The Inhibition of NS2B/NS3 Protease: A New Therapeutic Opportunity to Treat Dengue and Zika Virus Infection
Abstract
:1. Introduction
2. NS2B/NS3 DENV and ZIKV Protease: Structures and Functions
3. Interaction of Flavivirus NS2B/NS3 with Cellular Proteins
4. Crystal Structures of DENV and ZIKV NS2B/NS3 Protease
5. NS2B/NS3 Protease Inhibitors
Allosteric Inhibitors
6. Orthosteric Inhibitors
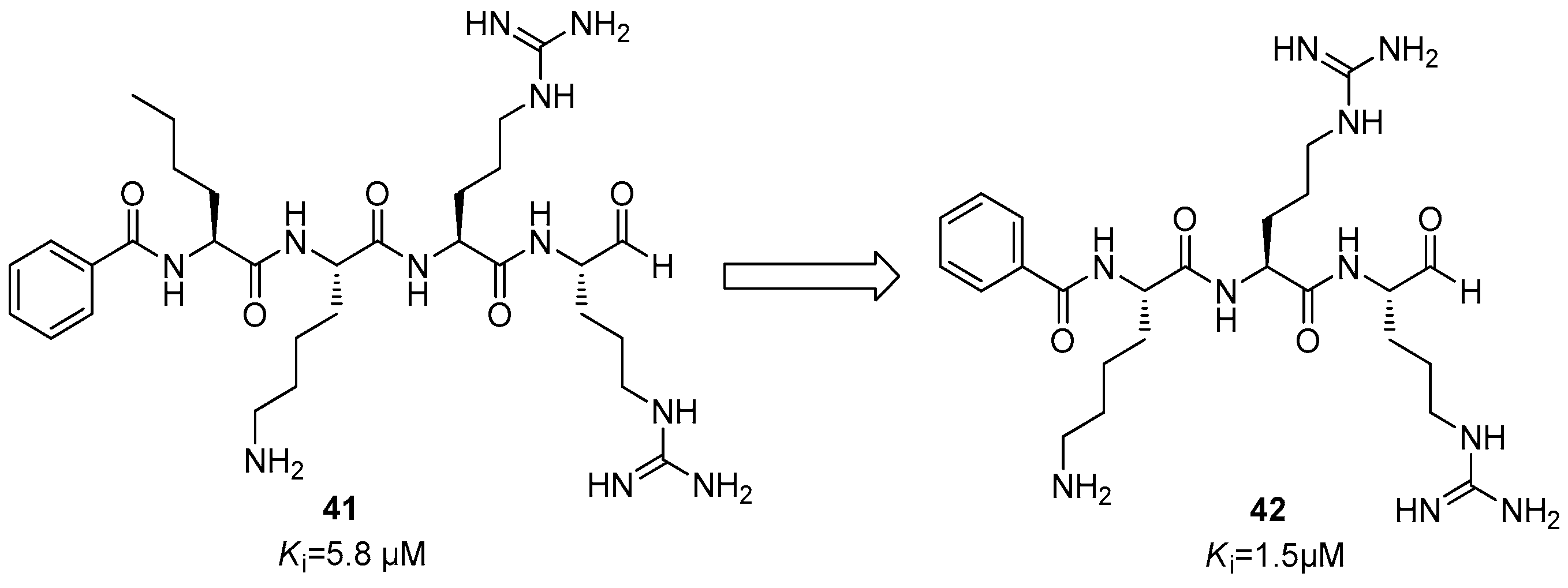
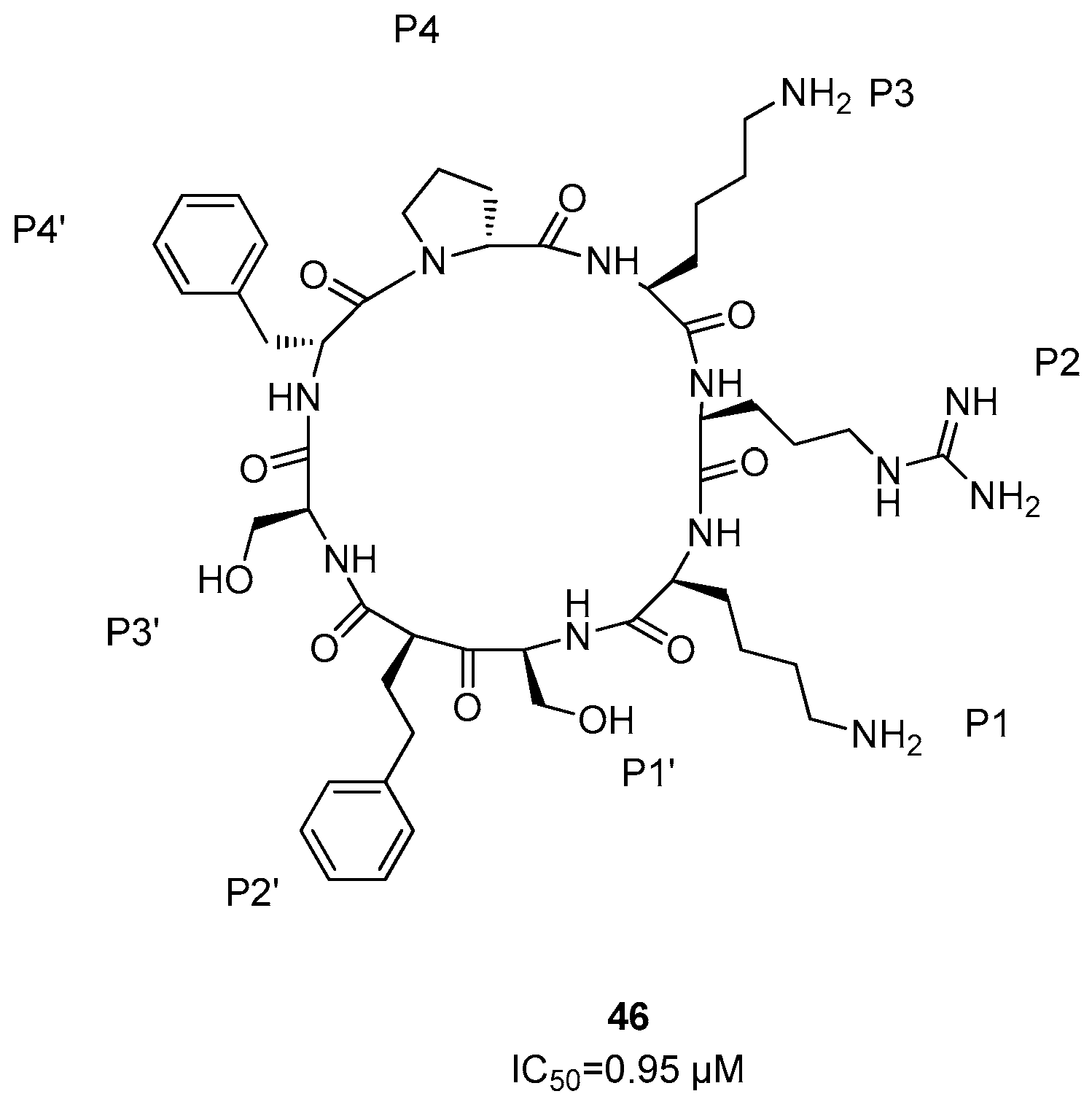
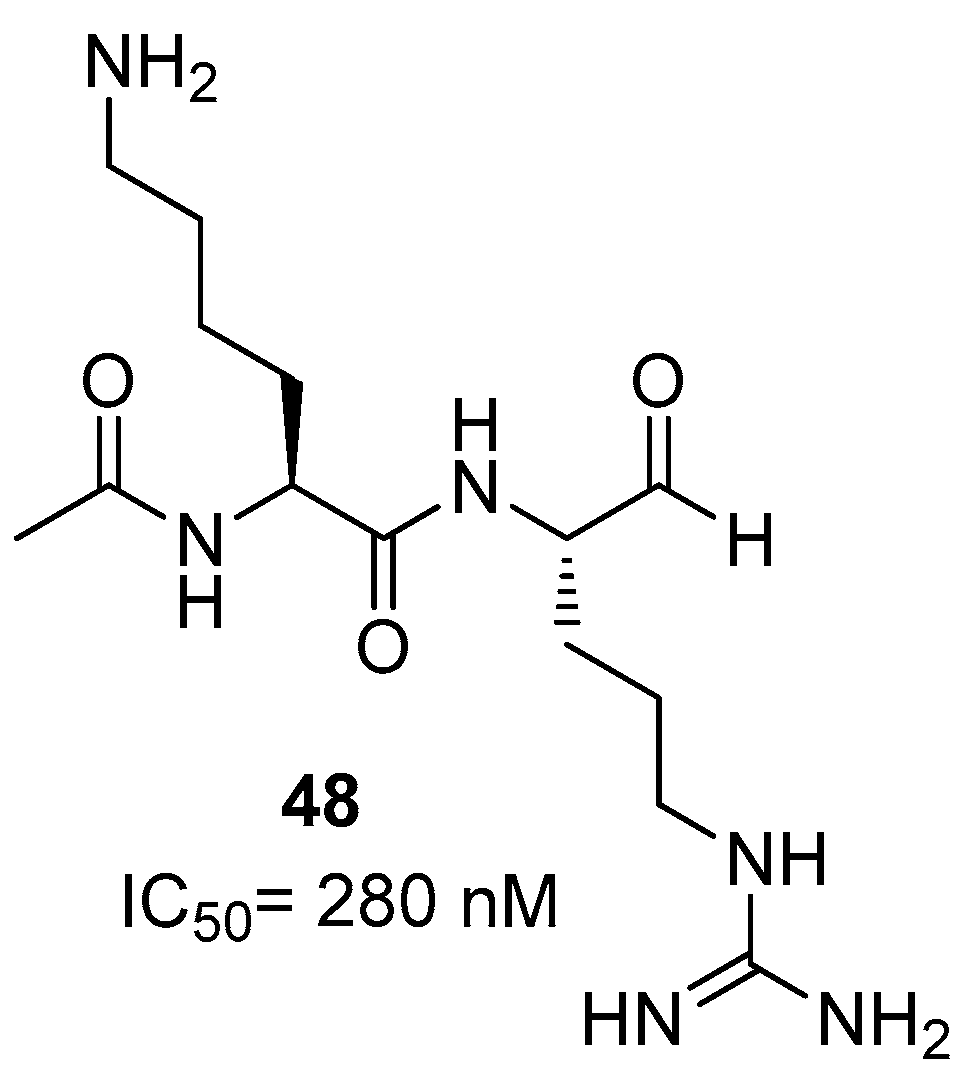
Dengue NS2B/NS3 Protease Inhibitors
7. Zika NS2B/NS3 Protease Inhibitors
8. Pharmacokinetic Properties, Antiviral Activity, and Cytotoxicity Evaluation of DENV and ZIKV Inhibitors
9. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Dengue and Severe Dengue. Available online: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue (accessed on 15 December 2023).
- da Silva-Junior, E.F.; de Araujo-Junior, J.X. Peptide derivatives as inhibitors of NS2B-NS3 protease from Dengue, West Nile, and Zika flaviviruses. Bioorg. Med. Chem. 2019, 27, 3963–3978. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, P.; Sabeena, S.P.; Varma, M.; Arunkumar, G. Current understanding of the pathogenesis of Dengue virus infection. Curr. Microbiol. 2021, 78, 17–32. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Zika Virus. Available online: https://www.who.int/news-room/fact-sheets/detail/zika-virus (accessed on 15 December 2023).
- Bhagat, R.; Kaur, G.; Seth, P. Molecular mechanisms of Zika virus pathogenesis: An update. Indian J. Med. Res. 2021, 154, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, M.; Ghosh, S.; Bell, J.A.; Sherman, W.; Hardy, J.A. Allosteric inhibition of the NS2B-NS3 protease from Dengue virus. ACS Chem. Biol. 2013, 8, 2744–2752. [Google Scholar] [CrossRef] [PubMed]
- Gangopadhyay, A.; Saha, A. Exploring allosteric hits of the NS2B-NS3 protease of DENV2 by structure-guided screening. Comput. Biol. Chem. 2023, 104, 107876. [Google Scholar] [CrossRef]
- Wahaab, A.; Mustafa, B.E.; Hameed, M.; Stevenson, N.J.; Anwar, M.N.; Liu, K.; Wei, J.; Qiu, Y.; Ma, Z. Potential role of flavivirus NS2B-NS3 proteases in viral pathogenesis and anti-flavivirus drug discovery employing animal cells and models: A review. Viruses 2021, 14, 44. [Google Scholar] [CrossRef]
- Noble, C.G.; Seh, C.C.; Chao, A.T.; Shi, P.Y. Ligand-bound structures of the dengue virus protease reveal the active conformation. J. Virol. 2012, 86, 438–446. [Google Scholar] [CrossRef]
- Coluccia, A.; Puxeddu, M.; Nalli, M.; Wei, C.K.; Wu, Y.H.; Mastrangelo, E.; Elamin, T.; Tarantino, D.; Bugert, J.J.; Schreiner, B.; et al. Discovery of Zika virus NS2B/NS3 inhibitors that prevent mice from life-threatening infection and brain damage. ACS Med. Chem. Lett. 2020, 11, 1869–1874. [Google Scholar] [CrossRef]
- Voss, S.; Nitsche, C. Inhibitors of the Zika virus protease NS2B-NS3. Bioorg. Med. Chem. Lett. 2020, 30, 126965. [Google Scholar] [CrossRef]
- Yao, Y.; Huo, T.; Lin, Y.L.; Nie, S.; Wu, F.; Hua, Y.; Wu, J.; Kneubehl, A.R.; Vogt, M.B.; Rico-Hesse, R.; et al. Discovery, X-ray crystallography and antiviral activity of allosteric inhibitors of flavivirus NS2B-NS3 protease. J. Am. Chem. Soc. 2019, 141, 6832–6836. [Google Scholar] [CrossRef]
- Meewan, I.; Shiryaev, S.A.; Kattoula, J.; Huang, C.T.; Lin, V.; Chuang, C.H.; Terskikh, A.V.; Abagyan, R. Allosteric inhibitors of Zika virus NS2B-NS3 protease targeting protease in “super-open” conformation. Viruses 2023, 15, 1106. [Google Scholar] [CrossRef] [PubMed]
- Heaton, N.S.; Perera, R.; Berger, K.L.; Khadka, S.; Lacount, D.J.; Kuhn, R.J.; Randall, G. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 17345–17350. [Google Scholar] [CrossRef] [PubMed]
- Silva, E.M.; Conde, J.N.; Allonso, D.; Ventura, G.T.; Coelho, D.R.; Carneiro, P.H.; Silva, M.L.; Paes, M.V.; Rabelo, K.; Weissmuller, G.; et al. Dengue virus nonstructural 3 protein interacts directly with human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and reduces its glycolytic activity. Sci. Rep. 2019, 9, 2651. [Google Scholar] [CrossRef] [PubMed]
- Bonafe, N.; Gilmore-Hebert, M.; Folk, N.L.; Azodi, M.; Zhou, Y.; Chambers, S.K. Glyceraldehyde-3-phosphate dehydrogenase binds to the AU-Rich 3’ untranslated region of colony-stimulating factor-1 (CSF-1) messenger RNA in human ovarian cancer cells: Possible role in CSF-1 posttranscriptional regulation and tumor phenotype. Cancer Res. 2005, 65, 3762–3771. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, Y.; Yamaji, R.; Irie, K.; Kioka, N.; Murakami, A. Glyceraldehyde-3-phosphate dehydrogenase regulates cyclooxygenase-2 expression by targeting mRNA stability. Arch. Biochem. Biophys. 2012, 528, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Lennemann, N.J.; Coyne, C.B. Dengue and Zika viruses subvert reticulophagy by NS2B3-mediated cleavage of FAM134B. Autophagy 2017, 13, 322–332. [Google Scholar] [CrossRef]
- Li, H.; Saucedo-Cuevas, L.; Yuan, L.; Ross, D.; Johansen, A.; Sands, D.; Stanley, V.; Guemez-Gamboa, A.; Gregor, A.; Evans, T.; et al. Zika virus protease cleavage of host protein septin-2 mediates mitotic defects in neural progenitors. Neuron 2019, 101, 1089–1098. [Google Scholar] [CrossRef] [PubMed]
- Neufeldt, C.J.; Cortese, M.; Acosta, E.G.; Bartenschlager, R. Rewiring cellular networks by members of the Flaviviridae family. Nat. Rev. Microbiol. 2018, 16, 125–142. [Google Scholar] [CrossRef]
- Brecher, M.; Zhang, J.; Li, H. The flavivirus protease as a target for drug discovery. Virol. Sin. 2013, 28, 326–336. [Google Scholar] [CrossRef]
- Phoo, W.W.; Zhang, Z.; Wirawan, M.; Chew, E.J.C.; Chew, A.B.L.; Kouretova, J.; Steinmetzer, T.; Luo, D. Structures of Zika virus NS2B-NS3 protease in complex with peptidomimetic inhibitors. Antivir. Res. 2018, 160, 17–24. [Google Scholar] [CrossRef]
- Shiryaev, S.A.; Farhy, C.; Pinto, A.; Huang, C.T.; Simonetti, N.; Elong Ngono, A.; Dewing, A.; Shresta, S.; Pinkerton, A.B.; Cieplak, P.; et al. Characterization of the Zika virus two-component NS2B-NS3 protease and structure-assisted identification of allosteric small-molecule antagonists. Antivir. Res. 2017, 143, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, Y.; Loh, Y.R.; Phoo, W.W.; Hung, A.W.; Kang, C.; Luo, D. Crystal structure of unlinked NS2B-NS3 protease from Zika virus. Science 2016, 354, 1597–1600. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Ji, X.; Yang, X.; Xie, W.; Yang, K.; Chen, C.; Wu, C.; Chi, H.; Mu, Z.; Wang, Z.; et al. The crystal structure of Zika virus helicase: Basis for antiviral drug design. Protein Cell 2016, 7, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Phoo, W.W.; Li, Y.; Zhang, Z.; Lee, M.Y.; Loh, Y.R.; Tan, Y.B.; Ng, E.Y.; Lescar, J.; Kang, C.; Luo, D. Structure of the NS2B-NS3 protease from Zika virus after self-cleavage. Nat. Commun. 2016, 7, 13410. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Hansen, G.; Nitsche, C.; Klein, C.D.; Zhang, L.; Hilgenfeld, R. Crystal structure of Zika virus NS2B-NS3 protease in complex with a boronate inhibitor. Science 2016, 353, 503–505. [Google Scholar] [CrossRef]
- Aleshin, A.E.; Shiryaev, S.A.; Strongin, A.Y.; Liddington, R.C. Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold. Protein Sci. 2007, 16, 795–806. [Google Scholar] [CrossRef] [PubMed]
- Chandramouli, S.; Joseph, J.S.; Daudenarde, S.; Gatchalian, J.; Cornillez-Ty, C.; Kuhn, P. Serotype-specific structural differences in the protease-cofactor complexes of the dengue virus family. J. Virol. 2010, 84, 3059–3067. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Chen, A.S.; Li, Q.; Kang, C. Expression, purification, and initial structural characterization of nonstructural protein 2B, an integral membrane protein of Dengue-2 virus, in detergent micelles. Protein Expr. Purif. 2011, 80, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Li, Q.; Joy, J.; Chen, A.S.; Ruiz-Carrillo, D.; Hill, J.; Lescar, J.; Kang, C. Lyso-myristoyl phosphatidylcholine micelles sustain the activity of Dengue non-structural (NS) protein 3 protease domain fused with the full-length NS2B. Protein Expr. Purif. 2013, 92, 156–162. [Google Scholar] [CrossRef]
- Liew, L.S.; Lee, M.Y.; Wong, Y.L.; Cheng, J.; Li, Q.; Kang, C. Selection of suitable detergents for obtaining an active dengue protease in its natural form from E. coli. Protein Expr. Purif. 2016, 121, 141–148. [Google Scholar] [CrossRef]
- Ng, E.Y.; Loh, Y.R.; Li, Y.; Li, Q.; Kang, C. Expression, purification of Zika virus membrane protein-NS2B in detergent micelles for NMR studies. Protein Expr. Purif. 2019, 154, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Shannon, A.E.; Chappell, K.J.; Stoermer, M.J.; Chow, S.Y.; Kok, W.M.; Fairlie, D.P.; Young, P.R. Simultaneous uncoupled expression and purification of the Dengue virus NS3 protease and NS2B co-factor domain. Protein Expr. Purif. 2016, 119, 124–129. [Google Scholar] [CrossRef]
- Poulsen, A.; Kang, C.; Keller, T.H. Drug design for flavivirus proteases: What are we missing? Curr. Pharm. Des. 2014, 20, 3422–3427. [Google Scholar] [CrossRef] [PubMed]
- Erbel, P.; Schiering, N.; D’Arcy, A.; Renatus, M.; Kroemer, M.; Lim, S.P.; Yin, Z.; Keller, T.H.; Vasudevan, S.G.; Hommel, U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat. Struct. Mol. Biol. 2006, 13, 372–373. [Google Scholar] [CrossRef] [PubMed]
- Radichev, I.; Shiryaev, S.A.; Aleshin, A.E.; Ratnikov, B.I.; Smith, J.W.; Liddington, R.C.; Strongin, A.Y. Structure-based mutagenesis identifies important novel determinants of the NS2B cofactor of the West Nile virus two-component NS2B-NS3 proteinase. J. Gen. Virol. 2008, 89, 636–641. [Google Scholar] [CrossRef] [PubMed]
- Chappell, K.J.; Stoermer, M.J.; Fairlie, D.P.; Young, P.R. Mutagenesis of the West Nile virus NS2B cofactor domain reveals two regions essential for protease activity. J. Gen. Virol. 2008, 89, 1010–1014. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Kang, C. Insights into Structures and Dynamics of Flavivirus Proteases from NMR Studies. Int. J. Mol. Sci. 2020, 21, 2527. [Google Scholar] [CrossRef]
- Millies, B.; von Hammerstein, F.; Gellert, A.; Hammerschmidt, S.; Barthels, F.; Goppel, U.; Immerheiser, M.; Elgner, F.; Jung, N.; Basic, M.; et al. Proline-based allosteric inhibitors of Zika and Dengue Virus NS2B/NS3 proteases. J. Med. Chem. 2019, 62, 11359–11382. [Google Scholar] [CrossRef] [PubMed]
- Maus, H.; Barthels, F.; Hammerschmidt, S.J.; Kopp, K.; Millies, B.; Gellert, A.; Ruggieri, A.; Schirmeister, T. SAR of novel benzothiazoles targeting an allosteric pocket of DENV and ZIKV NS2B/NS3 proteases. Bioorg. Med. Chem. 2021, 47, 116392. [Google Scholar] [CrossRef]
- Zuo, Z.; Liew, O.W.; Chen, G.; Chong, P.C.; Lee, S.H.; Chen, K.; Jiang, H.; Puah, C.M.; Zhu, W. Mechanism of NS2B-mediated activation of NS3pro in Dengue virus: Molecular dynamics simulations and bioassays. J. Virol. 2009, 83, 1060–1070. [Google Scholar] [CrossRef]
- Gruba, N.; Martinez, J.I.R.; Grzywa, R.; Wysocka, M.; Skorenski, M.; Dabrowska, A.; Lecka, M.; Suder, P.; Sienczyk, M.; Pyrc, K.; et al. One step beyond: Design of substrates spanning primed positions of Zika virus NS2B-NS3 protease. ACS Med. Chem. Lett. 2018, 9, 1025–1029. [Google Scholar] [CrossRef] [PubMed]
- Majerova, T.; Novotny, P.; Krysova, E.; Konvalinka, J. Exploiting the unique features of Zika and Dengue proteases for inhibitor design. Biochimie 2019, 166, 132–141. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Osswald, H.L.; Prato, G. Recent progress in the development of HIV-1 protease inhibitors for the treatment of HIV/AIDS. J. Med. Chem. 2016, 59, 5172–5208. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Patel, S.J.; Wang, W.L.; Wang, G.; Chan, W.L.; Rao, K.R.; Alam, J.; Jeyaraj, D.A.; Ngew, X.; Patel, V.; et al. Peptide inhibitors of Dengue virus NS3 protease. Part 1: Warhead. Bioorg. Med. Chem. Lett. 2006, 16, 36–39. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Patel, S.J.; Wang, W.L.; Chan, W.L.; Ranga Rao, K.R.; Wang, G.; Ngew, X.; Patel, V.; Beer, D.; Knox, J.E.; et al. Peptide inhibitors of Dengue virus NS3 protease. Part 2: SAR study of tetrapeptide aldehyde inhibitors. Bioorg. Med. Chem. Lett. 2006, 16, 40–43. [Google Scholar] [CrossRef] [PubMed]
- Nitsche, C.; Zhang, L.; Weigel, L.F.; Schilz, J.; Graf, D.; Bartenschlager, R.; Hilgenfeld, R.; Klein, C.D. Peptide-boronic acid inhibitors of flaviviral proteases: Medicinal chemistry and structural biology. J. Med. Chem. 2017, 60, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Steuer, C.; Gege, C.; Fischl, W.; Heinonen, K.H.; Bartenschlager, R.; Klein, C.D. Synthesis and biological evaluation of alpha-ketoamides as inhibitors of the Dengue virus protease with antiviral activity in cell-culture. Bioorg. Med. Chem. 2011, 19, 4067–4074. [Google Scholar] [CrossRef] [PubMed]
- Nitsche, C.; Steuer, C.; Klein, C.D. Arylcyanoacrylamides as inhibitors of the Dengue and West Nile virus proteases. Bioorg. Med. Chem. 2011, 19, 7318–7337. [Google Scholar] [CrossRef] [PubMed]
- Nitsche, C.; Behnam, M.A.; Steuer, C.; Klein, C.D. Retro peptide-hybrids as selective inhibitors of the Dengue virus NS2B-NS3 protease. Antivir. Res. 2012, 94, 72–79. [Google Scholar] [CrossRef]
- Chorev, M.; Goodman, M. Recent developments in retro peptides and proteins: An ongoing topochemical exploration. Trends Biotechnol. 1995, 13, 438–445. [Google Scholar] [CrossRef]
- Arranz-Gibert, P.; Ciudad, S.; Seco, J.; Garcia, J.; Giralt, E.; Teixido, M. Immunosilencing peptides by stereochemical inversion and sequence reversal: Retro-D-peptides. Sci. Rep. 2018, 8, 6446. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.C.; Weng, Z.; Shao, X.; Liu, F.; Nie, X.; Liu, J.; Wang, D.; Wang, C.; Guo, K. Discovery and SAR studies of methionine-proline anilides as Dengue virus NS2B-NS3 protease inhibitors. Bioorg. Med. Chem. Lett. 2013, 23, 6549–6554. [Google Scholar] [CrossRef] [PubMed]
- Weng, Z.; Shao, X.; Graf, D.; Wang, C.; Klein, C.D.; Wang, J.; Zhou, G.C. Identification of fused bicyclic derivatives of pyrrolidine and imidazolidinone as Dengue virus-2 NS2B-NS3 protease inhibitors. Eur. J. Med. Chem. 2017, 125, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Prusis, P.; Junaid, M.; Petrovska, R.; Yahorava, S.; Yahorau, A.; Katzenmeier, G.; Lapins, M.; Wikberg, J.E. Design and evaluation of substrate-based octapeptide and non substrate-based tetrapeptide inhibitors of dengue virus NS2B-NS3 proteases. Biochem. Biophys. Res. Commun. 2013, 434, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Prusis, P.; Lapins, M.; Yahorava, S.; Petrovska, R.; Niyomrattanakit, P.; Katzenmeier, G.; Wikberg, J.E. Proteochemometrics analysis of substrate interactions with Dengue virus NS3 proteases. Bioorg. Med. Chem. 2008, 16, 9369–9377. [Google Scholar] [CrossRef]
- Nitsche, C.; Schreier, V.N.; Behnam, M.A.; Kumar, A.; Bartenschlager, R.; Klein, C.D. Thiazolidinone-peptide hybrids as dengue virus protease inhibitors with antiviral activity in cell culture. J. Med. Chem. 2013, 56, 8389–8403. [Google Scholar] [CrossRef]
- Behnam, M.A.; Nitsche, C.; Vechi, S.M.; Klein, C.D. C-terminal residue optimization and fragment merging: Discovery of a potent peptide-hybrid inhibitor of Dengue protease. ACS Med. Chem. Lett. 2014, 5, 1037–1042. [Google Scholar] [CrossRef]
- Bastos Lima, A.; Behnam, M.A.; El Sherif, Y.; Nitsche, C.; Vechi, S.M.; Klein, C.D. Dual inhibitors of the dengue and West Nile virus NS2B-NS3 proteases: Synthesis, biological evaluation and docking studies of novel peptide-hybrids. Bioorg. Med. Chem. 2015, 23, 5748–5755. [Google Scholar] [CrossRef]
- Behnam, M.A.; Graf, D.; Bartenschlager, R.; Zlotos, D.P.; Klein, C.D. Discovery of nanomolar Dengue and West Nile virus protease inhibitors containing a 4-benzyloxyphenylglycine residue. J. Med. Chem. 2015, 58, 9354–9370. [Google Scholar] [CrossRef]
- Weigel, L.F.; Nitsche, C.; Graf, D.; Bartenschlager, R.; Klein, C.D. Phenylalanine and phenylglycine analogues as arginine mimetics in Dengue protease inhibitors. J. Med. Chem. 2015, 58, 7719–7733. [Google Scholar] [CrossRef]
- Li, J.; Lim, S.P.; Beer, D.; Patel, V.; Wen, D.; Tumanut, C.; Tully, D.C.; Williams, J.A.; Jiricek, J.; Priestle, J.P.; et al. Functional profiling of recombinant NS3 proteases from all four serotypes of Dengue virus using tetrapeptide and octapeptide substrate libraries. J. Biol. Chem. 2005, 280, 28766–28774. [Google Scholar] [CrossRef]
- Stoermer, M.J.; Chappell, K.J.; Liebscher, S.; Jensen, C.M.; Gan, C.H.; Gupta, P.K.; Xu, W.J.; Young, P.R.; Fairlie, D.P. Potent cationic inhibitors of West Nile virus NS2B/NS3 protease with serum stability, cell permeability and antiviral activity. J. Med. Chem. 2008, 51, 5714–5721. [Google Scholar] [CrossRef]
- Schuller, A.; Yin, Z.; Brian Chia, C.S.; Doan, D.N.; Kim, H.K.; Shang, L.; Loh, T.P.; Hill, J.; Vasudevan, S.G. Tripeptide inhibitors of Dengue and West Nile virus NS2B-NS3 protease. Antivir. Res. 2011, 92, 96–101. [Google Scholar] [CrossRef]
- Xu, S.; Li, H.; Shao, X.; Fan, C.; Ericksen, B.; Liu, J.; Chi, C.; Wang, C. Critical effect of peptide cyclization on the potency of peptide inhibitors against Dengue virus NS2B-NS3 protease. J. Med. Chem. 2012, 55, 6881–6887. [Google Scholar] [CrossRef]
- Terlau, H.; Olivera, B.M. Conus venoms: A rich source of novel ion channel-targeted peptides. Physiol. Rev. 2004, 84, 41–68. [Google Scholar] [CrossRef]
- Chu, J.J.; Lee, R.C.; Ang, M.J.; Wang, W.L.; Lim, H.A.; Wee, J.L.; Joy, J.; Hill, J.; Brian Chia, C.S. Antiviral activities of 15 Dengue NS2B-NS3 protease inhibitors using a human cell-based viral quantification assay. Antivir. Res. 2015, 118, 68–74. [Google Scholar] [CrossRef]
- Driggers, E.M.; Hale, S.P.; Lee, J.; Terrett, N.K. The exploration of macrocycles for drug discovery—an underexploited structural class. Nat. Rev. Drug Discov. 2008, 7, 608–624. [Google Scholar] [CrossRef] [PubMed]
- Matsson, P.; Doak, B.C.; Over, B.; Kihlberg, J. Cell permeability beyond the rule of 5. Adv. Drug Deliv. Rev. 2016, 101, 42–61. [Google Scholar] [CrossRef]
- Takagi, Y.; Matsui, K.; Nobori, H.; Maeda, H.; Sato, A.; Kurosu, T.; Orba, Y.; Sawa, H.; Hattori, K.; Higashino, K.; et al. Discovery of novel cyclic peptide inhibitors of Dengue virus NS2B-NS3 protease with antiviral activity. Bioorg. Med. Chem. Lett. 2017, 27, 3586–3590. [Google Scholar] [CrossRef] [PubMed]
- Steinkuhler, C.; Biasiol, G.; Brunetti, M.; Urbani, A.; Koch, U.; Cortese, R.; Pessi, A.; De Francesco, R. Product inhibition of the hepatitis C virus NS3 protease. Biochemistry 1998, 37, 8899–8905. [Google Scholar] [CrossRef]
- Ingallinella, P.; Altamura, S.; Bianchi, E.; Taliani, M.; Ingenito, R.; Cortese, R.; De Francesco, R.; Steinkuhler, C.; Pessi, A. Potent peptide inhibitors of human hepatitis C virus NS3 protease are obtained by optimizing the cleavage products. Biochemistry 1998, 37, 8906–8914. [Google Scholar] [CrossRef] [PubMed]
- Chanprapaph, S.; Saparpakorn, P.; Sangma, C.; Niyomrattanakit, P.; Hannongbua, S.; Angsuthanasombat, C.; Katzenmeier, G. Competitive inhibition of the dengue virus NS3 serine protease by synthetic peptides representing polyprotein cleavage sites. Biochem. Biophys. Res. Commun. 2005, 330, 1237–1246. [Google Scholar] [CrossRef]
- Lin, K.H.; Nalivaika, E.A.; Prachanronarong, K.L.; Yilmaz, N.K.; Schiffer, C.A. Dengue protease substrate recognition: Binding of the prime side. ACS Infect. Dis. 2016, 2, 734–743. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Z.; Phoo, W.W.; Loh, Y.R.; Wang, W.; Liu, S.; Chen, M.W.; Hung, A.W.; Keller, T.H.; Luo, D.; et al. Structural dynamics of Zika virus NS2B-NS3 protease binding to dipeptide inhibitors. Structure 2017, 25, 1242–1250. [Google Scholar] [CrossRef]
- Sylte, I.; Dawadi, R.; Malla, N.; von Hofsten, S.; Nguyen, T.M.; Solli, A.I.; Berg, E.; Adekoya, O.A.; Svineng, G.; Winberg, J.O. The selectivity of galardin and an azasugar-based hydroxamate compound for human matrix metalloproteases and bacterial metalloproteases. PLoS ONE 2018, 13, e0200237. [Google Scholar] [CrossRef] [PubMed]
- Passioura, T.; Katoh, T.; Goto, Y.; Suga, H. Selection-based discovery of druglike macrocyclic peptides. Annu. Rev. Biochem. 2014, 83, 727–752. [Google Scholar] [CrossRef]
- Gomes, B.; Augusto, M.T.; Felicio, M.R.; Hollmann, A.; Franco, O.L.; Goncalves, S.; Santos, N.C. Designing improved active peptides for therapeutic approaches against infectious diseases. Biotechnol. Adv. 2018, 36, 415–429. [Google Scholar] [CrossRef]

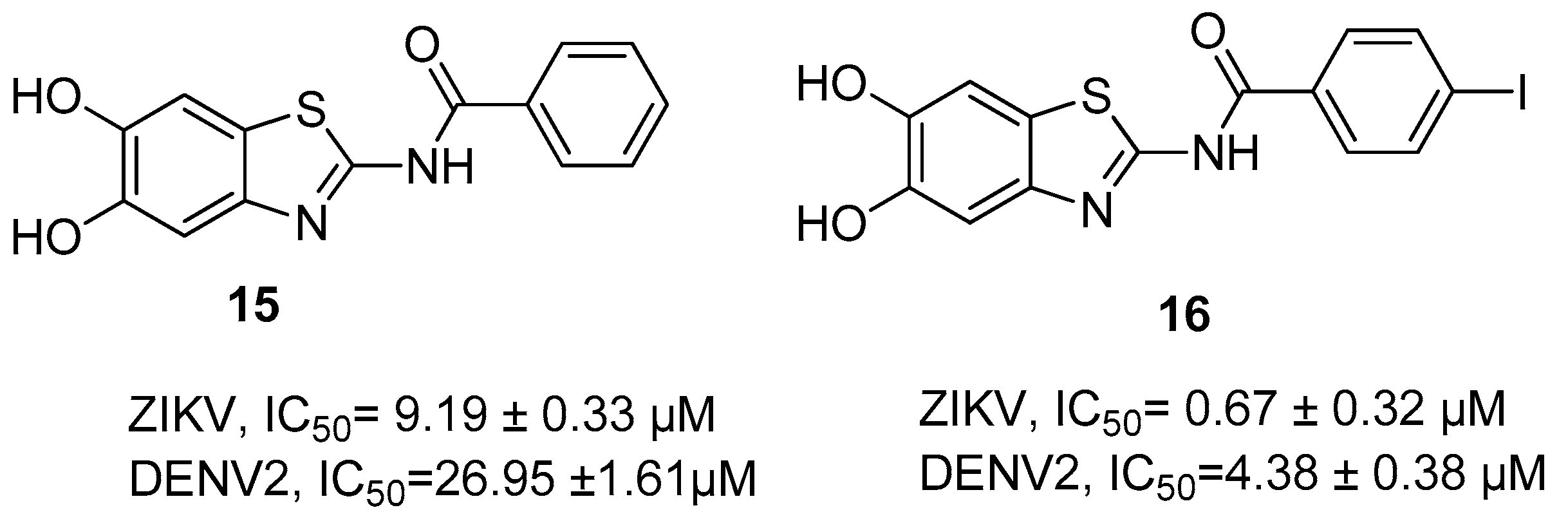

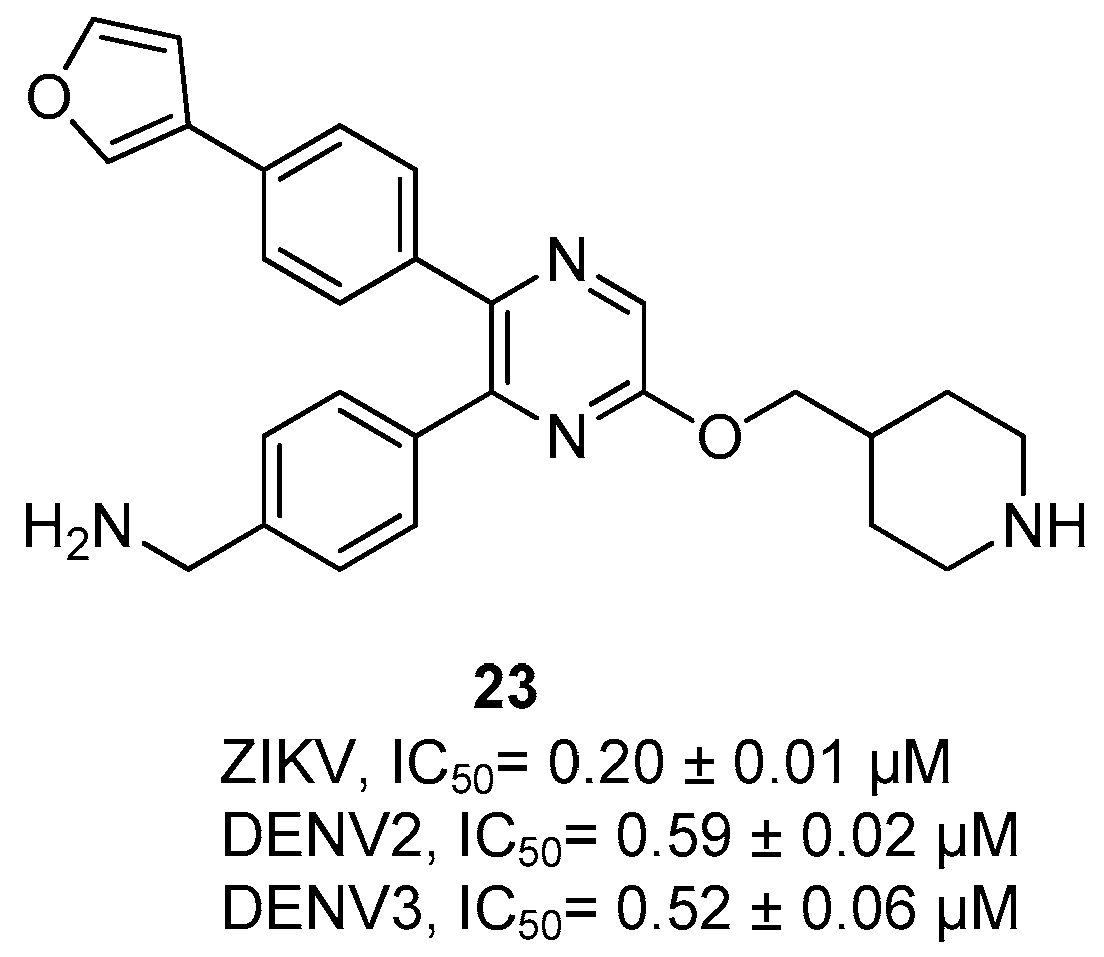
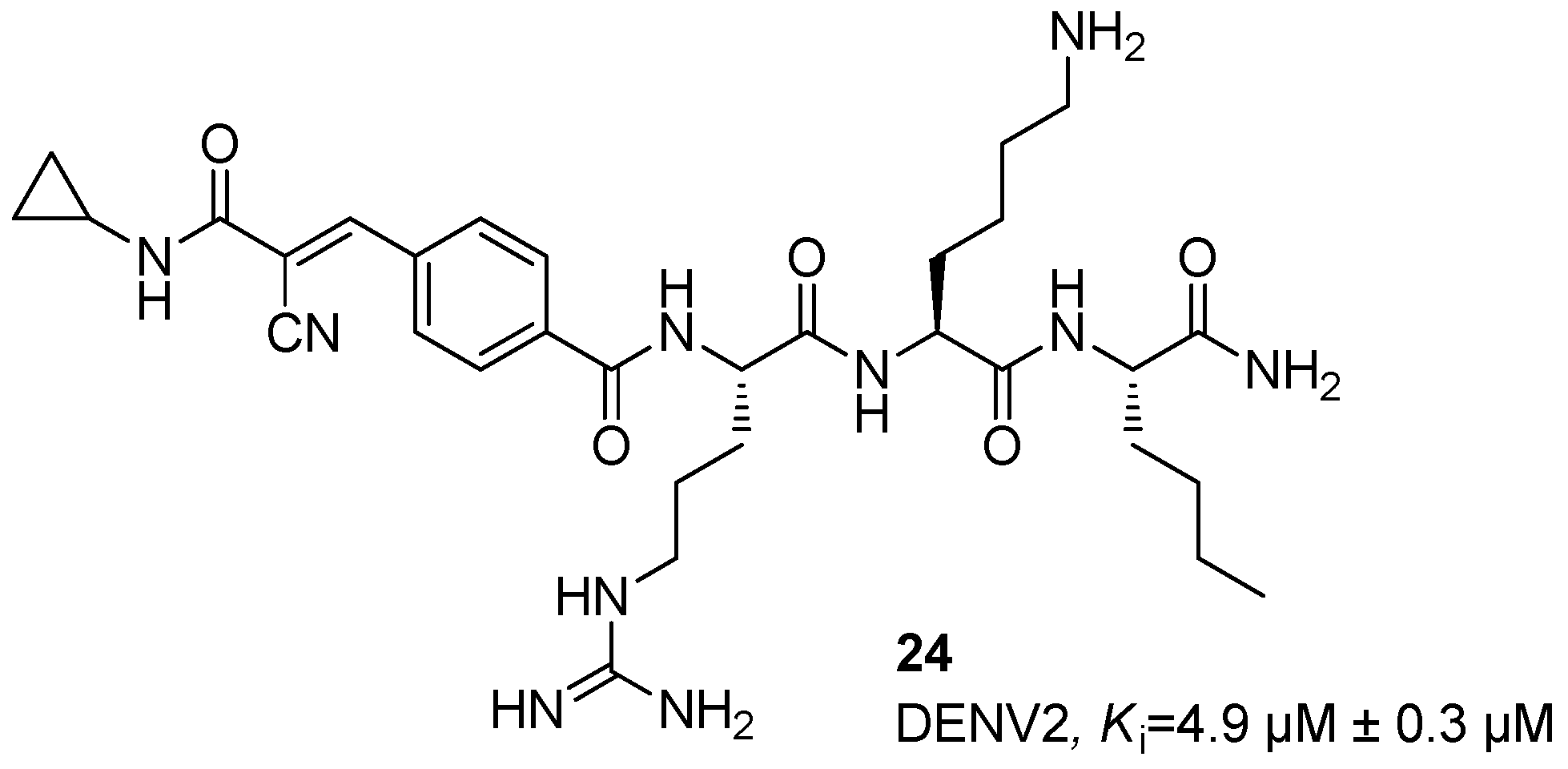
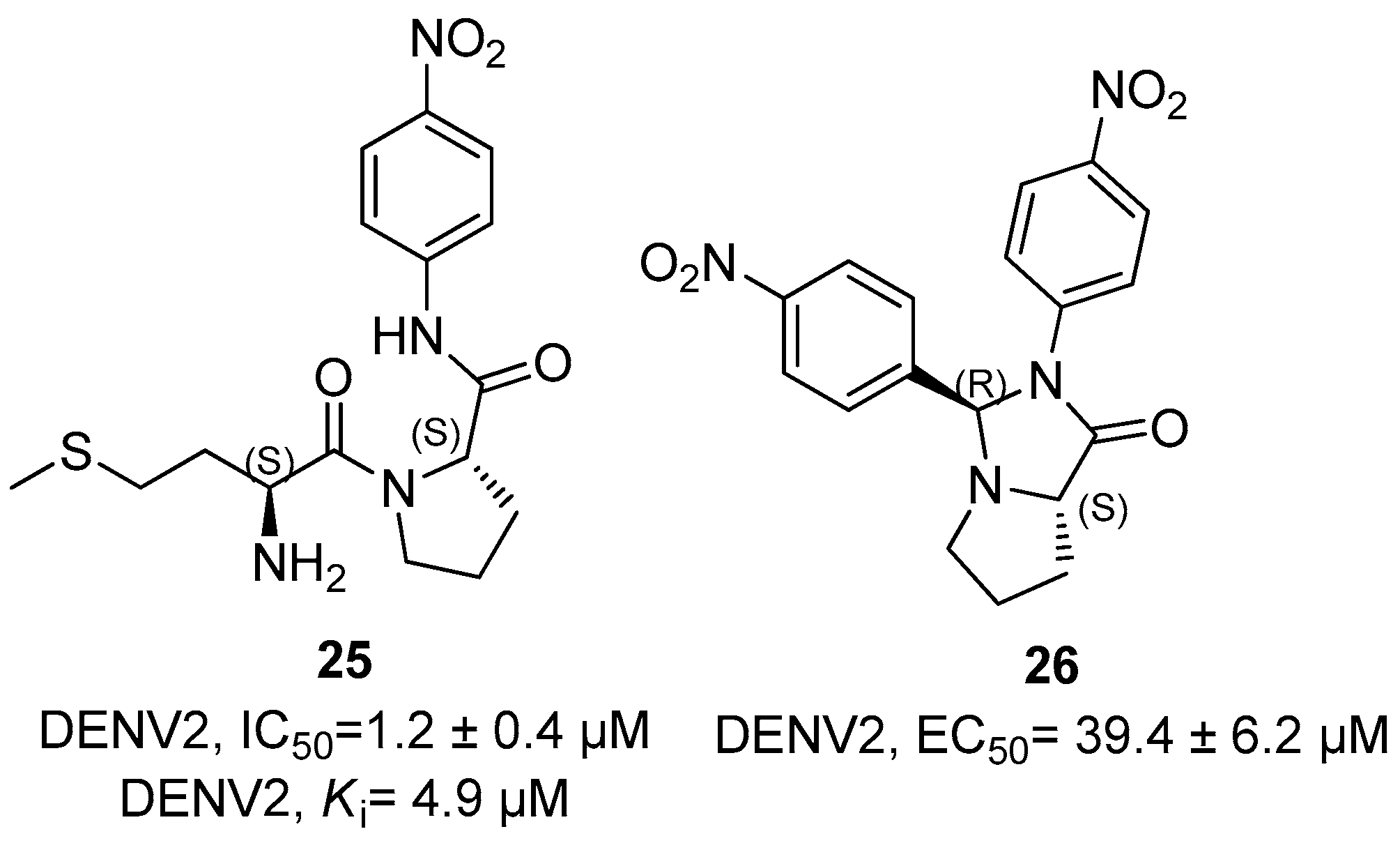

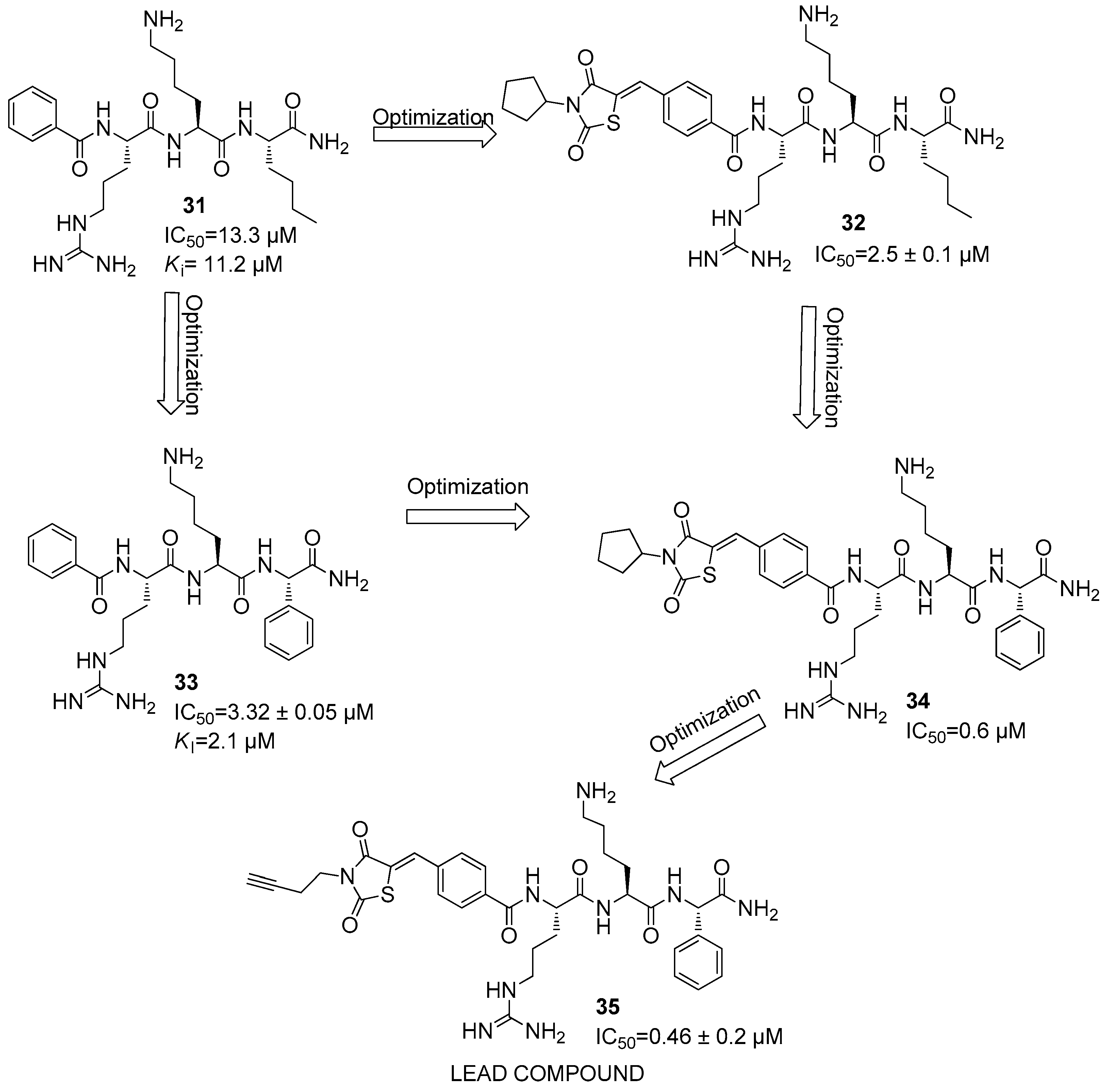
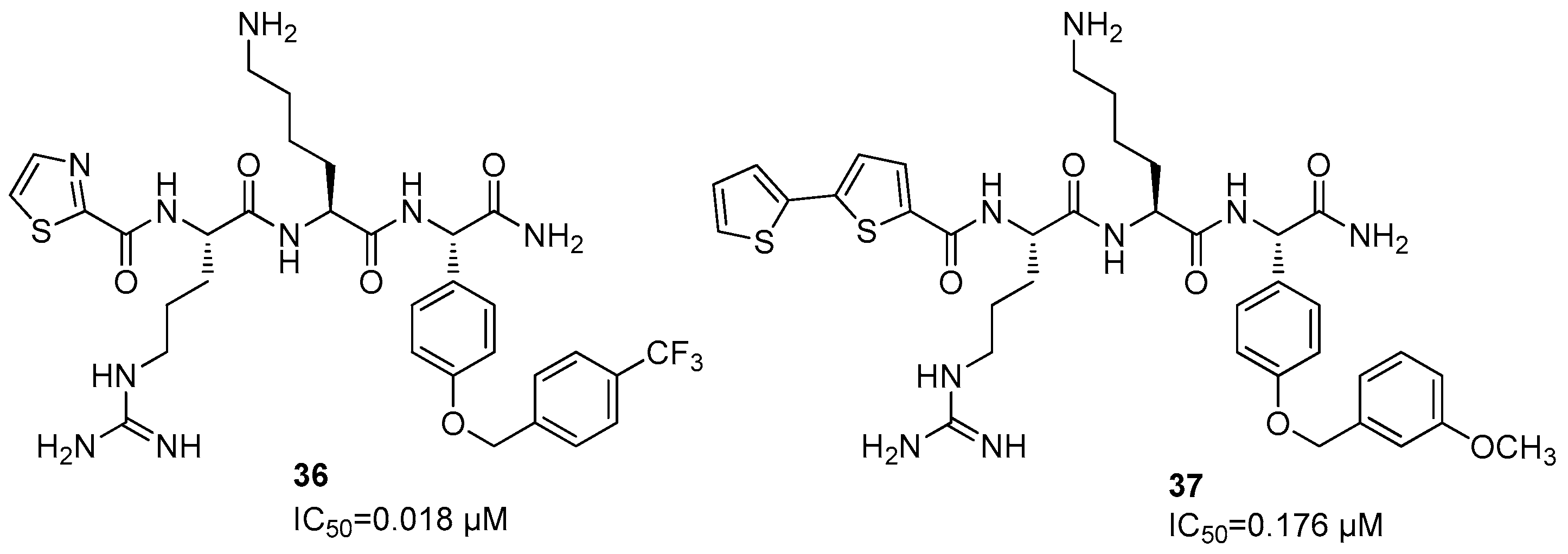
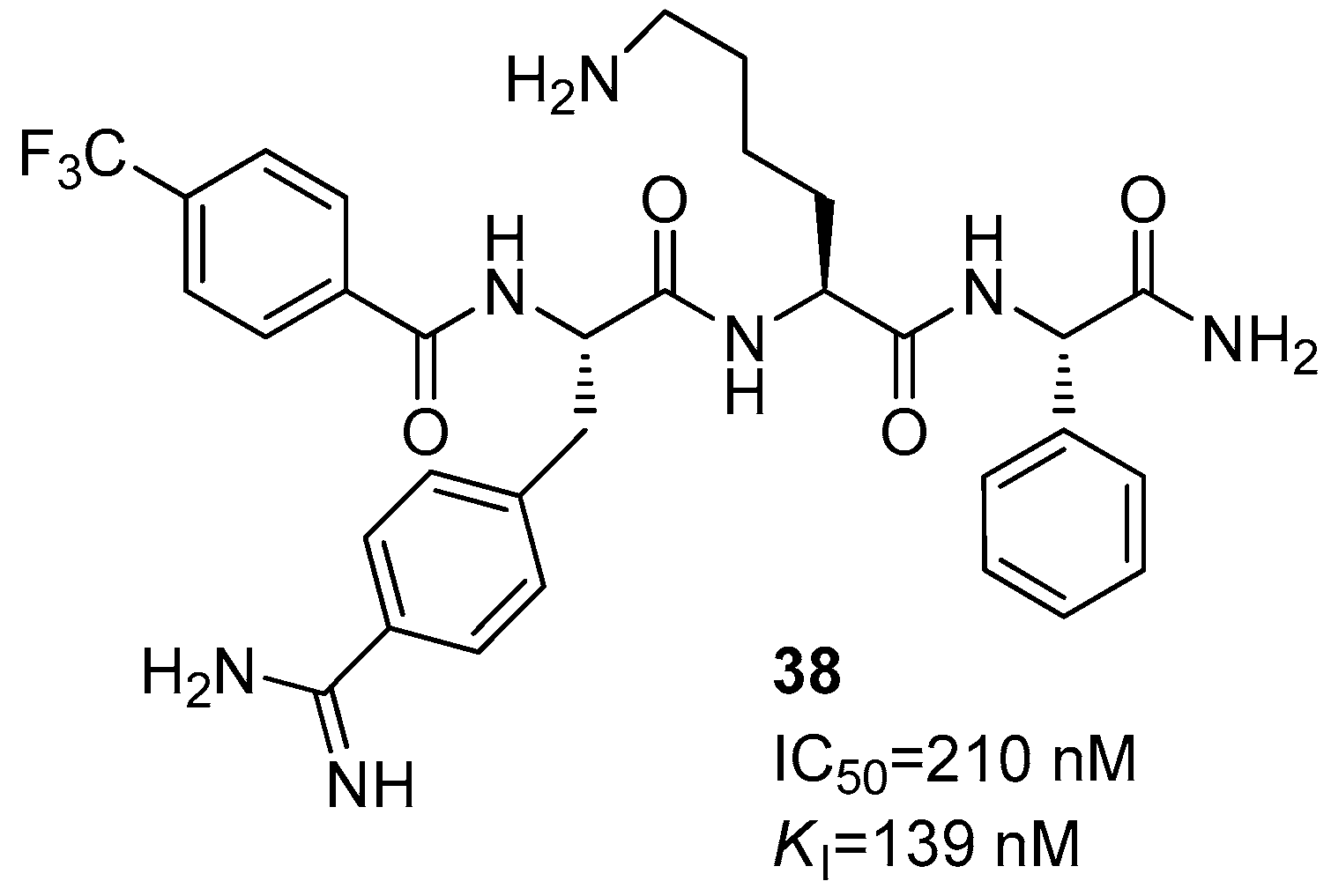
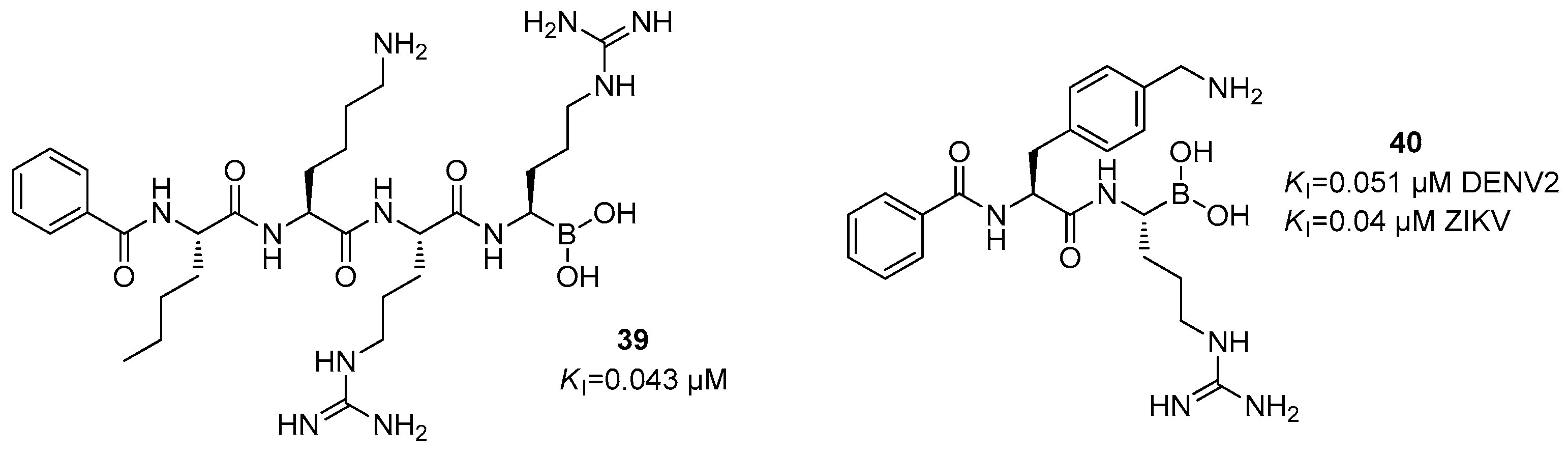





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Starvaggi, J.; Previti, S.; Zappalà, M.; Ettari, R. The Inhibition of NS2B/NS3 Protease: A New Therapeutic Opportunity to Treat Dengue and Zika Virus Infection. Int. J. Mol. Sci. 2024, 25, 4376. https://doi.org/10.3390/ijms25084376
Starvaggi J, Previti S, Zappalà M, Ettari R. The Inhibition of NS2B/NS3 Protease: A New Therapeutic Opportunity to Treat Dengue and Zika Virus Infection. International Journal of Molecular Sciences. 2024; 25(8):4376. https://doi.org/10.3390/ijms25084376
Chicago/Turabian StyleStarvaggi, Josè, Santo Previti, Maria Zappalà, and Roberta Ettari. 2024. "The Inhibition of NS2B/NS3 Protease: A New Therapeutic Opportunity to Treat Dengue and Zika Virus Infection" International Journal of Molecular Sciences 25, no. 8: 4376. https://doi.org/10.3390/ijms25084376





