Expression of MdMYB6-like in Response to Alternaria alternata Apple Pathotype Infection
2.4. Subcellular Localization of MdMYB6-like
2.5. MdMYB6-like Auto-Activation Activity Identification
2.6. Validation of Interactions between MdMYB6-like Variants
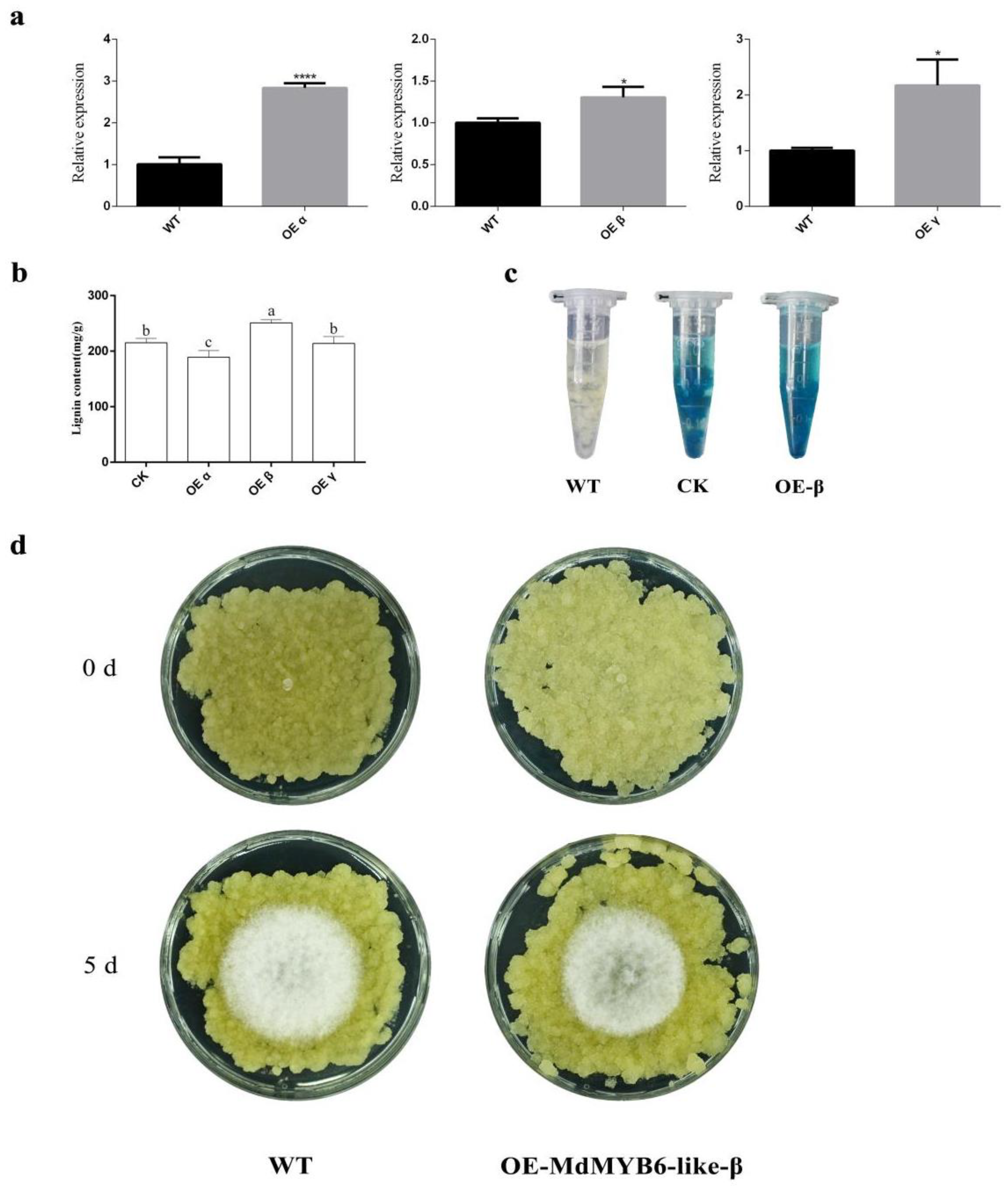
2.7. MdMYB6-like-β Can Impact the Lignin Content of Apple Leaves
2.8. Stable Overexpression of MdMYB6-like-β Isoforms Increases the Resistance of Apple Flesh Callus to Pathogens
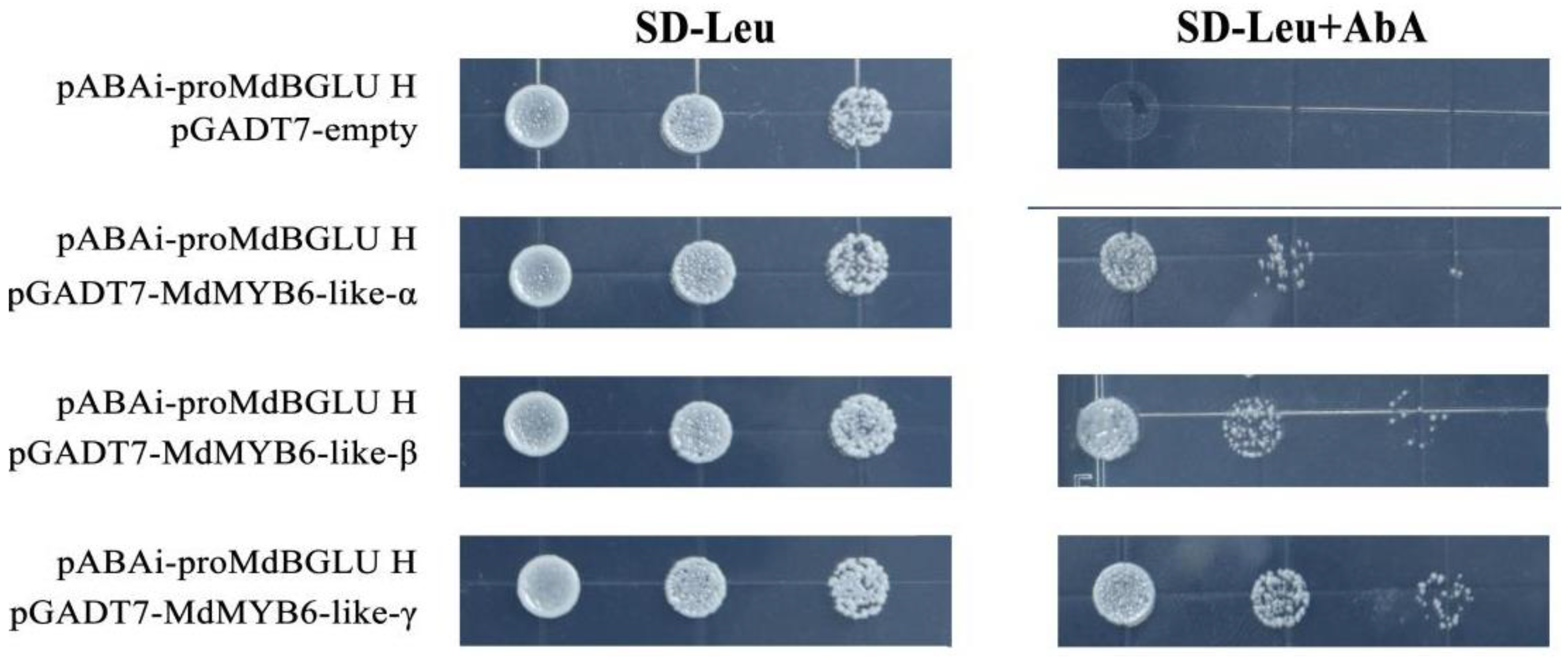
2.9. MdMYB6-like Can Bind to the Promoter of MdBGLU H
3. Discussion
4. Materials and Methods
4.1. Plant and Material
4.2. Pathogen Culture and Inoculation
4.3. Gene Cloning and Sequence Analysis
4.4. Subcellular Localization
4.5. Yeast Two-Hybrid Assays
4.6. Transient Transformation of Apple Leaves
4.7. Stable Transformation of Apple Flesh Callus
4.8. Total Lignin Extraction and Quantification
4.9. Real-Time Quantitative PCR Analysis
4.10. Yeast One-Hybrid Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arnold, M.; Gramza-Michalowska, A. Recent Development on the Chemical Composition and Phenolic Extraction Methods of Apple (Malus domestica)—A Review. Food Bioprocess Technol. 2023, 10, 1–42. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, Y.; He, D.; Li, Y. Identification of Apple Leaf Diseases Based on Deep Convolutional Neural Networks. Symmetry 2017, 10, 11. [Google Scholar] [CrossRef]
- Abe, K.; Iwanami, H.; Kotoda, N.; Moriya, S.; Takahashi, S. Evaluation of apple genotypes and Malus species for resistance to Alternaria blotch caused by Alternaria alternata apple pathotype using detached-leaf method. Plant Breed. 2010, 129, 208–218. [Google Scholar] [CrossRef]
- Qin, L.; Zhao, L.; Wu, C.; Qu, S.; Wang, S. Identification of microRNA transcriptome in apple response to Alternaria alternata infection and evidence that miR390 is negative regulator of defense response. Sci. Hortic. 2021, 289, 110435. [Google Scholar] [CrossRef]
- Saito, A.; Nakazawa, N.; Suzuki, M. Selection of mutants resistant to Alternaria blotch from in vitro-cultured apple shoots irradiated with X- and γ-rays. J. Plant Physiol. 2001, 158, 391–400. [Google Scholar] [CrossRef]
- Li, Y.; Aldwinckle, H.S.; Sutton, T.; Tsuge, T.; Kang, G.; Cong, P.-H.; Cheng, Z.-M. Interactions of Apple and the Alternaria alternate Apple Pathotype. Crit. Rev. Plant Sci. 2012, 32, 141–150. [Google Scholar] [CrossRef]
- Li, Y.; Hirst, P.M.; Wan, Y.; Liu, Y.; Zhou, Q.; Gao, H.; Guo, Y.; Zhao, Z.; Wang, L.; Han, M. Resistance to Marssonina coronaria and Alternaria alternata Apple Pathotype in the Major Apple Cultivars and Rootstocks Used in China. HortScience 2012, 47, 1241–1244. [Google Scholar] [CrossRef]
- Zhu, L.; Ni, W.; Liu, S.; Cai, B.; Xing, H.; Wang, S. Transcriptomics Analysis of Apple Leaves in Response to Alternaria alternata Apple Pathotype Infection. Front. Plant Sci. 2017, 8, 22. [Google Scholar] [CrossRef]
- Liu, K.; Liang, Z.; Yang, A.; Yan, J.; Cong, P.; Han, X.; Zhang, C. Comparative transcriptome analysis of apple cultivars reveals key genes and pathways in response to Alternaria alternata apple pathotype infection. Hortic. Plant J. 2023, in press. [Google Scholar] [CrossRef]
- Singh, K. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002, 5, 430–436. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Zou, B.; Jia, Z.; Tian, S.; Wang, X.; Gou, Z.; Lü, B.; Dong, H. AtMYB44 positively modulates disease resistance to Pseudomonas syringae through the salicylic acid signalling pathway in Arabidopsis. Funct. Plant Biol. 2013, 40, 304–313. [Google Scholar] [CrossRef]
- Seo, P.J.; Park, C.M. MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol. 2010, 186, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Guo, X.; Guo, X.; Zhao, D.; Zhao, W.; Chen, J.; Li, T. PacMYBA, a sweet cherry R2R3-MYB transcription factor, is a positive regulator of salt stress tolerance and pathogen resistance. Plant Physiol. Biochem. 2017, 112, 302–311. [Google Scholar] [CrossRef]
- Qiu, B.; Chen, H.; Zheng, L.; Su, L.; Cui, X.; Ge, F.; Liu, D. An MYB Transcription Factor Modulates Panax notoginseng Resistance Against the Root Rot Pathogen Fusarium solani by Regulating the Jasmonate Acid Signaling Pathway and Photosynthesis. Phytopathology 2022, 112, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Vannini, C.; Campa, M.; Iriti, M.; Genga, A.; Faoro, F.; Carravieri, S.; Rotino, G.L.; Rossoni, M.; Spinardi, A.; Bracale, M. Evaluation of transgenic tomato plants ectopically expressing the rice Osmyb4 gene. Plant Sci. 2007, 173, 231–239. [Google Scholar] [CrossRef]
- Li, J.-B.; Luan, Y.-S.; Yin, Y.-L. SpMYB overexpression in tobacco plants leads to altered abiotic and biotic stress responses. Gene 2014, 547, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Guo, D.; Li, G.; Yang, Y.; Zhang, G.; Li, S.; Liang, Z. The grapevine R2R3-type MYB transcription factor VdMYB1 positively regulates defense responses by activating the stilbene synthase gene 2 (VdSTS2). BMC Plant Biol. 2019, 19, 478. [Google Scholar] [CrossRef] [PubMed]
- Staiger, D.; Brown, J.W.S. Alternative Splicing at the Intersection of Biological Timing, Development, and Stress Responses. Plant Cell 2013, 25, 3640–3656. [Google Scholar] [CrossRef]
- Gao, L.; Wang, W.; Li, H.; Li, H.; Yang, Y.; Zheng, H.; Tao, J. Anthocyanin accumulation in grape berry flesh is associated with an alternative splicing variant of VvMYBA1. Plant Physiol. Biochem. 2023, 195, 1–13. [Google Scholar] [CrossRef]
- Graveley, B.R. Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 2001, 17, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Stamm, S.; Ben-Ari, S.; Rafalska, I.; Tang, Y.; Zhang, Z.; Toiber, D.; Thanaraj, T.A.; Soreq, H. Function of alternative splicing. Gene 2005, 344, 1–20. [Google Scholar] [CrossRef]
- Black, D.L. Mechanisms of Alternative Pre-Messenger RNA Splicing. Annu. Rev. Biochem. 2003, 72, 291–336. [Google Scholar] [CrossRef] [PubMed]
- Syed, N.H.; Kalyna, M.; Marquez, Y.; Barta, A.; Brown, J.W.S. Alternative splicing in plants—Coming of age. Trends Plant Sci. 2012, 17, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Marquez, Y.; Brown, J.W.S.; Simpson, C.; Barta, A.; Kalyna, M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012, 22, 1184–1195. [Google Scholar] [CrossRef] [PubMed]
- Kalyna, M.; Simpson, C.G.; Syed, N.H.; Lewandowska, D.; Marquez, Y.; Kusenda, B.; Marshall, J.; Fuller, J.; Cardle, L.; McNicol, J.; et al. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 2012, 40, 2454–2469. [Google Scholar] [CrossRef]
- Aleynova, O.A.; Kiselev, K.V.; Ogneva, Z.V.; Dubrovina, A.S. The Grapevine Calmodulin-Like Protein Gene CML21 Is Regulated by Alternative Splicing and Involved in Abiotic Stress Response. Int. J. Mol. Sci. 2020, 21, 7939. [Google Scholar] [CrossRef]
- Lim, S.H.; Kim, D.H.; Jung, J.A.; Lee, J.Y. Alternative Splicing of the Basic Helix–Loop–Helix Transcription Factor Gene CmbHLH2 Affects Anthocyanin Biosynthesis in Ray Florets of Chrysanthemum (Chrysanthemum morifolium). Front. Plant Sci. 2021, 12, 669315. [Google Scholar] [CrossRef]
- Li, J.; Yan, X.; Ahmad, M.; Yu, W.; Song, Z.; Ni, J.; Yang, Q.; Teng, Y.; Zhang, H.; Bai, S. Alternative splicing of the dormancy-associated MADS-box transcription factor gene PpDAM1 is associated with flower bud dormancy in ‘Dangshansu’ pear (Pyrus pyrifolia white pear group). Plant Physiol. Biochem. 2021, 166, 1096–1108. [Google Scholar] [CrossRef]
- Wang, H.L.; Zhang, Y.; Wang, T.; Yang, Q.; Li, Z. An Alternative Splicing Variant of PtRD26 Delays Leaf Senescence by Regulating Multiple NAC Transcription Factors in Populus. Plant Cell 2021, 33, 1594–1614. [Google Scholar] [CrossRef]
- Yoon, J.T.; Lee, J.T. Effect of Calcium on the Apple Varieties Resistance to Alternaria Leaf Spot and Mycelial Growth of Alternaria mali Roberts. Korean J. Appl. Entomol. 1987, 26, 239–244. [Google Scholar]
- Heng, W.; Liu, L.; Wang, M.-d.; Jia, B.; Liu, P.; Ye, Z.-f.; Zhu, L.-w. Differentially expressed genes related to the formation of russet fruit skin in a mutant of ‘Dangshansuli’ pear (Pyrus bretchnederi Rehd.) determined by suppression subtractive hybridization. Euphytica 2013, 196, 285–297. [Google Scholar] [CrossRef]
- Yu, J.; Li, Y.; Ishizuka, T.; Guenther, M.G.; Lazar, M.A. A SANT motif in the SMRT corepressor interprets the histone code and promotes histone deacetylation. Embo J. 2003, 22, 3403–3410. [Google Scholar] [CrossRef] [PubMed]
- Ninkuu, V.; Yan, J.; Fu, Z.; Yang, T.; Ziemah, J.; Ullrich, M.S.; Kuhnert, N.; Zeng, H. Lignin and Its Pathway-Associated Phytoalexins Modulate Plant Defense against Fungi. J. Fungi 2022, 9, 52. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Xu, L.; Sun, H.; Yue, Q.; Zhai, H.; Yao, Y. VvVHP1; 2 Is Transcriptionally Activated by VvMYBA1 and Promotes Anthocyanin Accumulation of Grape Berry Skins via Glucose Signal. Front. Plant Sci. 2017, 8, 1811. [Google Scholar] [CrossRef]
- Li, J.; Zhou, H.; Xiong, C.; Peng, Z.; Du, W.; Li, H.; Wang, L.; Ruan, C. Genome-wide analysis R2R3-MYB transcription factors in Xanthoceras sorbifolium Bunge and functional analysis of XsMYB30 in drought and salt stresses tolerance. Ind. Crops Prod. 2022, 178, 114597. [Google Scholar] [CrossRef]
- Hatton, D.; Sablowski, R.; Yung, M.H.; Smith, C.; Schuch, W.; Bevan, M. Two classes of cis sequences contribute to tissue-specific expression of a PAL2 promoter in transgenic tobacco. Plant J. 1995, 7, 859–876. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Jia, H.; Yang, Z.; Wang, S.; Liu, Y.; Ma, H.; Liang, X.; Wang, B.; Zhu, M.; Meng, Y.; et al. Sufficient coumarin accumulation improves apple resistance to Cytospora maliunder high-potassium status. Plant Physiol. 2023, 192, 1396–1419. [Google Scholar] [CrossRef]
- Zhao, X.-Y.; Qi, C.-H.; Jiang, H.; Zheng, P.-F.; Zhong, M.-S.; Zhao, Q.; You, C.-X.; Li, Y.-Y.; Hao, Y.-J. Functional identification of apple on MdHIR4 in biotic stress. Plant Sci. 2019, 283, 396–406. [Google Scholar] [CrossRef]
- Liu, J.; Chen, X.; Liang, X.; Zhou, X.; Yang, F.; Liu, J.; He, S.Y.; Guo, Z. Alternative splicing of rice WRKY62 and WRKY76 transcription factor genes in pathogen defense. Plant Physiol. 2016, 171, 1427–1442. [Google Scholar] [CrossRef]
- Century, K.S.; Holub, E.B.; Staskawicz, B.J. NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc. Natl. Acad. Sci. USA 1995, 92, 6597–6601. [Google Scholar] [CrossRef] [PubMed]
- Miedes, E.; Vanholme, R.; Boerjan, W.; Molina, A. The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 2014, 5, 358. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, P.; Bosneaga, E.; Auer, M. Plant cell walls throughout evolution: Towards a molecular understanding of their design principles. J. Exp. Bot. 2009, 60, 3615–3635. [Google Scholar] [CrossRef]
- Hématy, K.; Cherk, C.; Somerville, S. Host–pathogen warfare at the plant cell wall. Curr. Opin. Plant Biol. 2009, 12, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Heazlewood, J.L.; Zhu, L.; Shan, H.; Chen, S.; Jiang, J.; Gu, C.; Zhou, G.; Chen, Y.; Song, A.; Chen, F. The Heterologous Expression of the Chrysanthemum R2R3-MYB Transcription Factor CmMYB1 Alters Lignin Composition and Represses Flavonoid Synthesis in Arabidopsis thaliana. PLoS ONE 2013, 8, e65680. [Google Scholar]
- Li, C.; Wang, X.; Lu, W.; Liu, R.; Tian, Q.; Sun, Y. A poplar R2R3-MYB transcription factor, PtrMYB152, is involved in regulation of lignin biosynthesis during secondary cell wall formation. Plant Cell Tissue Organ Cult. 2014, 119, 553–563. [Google Scholar] [CrossRef]
- Yang, J.; Xu, J.; Zhang, Y.; Cui, J.; Hu, H.; Xue, J.; Zhu, L. Two R2R3-MYB transcription factors from Chinese cedar (Cryptomeria fortunei Hooibrenk) are involved in the regulation of secondary cell wall formation. Plant Physiol. Biochem. 2023, 201, 107879. [Google Scholar] [CrossRef] [PubMed]
- Treisman, R.; Orkin, S.H.; Maniatis, T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature 1983, 302, 591–596. [Google Scholar] [CrossRef]
- Li, X.; He, G.; Jiang, S.; Yang, C.; Yang, B.; Ming, F. Function of two splicing variants of RcCPR5 in the resistance of Rosa chinensis to powdery mildew. Plant Sci. 2023, 335, 111678. [Google Scholar] [CrossRef]
- Zhou, J.; Zheng, X.; Shen, H. Targeting RNA-Splicing for SMA Treatment. Mol. Cells 2012, 33, 223–228. [Google Scholar] [CrossRef]
- Qiao, D.; Yang, C.; Chen, J.; Guo, Y.; Li, Y.; Niu, S.; Cao, K. Comprehensive identification of the full-length transcripts and alternative splicing related to the secondary metabolism pathways in the tea plant (Camellia sinensis). Sci. Rep. 2019, 9, 2709. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Constabel, C.P. MYB Repressors as Regulators of Phenylpropanoid Metabolism in Plants. Trends Plant Sci. 2019, 24, 275–289. [Google Scholar] [CrossRef]
- Liu, J.; Osbourn, A.; Ma, P. MYB Transcription Factors as Regulators of Phenylpropanoid Metabolism in Plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef] [PubMed]
- Busche, M.; Pucker, B.; Weisshaar, B.; Stracke, R. Three R2R3-MYB transcription factors from banana (Musa acuminata) activate structural anthocyanin biosynthesis genes as part of an MBW complex. BMC Res. Notes 2023, 16, 103. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Hou, Y.; Cao, L.; Zhou, T.; Wang, S.; Hu, K.; Chen, J.; Qu, S. MicroRNA candidate miRcand137 in apple is induced by Botryosphaeria dothidea for impairing host defense. Plant Physiol. 2022, 189, 1814–1832. [Google Scholar] [CrossRef] [PubMed]
- Abdullah-Zawawi, M.R.; Ahmad-Nizammuddin, N.F.; Govender, N.; Harun, S.; Mohd-Assaad, N.; Mohamed-Hussein, Z.A. Comparative genome-wide analysis of WRKY, MADS-box and MYB transcription factor families in Arabidopsis and rice. Sci. Rep. 2021, 11, 19678. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, Y.; Wu, Y.; Cheng, H.; Li, Y.; Zhao, Y.; Li, Y. Transgenic plants from fragmented shoot tips of apple (Malus baccata (L.) Borkhausen) via agrobacterium-mediated transformation. Sci. Hortic. 2011, 128, 450–456. [Google Scholar] [CrossRef]
- Janshekar, H.; Brown, C.; Fiechter, A. Determination of biodegraded lignin by ultraviolet spectrophotometry. Anal. Chim. Acta 1981, 130, 81–91. [Google Scholar] [CrossRef]
- Huis, R.; Hawkins, S.; Neutelings, G. Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.). BMC Plant Biol. 2010, 10, 71. [Google Scholar] [CrossRef]
- Zheng, J.; Liu, F.; Zhu, C.; Li, X.; Dai, X.; Yang, B.; Zou, X.; Ma, Y. Identification, expression, alternative splicing and functional analysis of pepper WRKY gene family in response to biotic and abiotic stresses. PLoS ONE 2019, 14, e0219775. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, X.; Wu, C.; Zhang, L.; Lan, L.; Fu, W.; Wang, S. Molecular Mechanism of Resistance to Alternaria alternata Apple Pathotype in Apple by Alternative Splicing of Transcription Factor MdMYB6-like. Int. J. Mol. Sci. 2024, 25, 4353. https://doi.org/10.3390/ijms25084353
Zeng X, Wu C, Zhang L, Lan L, Fu W, Wang S. Molecular Mechanism of Resistance to Alternaria alternata Apple Pathotype in Apple by Alternative Splicing of Transcription Factor MdMYB6-like. International Journal of Molecular Sciences. 2024; 25(8):4353. https://doi.org/10.3390/ijms25084353
Chicago/Turabian StyleZeng, Xianqi, Chao Wu, Lulu Zhang, Liming Lan, Weihong Fu, and Sanhong Wang. 2024. "Molecular Mechanism of Resistance to Alternaria alternata Apple Pathotype in Apple by Alternative Splicing of Transcription Factor MdMYB6-like" International Journal of Molecular Sciences 25, no. 8: 4353. https://doi.org/10.3390/ijms25084353
APA StyleZeng, X., Wu, C., Zhang, L., Lan, L., Fu, W., & Wang, S. (2024). Molecular Mechanism of Resistance to Alternaria alternata Apple Pathotype in Apple by Alternative Splicing of Transcription Factor MdMYB6-like. International Journal of Molecular Sciences, 25(8), 4353. https://doi.org/10.3390/ijms25084353




