Establishment of a Homologous Silencing System with Intact-Plant Infiltration and Minimized Operation for Studying Gene Function in Herbaceous Peonies
Abstract
1. Introduction
2. Results
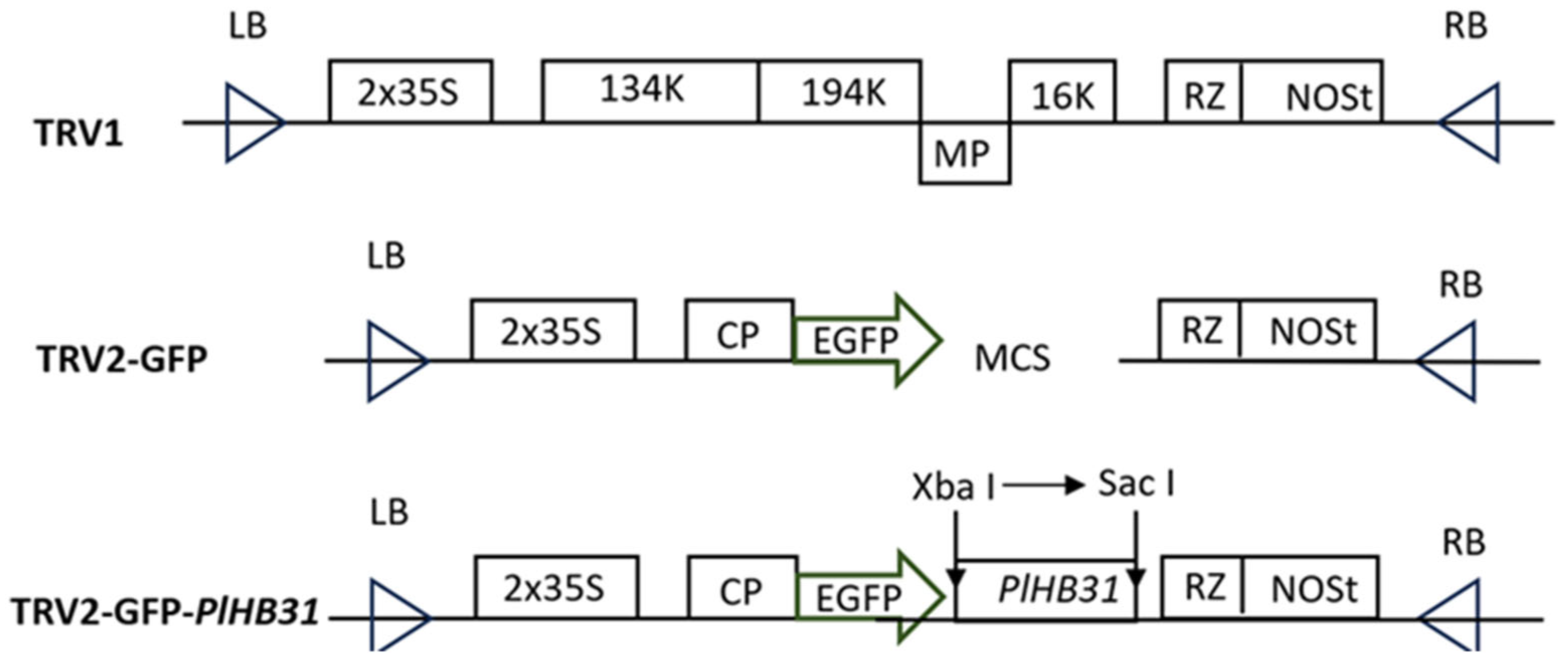
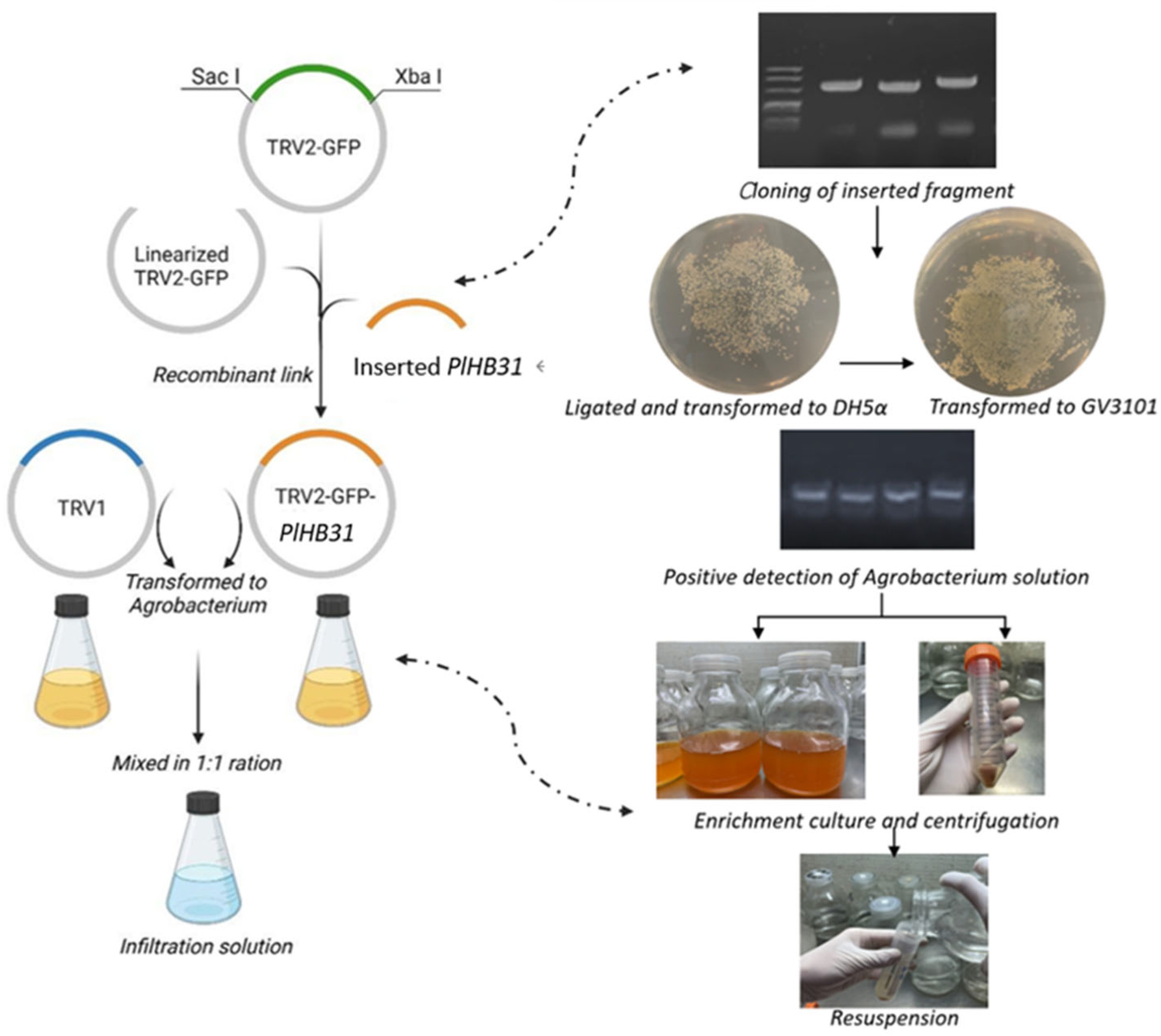
2.1. Construction of the TRV2-GFP-PlHB31 Recombinant Vector
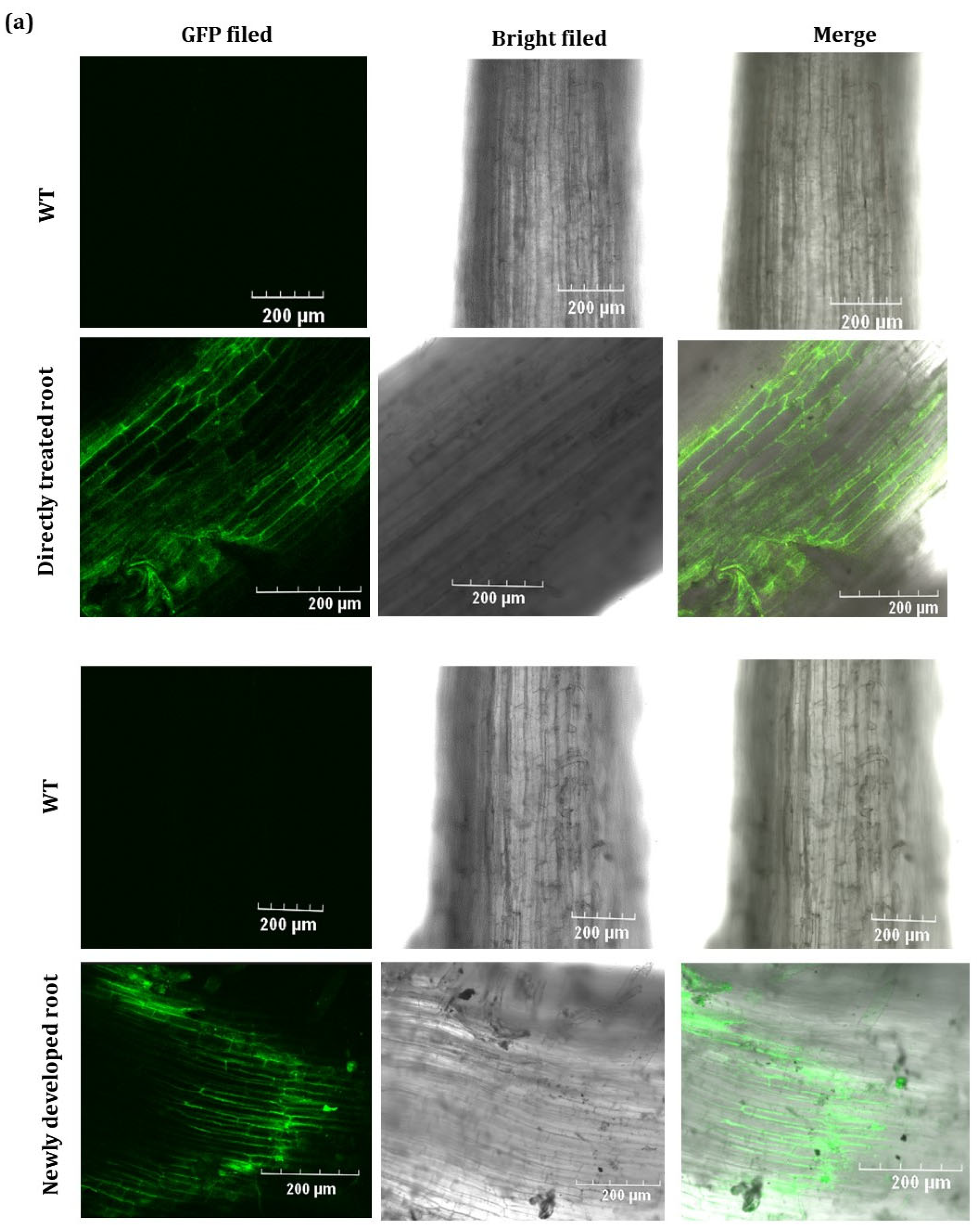
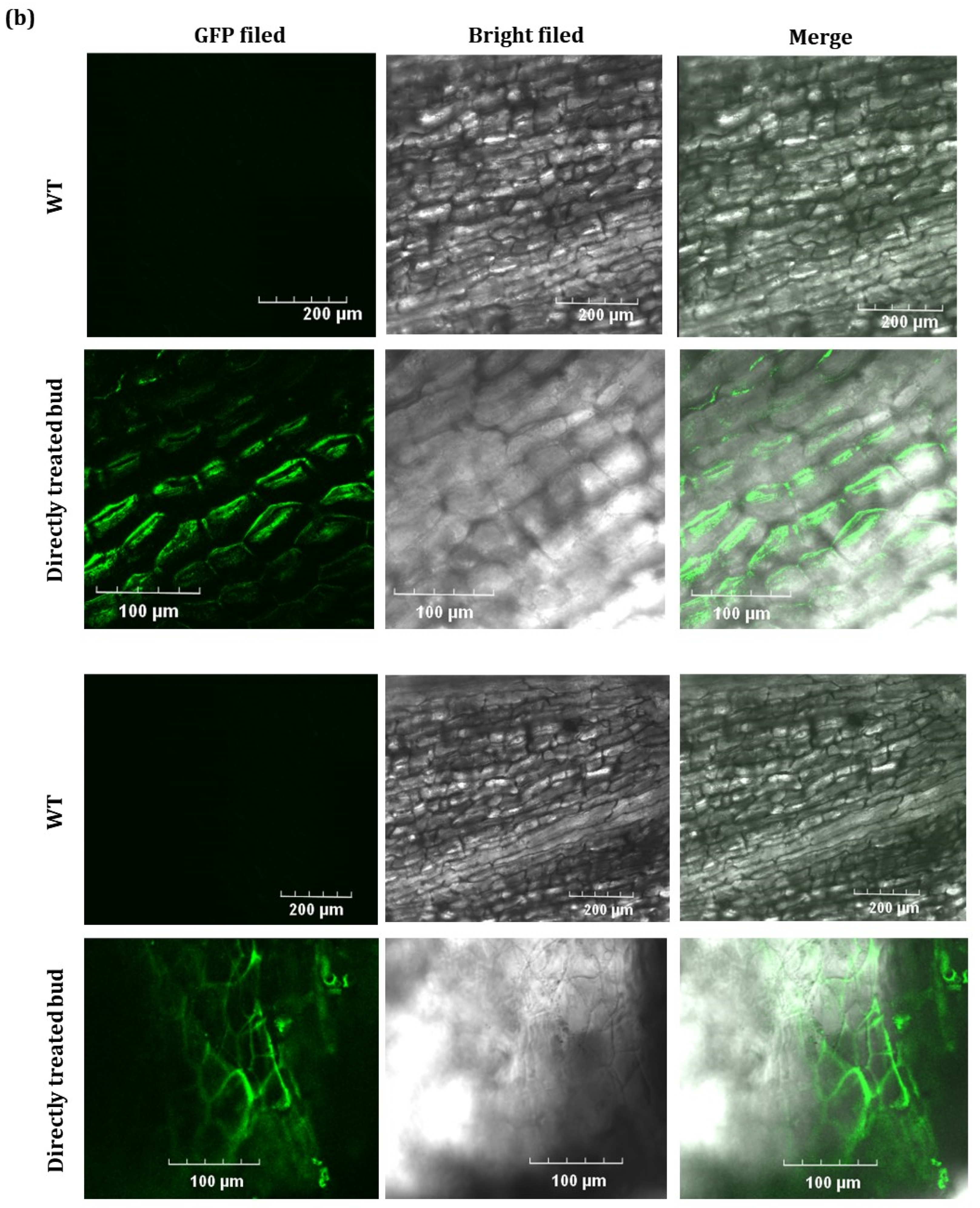
2.2. GFP Expressed from TRV2-GFP-PlHB31 in Infiltrated Organs
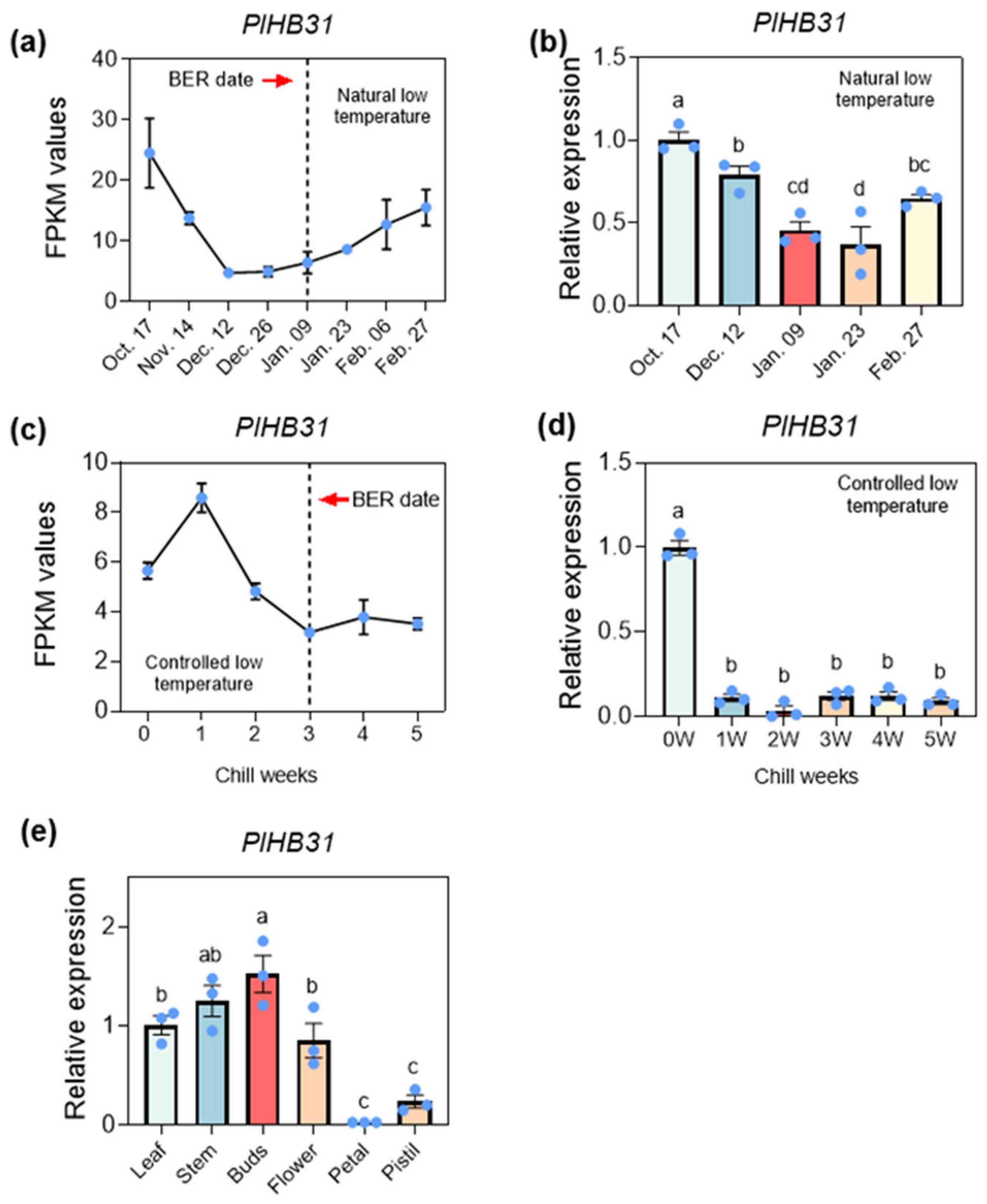
2.3. Screening and Silencing of PlHB31 in P. lactiflora
3. Discussion
3.1. VIGS Is an Ideal Technique for Identifying Gene Function in Herbaceous Peony but Needs to Be Improved Further
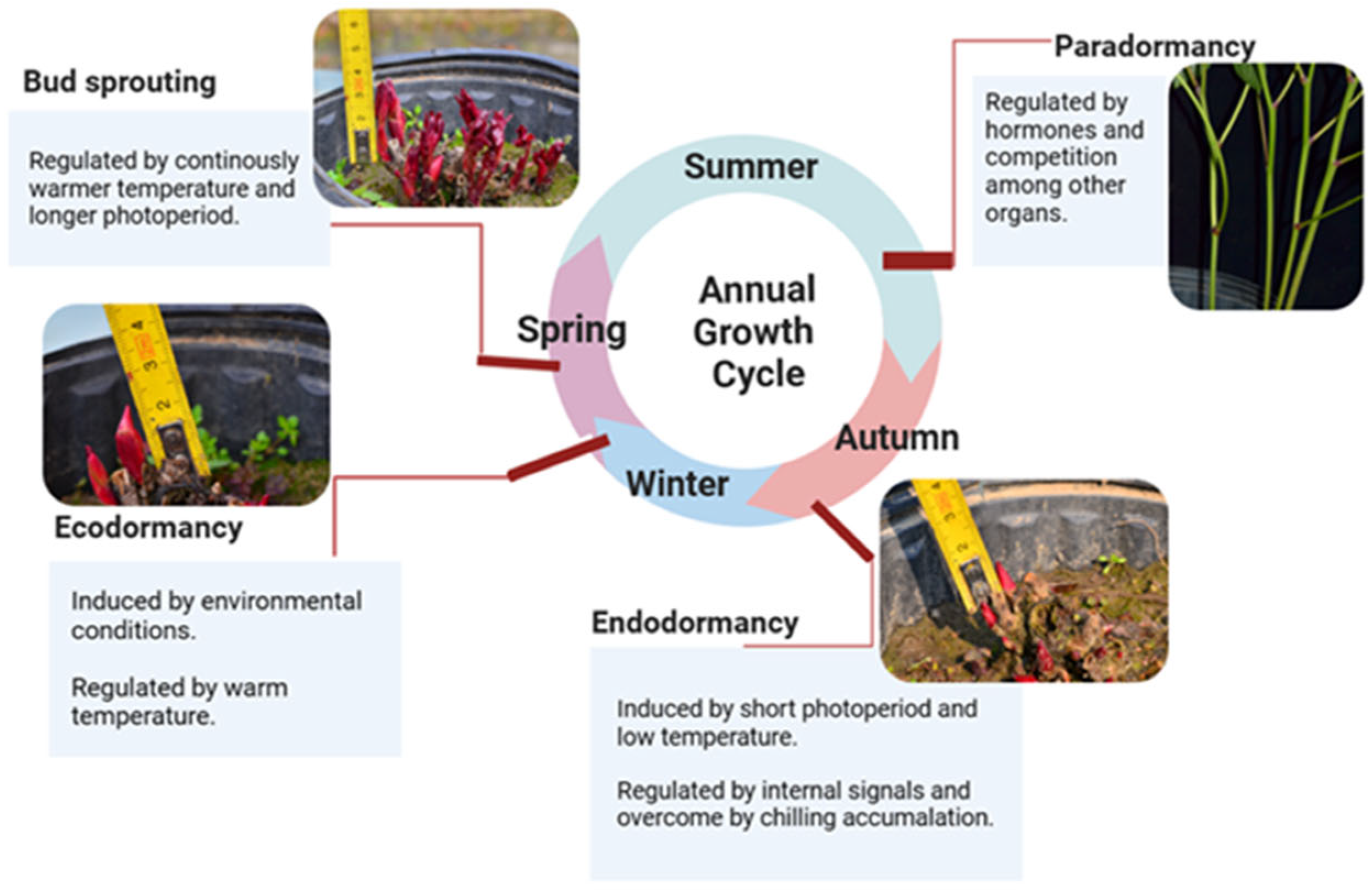
3.2. Selection One-Year-Old Roots of Herbaceous Peony for VIGS: Realizing the Intact-Plant Infiltration and Achieving the Minimization of the Operation Space, Material, Reagent and Manpower
3.3. Ensuring a Year-Round Abundant Supply of Robust Materials for Infection and Silencing
3.4. Determination of the Crucial Prechilling Treatment Duration Depends on Specific Scientific Questions
3.5. Functional Identification of the Template Gene PlHB31 via the Established VIGS System
3.6. The Established System Is Suitable for Research on the Functions of Genes Regulating Various Physiological Activities, except Blooming and Fruiting, in Herbaceous Peony
3.7. The Established VIGS System Is Suitable for Gene Function Research in All Herbaceous Peonies and Provides a Reference for Gene Silencing in Other Geophytes
4. Materials and Methods
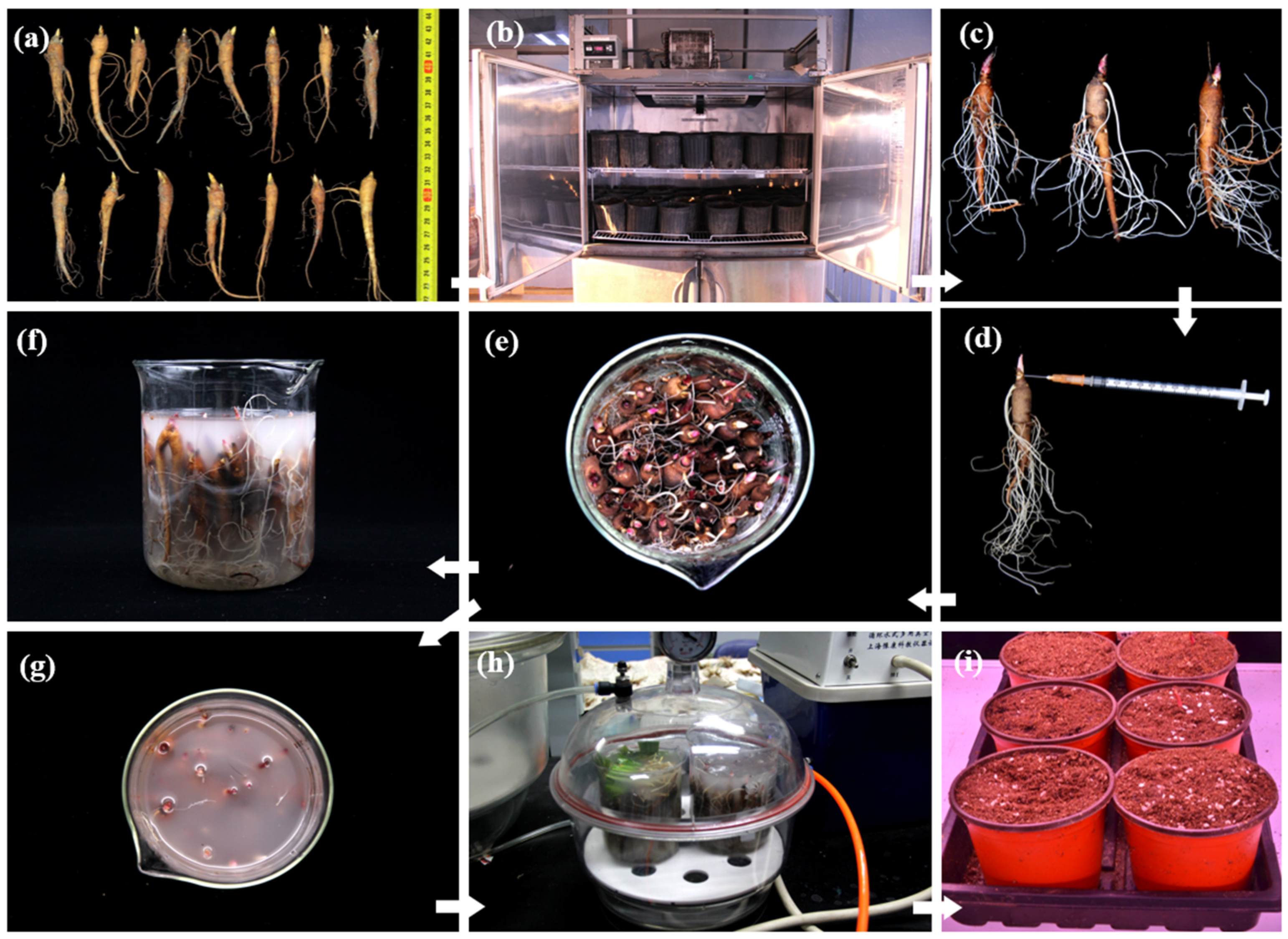
4.1. Plant Material and Selection Standard
4.2. Prechilling Treatment
4.3. Target Gene Identification and Isolation
4.4. Vector Construction
4.5. Plasmid Transformation and Preparation for Infection
4.6. Vacuum Infiltration of the Intact-Plant with One-Year-Old Roots
4.7. GFP Imaging Detection
4.8. Phenotypic Observation
4.9. Determination of Gene Expression
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, W.; Yuan, J.S.; Stewart, C.N., Jr. Advanced genetic tools for plant biotechnology. Nat. Rev. Genet. 2013, 14, 781–793. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liang, Z.; Zong, Y.; Wang, Y.; Liu, J.; Chen, K.; Qiu, J.-L.; Gao, C. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016, 7, 12617. [Google Scholar] [CrossRef] [PubMed]
- Butt, H.I.; Yang, Z.E.; Gong, Q.; Chen, E.Y.; Wang, X.Q.; Zhao, G.; Ge, X.Y.; Zhang, X.Y.; Li, F.G. GaMYB85, an R2R3 MYB gene, in transgenic Arabidopsis plays an important role in drought tolerance. BMC Plant Biol. 2017, 17, 142. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.L.; Yao, L.J.; Zhang, Y.; Baskin, J.M.; Baskin, C.C.; Xiong, Z.M.; Tao, J. A review of the seed biology of Paeonia species (Paeoniaceae), with particular reference to dormancy and germination. Planta 2019, 249, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Kamenetski, R.; Barzilay, A.; Cohen, M. Herbaceous peony for cut flower production: Flowering physiologation techniques and cultivation techniques. In Proceedings of the International Conference on Quality Management in Supply Chains of Ornamentals, King Mongkuts Univ Technol Thonburi, Bangkok, Thailand, 3–6 December 2007; pp. 121–126. [Google Scholar]
- Falavigna, V.D.S.; Guitton, B.; Costes, E.; Andres, F. I Want to (Bud) Break Free: The Potential Role of DAM and SVP-Like Genes in Regulating Dormancy Cycle in Temperate Fruit Trees. Front. Plant Sci. 2019, 9, 1990. [Google Scholar] [CrossRef] [PubMed]
- Miotto, Y.E.; Tessele, C.; Czermainski, A.B.C.; Porto, D.D.; Falavigna, V.D.; Sartor, T.; Cattani, A.M.; Delatorre, C.A.; de Alencar, S.A.; da Silva, O.B.; et al. Spring Is Coming: Genetic Analyses of the Bud Break Date Locus Reveal Candidate Genes from the Cold Perception Pathway to Dormancy Release in Apple (Malus × domestica Borkh.). Front. Plant Sci. 2019, 10, 33. [Google Scholar] [CrossRef]
- Kamenetsky-Goldstein, R.; Yu, X. Cut peony industry: The first 30 years of research and new horizons. Hortic. Res. 2022, 9, uhac079. [Google Scholar] [CrossRef] [PubMed]
- Barzilay, A.; Zemah, H.; Kamenetsky, R.; Ran, I. Annual life cycle and floral development of ‘Sarah Bernhardt’ peony in Israel. Hortscience 2002, 37, 300–303. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, D.; Wei, J.; Shi, X.; Ding, H.; Qiu, S.; Guo, J.; Li, D.; Zhu, K.; Horvath, D.P.; et al. Annual growth cycle observation, hybridization and forcing culture for improving the ornamental application of Paeonia lactiflora Pall. in the low-latitude regions. PLoS ONE 2019, 14, e0218164. [Google Scholar] [CrossRef]
- Ji, L.J.; Wang, Q.; da Silva, J.A.T.; Yu, X.N. The genetic diversity of Paeonia L. Sci. Hortic. 2012, 143, 62–74. [Google Scholar] [CrossRef]
- Duan, S.Y.; Xin, R.J.; Guan, S.X.; Li, X.T.; Fei, R.W.; Cheng, W.; Pan, Q.; Sun, X.M. Optimization of callus induction and proliferation of Paeonia lactiflora Pall. and Agrobacterium-mediated genetic transformation. Front. Plant Sci. 2022, 13, 996690. [Google Scholar] [CrossRef]
- Cai, X.; Wei, H.; Liu, C.; Ren, X.X.; Luc, H.; Jeong, Y.R.Y. Synergistic Effect of NaCl Pretreatment and PVP on Browning Suppression and Callus Induction from Petal Explants of Paeonia Lactiflora Pall. ‘Festival Maxima’. Plants 2020, 9, 346. [Google Scholar] [CrossRef]
- Shen, M.M.; Tang, Z.J.; da Silva, J.A.T.; Yu, X.N. Induction and proliferation of axillary shoots from in vitro culture of Paeonia lactiflora Pall. mature zygotic embryos. N. Z. J. Crop Hortic. Sci. 2015, 43, 42–52. [Google Scholar] [CrossRef]
- Shen, M.M.; Wang, Q.; Yu, X.N.; da Silva, J.A.T. Micropropagation of herbaceous peony (Paeonia lactiflora Pall.). Sci. Hortic. 2012, 148, 30–38. [Google Scholar] [CrossRef]
- Liu, Y.L.; Schiff, M.; Dinesh-Kumar, S.P. Virus-induced gene silencing in tomato. Plant J. 2002, 31, 777–786. [Google Scholar] [CrossRef]
- Liu, Q.L.; Xu, K.D.; Yi, L.; Hou, Y.L.; Li, D.X.; Hu, H.Y.; Zhou, F.; Song, P.W.; Yu, Y.G.; Wei, Q.C.; et al. A rapid, simple, and highly efficient method for VIGS and in vitro-inoculation of plant virus by INABS applied to crops that develop axillary buds and can survive from cuttings. BMC Plant Biol. 2021, 21, 545. [Google Scholar] [CrossRef] [PubMed]
- Burch-Smith, T.M.; Anderson, J.C.; Martin, G.B.; Dinesh-Kumar, S.P. Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 2004, 39, 734–746. [Google Scholar] [CrossRef]
- Purkayastha, A.; Mathur, S.; Verma, V.; Sharma, S.; Dasgupta, I. Virus-induced gene silencing in rice using a vector de-rived from a DNA virus. Planta 2010, 232, 1531–1540. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Xie, K.; Jia, Q.; Zhao, J.; Chen, T.; Li, H.; Wei, X.; Diao, X.; Hong, Y.; Liu, Y. Foxtail Mosaic Virus-Induced Gene Silencing in Monocot Plants. Plant Physiol. 2016, 171, 1801–1807. [Google Scholar] [CrossRef]
- Li, Y.; Kong, F.; Liu, Z.; Peng, L.; Shu, Q. PhUGT78A22, a novel glycosyltransferase in Paeonia ‘He Xie’, can catalyze the transfer of glucose to glucosylated anthocyanins during petal blotch formation. BMC Plant Biol. 2022, 22, 405. [Google Scholar] [CrossRef]
- Sun, Y.; Shang, W.; Yuan, J.; Wang, Z.; He, S.; Song, Y.; Shi, L.; Shen, Y.; Ma, J.; Xu, Y.; et al. Functional Analysis of PsARRO−1 in Root Development of Paeonia suffruticosa. Horticulturae 2022, 8, 903. [Google Scholar] [CrossRef]
- Tang, Y.H.; Lu, L.L.; Huang, X.Q.; Zhao, D.Q.; Tao, J. The herbaceous peony transcription factor WRKY41a promotes secondary cell wall thickening to enhance stem strength. Plant Physiol. 2023, 191, 428–445. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, J.; Guo, J.; Qiao, Q.; Guo, X.; Ma, Y. The WRKY transcription factor PlWRKY65 enhances the resistance of Paeonia lactiflora (herbaceous peony) to Alternaria tenuissima. Hortic. Res. 2020, 7, 57. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.Q.; Luan, Y.T.; Shi, W.B.; Tang, Y.H.; Huang, X.Q.; Tao, J. Melatonin enhances stem strength by increasing lignin content and secondary cell wall thickness in herbaceous peony. J. Exp. Bot. 2022, 73, 5974–5991. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Cheng, Z.; Meng, J.; Tao, J.; Zhao, D. PlMAPK1 facilitates growth and photosynthesis of herbaceous peony (Paeonia lactiflora Pall.) under high-temperature stress. Sci. Hortic. 2023, 310, 111701. [Google Scholar] [CrossRef]
- Bian, T.; Ma, Y.; Guo, J.; Wu, Y.; Shi, D.; Guo, X. Herbaceous peony (Paeonia lactiflora Pall.) PlDELLA gene negatively regulates dormancy release and plant growth. Plant Sci. 2020, 297, 110539. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gao, L.; Wang, Y.; Niu, D.; Yuan, Y.; Liu, C.; Zhan, X.; Gai, S. Dual functions of PsmiR172b-PsTOE3 module in dormancy release and flowering in tree peony (Paeonia suffruticosa). Hortic. Res. 2023, 10, uhad033. [Google Scholar] [CrossRef]
- Gao, L.; Niu, D.; Chi, T.; Yuan, Y.; Liu, C.; Gai, S.; Zhang, Y. PsRGL1 negatively regulates chilling- and gibberellin-induced dormancy release by PsF-box1-mediated targeting for proteolytic degradation in tree peony. Hortic. Res. 2023, 10, uhad044. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Duan, X.; Wang, S.; Wu, W.; Zhang, X. Virus-induced gene silencing (VIGS) for functional analysis of MYB80 gene involved in Solanum lycopersicum cold tolerance. Protoplasma 2019, 256, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wu, Y.; Shi, M.; Zhao, D.; Tao, J. Isolation of PlANS and PlDFR genes from herbaceous peony (Paeonia lactiflora Pall.) and its functional characterization in Arabidopsis and tobacco. Plant Cell Tissue Organ Cult. 2020, 141, 435–445. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, S.; Cheng, Y.; Wang, J.; Wang, X.; Liu, R.; Han, J. Functional identification of PsMYB57 involved in an-thocyanin regulation of tree peony. BMC Genet. 2020, 21, 124. [Google Scholar] [CrossRef]
- Meng, J.; Guo, J.; Li, T.; Chen, Z.; Li, M.; Zhao, D.; Tao, J. Analysis and Functional Verification of PlPM19L Gene Associated with Drought-Resistance in Paeonia lactiflora Pall. Int. J. Mol. Sci. 2022, 23, 15695. [Google Scholar] [CrossRef] [PubMed]
- Zulfiqar, S.; Farooq, M.A.; Zhao, T.T.; Wang, P.P.; Tabusam, J.; Wang, Y.H.; Xuan, S.X.; Zhao, J.J.; Chen, X.P.; Shen, S.X.; et al. Virus-Induced Gene Silencing (VIGS): A Powerful Tool for Crop Improvement and Its Advancement towards Epigenetics. Int. J. Mol. Sci. 2023, 24, 5608. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA targeting with CRISPR-Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.A.T.d.; Shen, M.; Yu, X. Tissue Culture and Micropropagation of Tree Peony (Paeonia suffruticosa Andr.). J. Crop Sci. Biotechnol. 2012, 15, 159–168. [Google Scholar] [CrossRef]
- Wang, X.B.; Li, D.Q.; Zhang, D.; Shi, X.H.; Wu, Y.; Qi, Z.Y.; Ding, H.Q.; Zhu, K.Y.; Xia, Y.P.; Zhang, J.P. Improving crucial details and selecting the optimal model for evaluating the chilling requirement of Paeonia lactiflora Pall. at low lati-tudes during four winters. Sci. Hortic. 2020, 265, 109175. [Google Scholar] [CrossRef]
- Ni, W.; Zhao, H.; Xu, W.; Xu, F.; Huang, S.; Wang, R.; Wu, D. First Report of Root Rot Caused by Pleiocarpon algeriense on Peony (Paeonia suffruticosa) in China. Plant Dis. 2023, 107, 2858. [Google Scholar] [CrossRef]
- Zhang, J.P.; Li, D.Q.; Shi, X.H.; Zhang, D.; Qiu, S.; Wei, J.F.; Zhang, J.; Zhou, J.H.; Zhu, K.Y.; Xia, Y.P. Mining and expression analysis of candidate genes involved in regulating the chilling requirement fulfillment of Paeonia lactiflora ‘Hang Baishao’. BMC Plant Biol. 2017, 17, 262. [Google Scholar] [CrossRef]
- Wang, X.B.; Zhang, R.L.; Huang, Q.Y.; Shi, X.H.; Li, D.Q.; Shao, L.M.; Xu, T.; Horvath, D.P.; Xia, Y.P.; Zhang, J.P. Comparative Study on Physiological Responses and Gene Expression of Bud Endodormancy Release Between Two Her-baceous Peony Cultivars (Paeonia lactiflora Pall.) with Contrasting Chilling Requirements. Front. Plant Sci. 2022, 12, 772285. [Google Scholar] [CrossRef]
- Zhang, R.L.; Wang, X.B.; Shi, X.H.; Shao, L.M.; Xu, T.; Xia, Y.P.; Li, D.Q.; Zhang, J.P. Chilling Requirement Validation and Physiological and Molecular Responses of the Bud Endodormancy Release in Paeonia lactiflora ‘Meiju’. Int. J. Mol. Sci. 2021, 22, 8382. [Google Scholar] [CrossRef]
- Park, J.H.; Rhie, Y.H.; Lee, S.Y.; Kim, K.S. Pre-chilling Promotes Flowering in Paeonia lactiflora ‘Taebaek’ without Flower Bud Abortion. Hortic. Environ. Biotechnol. 2015, 56, 1–8. [Google Scholar] [CrossRef]
- Kumagai, M.H.; Donson, J.; Dellacioppa, G.; Harvey, D.; Hanley, K.; Grill, L.K. Cytoplasmic Inhibition of Carotenoid Biosynthesis with Virus-Derived RNA. Proc. Natl. Acad. Sci. USA 1995, 92, 1679–1683. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Zhang, Q.; Sun, D.; Yang, W.; Hu, J.; Niu, L.; Zhang, Y. Virus-induced gene silencing in the perennial woody Paeonia ostii. PeerJ 2019, 7, e7001. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Fang, Z.; Liu, M.; Zhao, D.; Tao, J. Color characteristics, pigment accumulation and biosynthetic analyses of leaf color variation in herbaceous peony Paeonia lactiflora Pall.). 3 Biotech 2020, 10, 76. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, S.; Ma, H.; Duan, X.; Gao, S.; Zhou, X.; Cheng, Y. The R2R3-MYB gene PsMYB58 positively regulates an-thocyanin biosynthesis in tree peony flowers. Plant Physiol. Biochem. 2021, 164, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.S.; Niu, Q.F.; Li, J.Z.; Zheng, X.Y.; Ma, Y.J.; Bai, S.L.; Teng, Y.W. PpHB22, a member of HD-Zip proteins, activates PpDAM1 to regulate bud dormancy transition in ‘Suli’ pear Pyres pyrifolia White Pear Group). Plant Physiol. Biochem. 2018, 127, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Gao, S.; Xiong, C.; Yu, G.; Chang, J.; Ye, Z.; Yang, C. Comprehensive analysis and expression profile of the homeodomain leucine zipper IV transcription factor family in tomato. Plant Physiol. Biochem. 2015, 96, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Sun, M.; Zhao, G.; Yang, F.; Long, X.; Li, J.; Wang, J.; Yu, Y. Origin of the mafic microgranular enclaves (MMEs) and their host granitoids from the Tagong pluton in Songpan-Ganze terrane: An igneous response to the closure of the Paleo-Tethys Ocean. Lithos 2017, 290, 1–17. [Google Scholar] [CrossRef]
- Zhong, X.H.; Yuan, X.; Wu, Z.; Khan, M.A.; Chen, J.; Li, X.X.; Gong, B.H.; Zhao, Y.; Wu, J.; Wu, C.; et al. Virus-induced gene silencing for comparative functional studies in Gladiolus hybridus. Plant Cell Rep. 2014, 33, 301–312. [Google Scholar] [CrossRef]
- Tian, J.; Pei, H.; Zhang, S.; Chen, J.; Chen, W.; Yang, R.; Meng, Y.; You, J.; Gao, J.; Ma, N. TRVGFP: A modified Tobacco rattle virus vector for efficient and visualizable analysis of gene function. J. Exp. Bot. 2014, 65, 311–322. [Google Scholar] [CrossRef]
- Tian, J.; Cheng, L.; Han, Z.-y.; Yao, Y.-c. Tobacco rattle virus mediated gene silencing in strawberry plants. Plant Cell Tissue Organ Cult. 2015, 120, 1131–1138. [Google Scholar] [CrossRef]
- Halevy, A.H.; Barzilay, A.; Kamenetsky, R. Flowering advancement in herbaceous peony. In Proceedings of the 9th International Symposium on Flower Bulbs, Niigata, Japan, 19–22 April 2004; pp. 279–285. [Google Scholar]
- Ryu, C.M.; Anand, A.; Kang, L.; Mysore, K.S. Agrodrench: A novel and effective Agroinoculation method for virus-induced gene silencing in roots and diverse Solanaceous species. Plant J. 2004, 40, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.H.; Shu, X.C.; Wang, Z.; Wang, N.; Zhang, F.J. Establishing a Virus-Induced Gene Silencing System in Lycoris chinensis. Plants 2023, 12, 2458. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Kumar, P.; Jiang, C.Z.; Reid, M.S. TRV Based Virus Induced Gene Silencing in Gladiolus grandiflorus L.), A Monocotyledonous Ornamental Plant. Vegetos 2013, 26, 170–174. [Google Scholar] [CrossRef]
- Pan, W.Q.; Li, J.R.; Du, Y.P.; Zhao, Y.J.; Xin, Y.; Wang, S.K.; Liu, C.; Lin, Z.M.; Fang, S.Z.; Yang, Y.D.; et al. Epigenetic silencing of callose synthase by VIL1 promotes bud-growth transition in lily bulbs. Nature Plants 2023, 9, 1451–1467. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Jin, Y.J.; Liu, C.; Vonapartis, E.; Liang, J.H.; Wu, W.J.; Gazzarrini, S.; He, J.N.; Yi, M.F. GhNAC83 inhibits corm dormancy release by regulating ABA signaling and cytokinin biosynthesis in Gladiolus hybridus. J. Exp. Bot. 2019, 70, 1221–1237. [Google Scholar] [CrossRef]
- Guan, S.X.; Kang, X.N.; Ge, J.Y.; Fei, R.W.; Duan, S.Y.; Sun, X.M. An efficient Agrobacterium-mediated transient transformation system and its application in gene function elucidation in Paeonia lactiflora Pall. Front. Plant Sci. 2022, 13, 999433. [Google Scholar] [CrossRef]



| Primer Names | Nucleotide Sequences (5′-3′) | Purposes | Sizes |
|---|---|---|---|
| PlHB31 | F: AAGGTTACCGAATTCTCTAGA CTGCAACTGCCACAGGAACT R: GGCCTCGAGACGCGTGAGCTC TGTCAGTATCTGATCCGCCG | Cloning inserted fragment and q-RT PCR analysis | 247 bp |
| TRV1 | F: AAGGTTACCGAATTCTCTAGA TTACAGGTTATTTGGGCTAG R: GGCCTCGAGACGCGTGAGCTC CCGGGTTCAATTCCTTATC | RT-PCR analysis | 647 bp |
| TRV2 | F: AAGGTTACCGAATTCTCTAGA TGGGAGATGATACGCTGTT R: GGCCTCGAGACGCGTGAGCTC CCTAAAACTTCAGACACG | RT-PCR analysis (containing inserted fragment) | >281 bp |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, K.; Wang, X.; Chen, X.; Zhang, R.; Guo, J.; Wang, Q.; Li, D.; Shao, L.; Shi, X.; Han, J.; et al. Establishment of a Homologous Silencing System with Intact-Plant Infiltration and Minimized Operation for Studying Gene Function in Herbaceous Peonies. Int. J. Mol. Sci. 2024, 25, 4412. https://doi.org/10.3390/ijms25084412
Zhang K, Wang X, Chen X, Zhang R, Guo J, Wang Q, Li D, Shao L, Shi X, Han J, et al. Establishment of a Homologous Silencing System with Intact-Plant Infiltration and Minimized Operation for Studying Gene Function in Herbaceous Peonies. International Journal of Molecular Sciences. 2024; 25(8):4412. https://doi.org/10.3390/ijms25084412
Chicago/Turabian StyleZhang, Kaijing, Xiaobin Wang, Xiaoxuan Chen, Runlong Zhang, Junhong Guo, Qiyao Wang, Danqing Li, Lingmei Shao, Xiaohua Shi, Jingtong Han, and et al. 2024. "Establishment of a Homologous Silencing System with Intact-Plant Infiltration and Minimized Operation for Studying Gene Function in Herbaceous Peonies" International Journal of Molecular Sciences 25, no. 8: 4412. https://doi.org/10.3390/ijms25084412
APA StyleZhang, K., Wang, X., Chen, X., Zhang, R., Guo, J., Wang, Q., Li, D., Shao, L., Shi, X., Han, J., Liu, Z., Xia, Y., & Zhang, J. (2024). Establishment of a Homologous Silencing System with Intact-Plant Infiltration and Minimized Operation for Studying Gene Function in Herbaceous Peonies. International Journal of Molecular Sciences, 25(8), 4412. https://doi.org/10.3390/ijms25084412








