Multi-System-Level Analysis with RNA-Seq on Pterygium Inflammation Discovers Association between Inflammatory Responses, Oxidative Stress, and Oxidative Phosphorylation
Abstract
:1. Introduction
2. Results
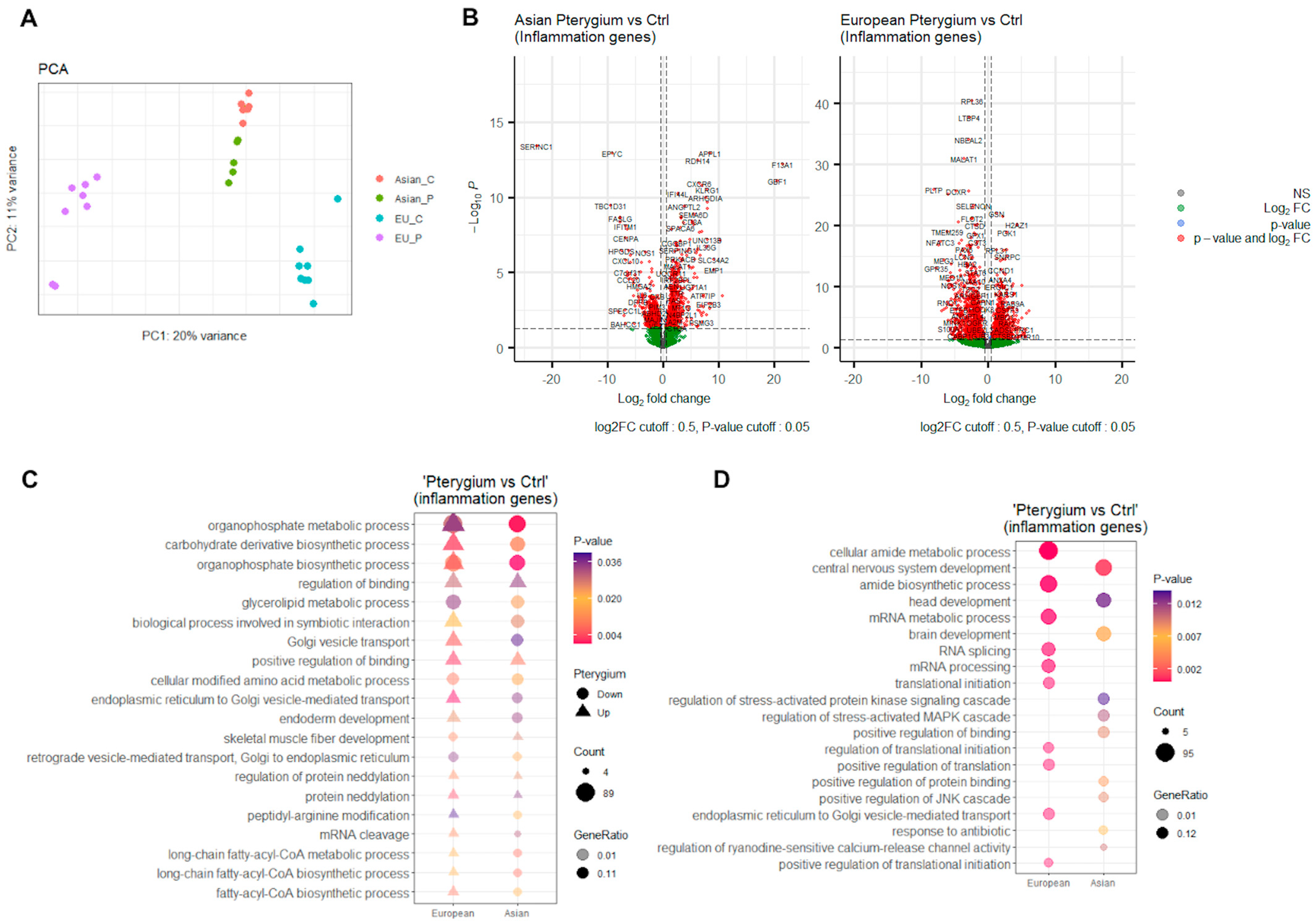
2.1. Inflammation-Associated Differentially Expressed Genes Might Have Significant Roles
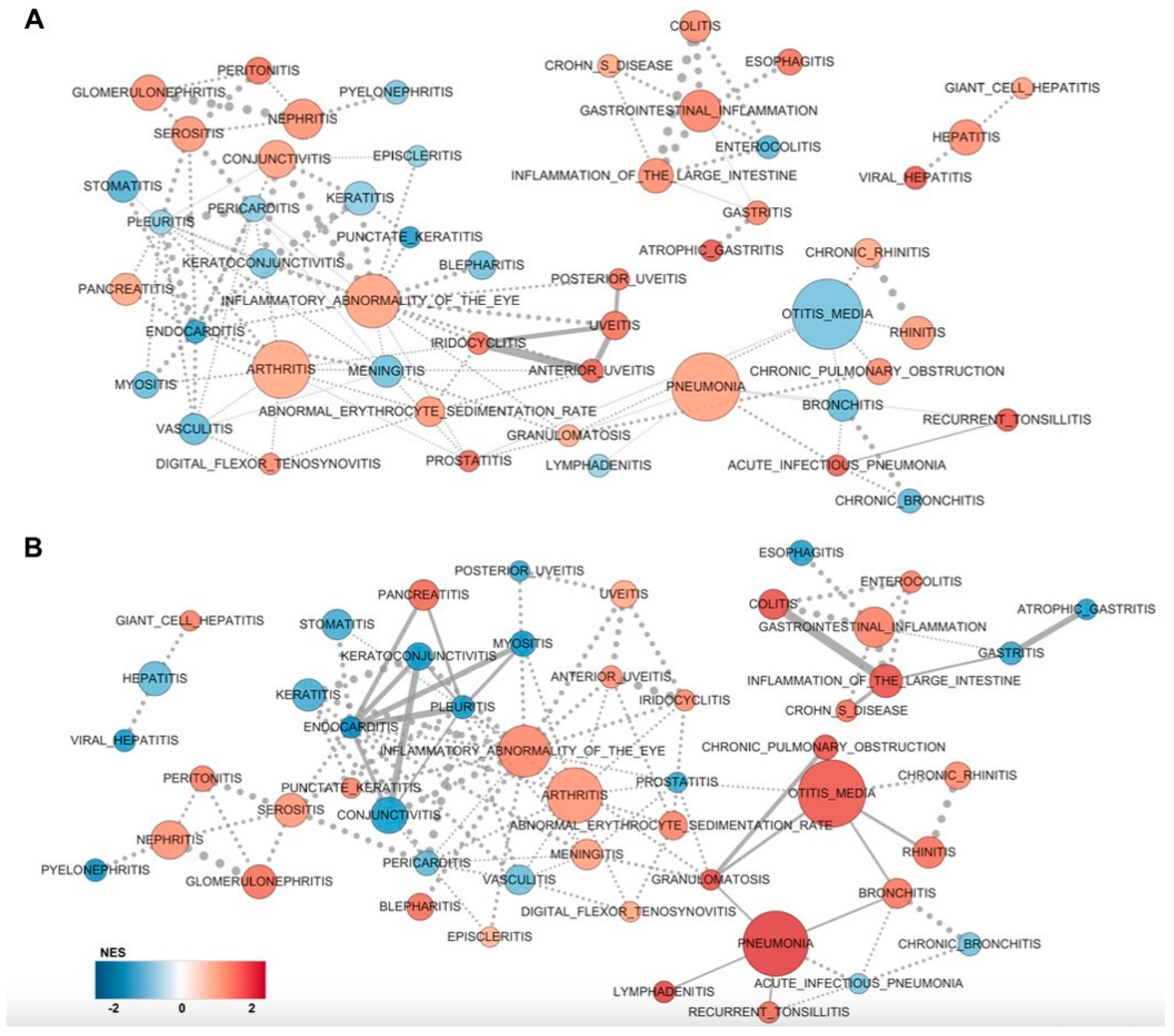
2.2. GSEA Clusters Revealed Eye-Associated Inflammatory Responses
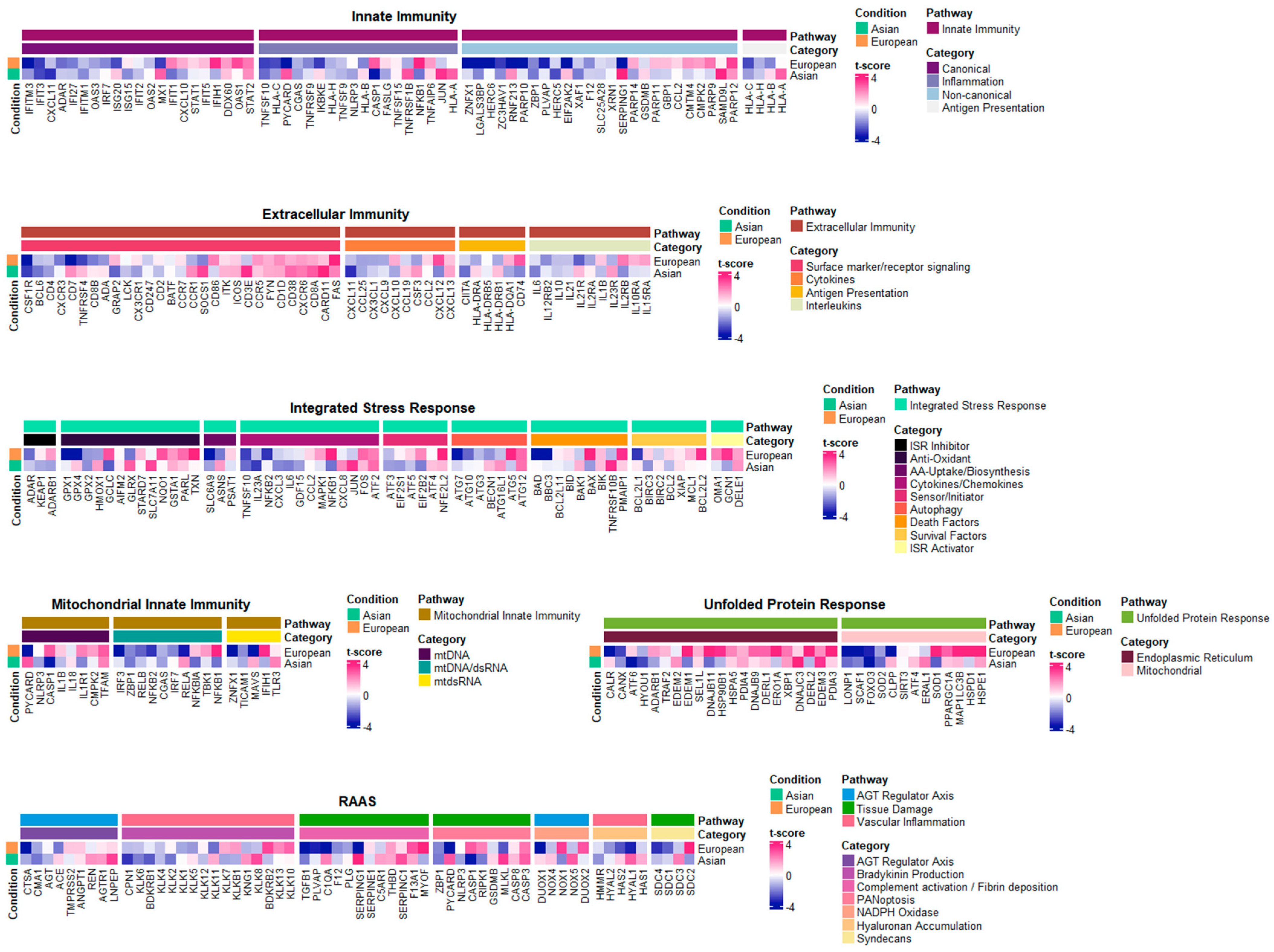
2.3. Core Inflammatory Genes of the Asian Pterygiums Demonstrated Significant Alterations on Inflammatory Responses and Oxidative Stress
2.4. Inflammatory Pathways Displayed Dynamic Alterations on Asian Pterygiums
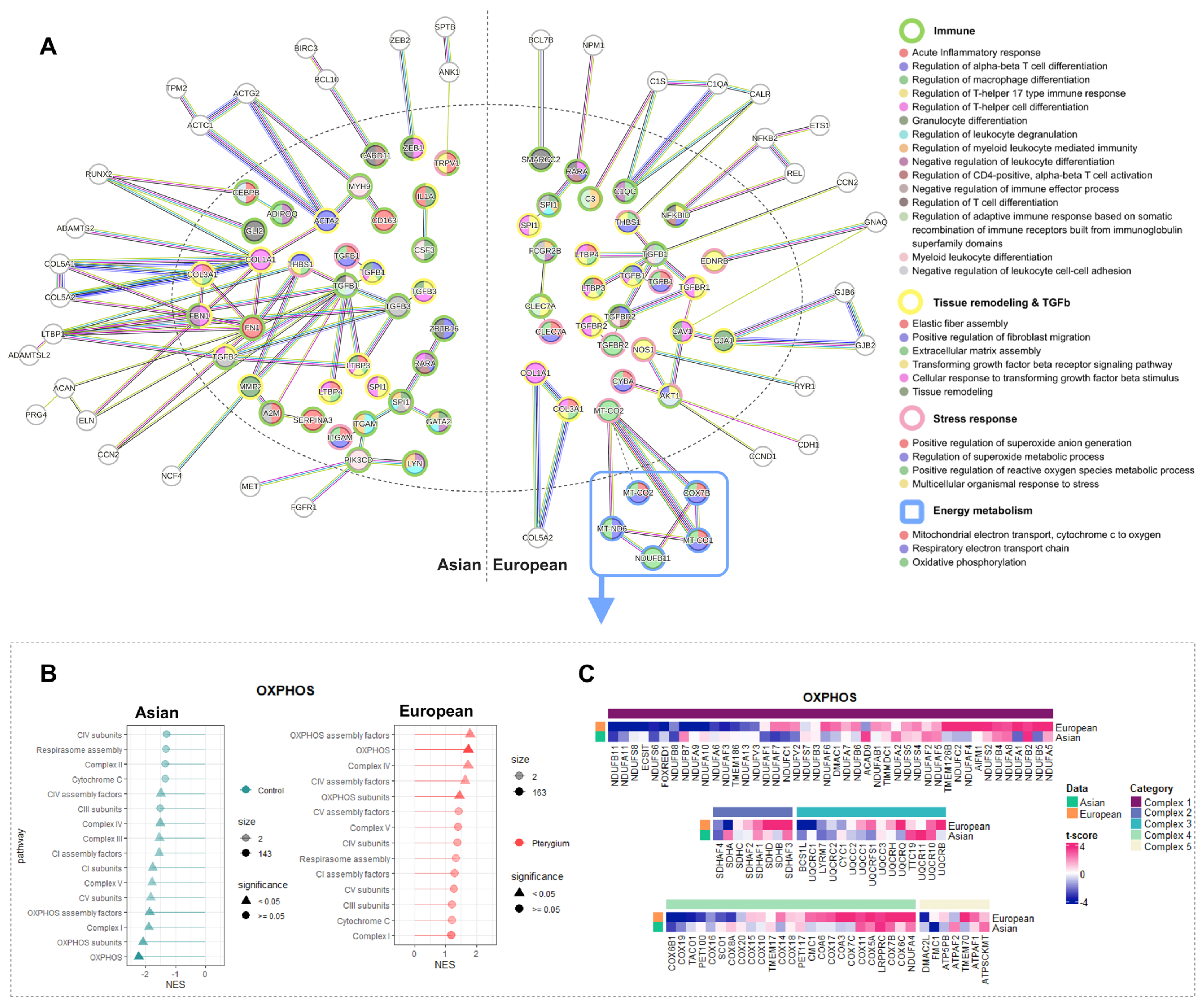
2.5. Compatible Patterns between the Asian and European Pterygiums Are Associated with Oxidative Stress
2.6. Oxidative Phosphorylation Might Cause the Different Responses against Oxidative Stress in European Pterygiums
2.7. Correlated Inflammatory Modules Might Play an Important Role
3. Discussion
4. Materials and Methods
4.1. Patient Samples
4.2. RNA-Seq Preparation and Preprocessing
4.3. Differential and Enrichment Analysis
4.4. Correlation Analysis between Inflammatory-Modules
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chui, J.; Di Girolamo, N.; Wakefield, D.; Coroneo, M.T. The pathogenesis of pterygium: Current concepts and their therapeutic implications. Ocul. Surf. 2008, 6, 24–43. [Google Scholar] [CrossRef] [PubMed]
- Dushku, N.; John, M.K.; Schultz, G.S.; Reid, T.W. Pterygia pathogenesis: Corneal invasion by matrix metalloproteinase expressing altered limbal epithelial basal cells. Arch. Ophthalmol. 2001, 119, 695–706. [Google Scholar] [CrossRef] [PubMed]
- Chui, J.; Coroneo, M.T.; Tat, L.T.; Crouch, R.; Wakefield, D.; Girolamo, N.D. Ophthalmic pterygium: A stem cell disorder with premalignant features. Am. J. Pathol. 2011, 178, 817–827. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, F.D.; Hirst, L.W.; Battistutta, D.; Green, A. Risk analysis in the development of pterygia. Ophthalmology 1992, 99, 1056–1061. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, J.; Nie, D.; Zeng, K.; Hu, H.; Tie, J.; Sun, L.; Peng, L.; Liu, X.; Wang, J. Comparative Transcriptomic Analysis to Identify the Important Coding and Non-coding RNAs Involved in the Pathogenesis of Pterygium. Front. Genet. 2021, 12, 646550. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Qiao, C.; He, S.; Lu, C.; Dong, S.; Wu, X.; Yan, M.; Zheng, F. Identification of Functional Genes in Pterygium Based on Bioinformatics Analysis. Biomed. Res. Int. 2020, 2020, 2383516. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ding, X.; Yuan, L.; Zhang, X. Identification of pterygium-related mRNA expression profiling by microarray analysis. Eye 2017, 12, 1733–1739. [Google Scholar] [CrossRef] [PubMed]
- Engelsvold, D.H.; Utheim, T.P.; Olstad, O.K.; Gonzalez, P.; Eidet, J.R.; Lyberg, T.; Trøseid, A.M.; Dartt, D.A.; Raeder, S. miRNA and mRNA expression profiling identifies members of the miR-200 family as potential regulators of epithelial-mesenchymal transition in pterygium. Exp. Eye Res. 2013, 115, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Cho, W.; Elbasiony, E.; Singh, R.B.; Mittal, S.K.; Chauhan, S.K. Non-immune and immune functions of interleukin-36γ suppress epithelial repair at the ocular surface. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2022, 36, 1–13. [Google Scholar] [CrossRef]
- Yu, F.; Zhang, W.; Yan, C.; Yan, D.; Zhou, M.; Chen, J.; Zhao, X.; Zhu, A.; Zhou, J.; Liu, H.; et al. PAX6, modified by SUMOylation, plays a protective role in corneal endothelial injury. Cell Death Dis. 2020, 11, 683. [Google Scholar] [CrossRef]
- Huang, J.; Yang, Y.; Fang, F.; Liu, K. MALAT1 modulates the autophagy of retinoblastoma cell through miR-124-mediated stx17 regulation. J. Cell Biochem. 2018, 119, 3853–3863. [Google Scholar] [CrossRef] [PubMed]
- Guarnieri, J.W.; Topper, M.J.; Beigel, K.; Haltom, J.A.; Chadburn, A.; Frere, J.; An, J.; Cope, H.; Borczuk, A.; Sinha, S.; et al. Lethal COVID-19 Associates with RAAS-Induced Inflammation for Multiple Organ Damage Including Mediastinal Lymph Nodes. bioRxiv 2023. [Google Scholar] [CrossRef]
- Fan, Z.; Fan, C.; Qi, B.; Zhang, B.; Li, W.; Qi, X.; Liu, X.; Zhang, B.N.; Huang, Y. Sympathetic Nerve-Mediated Fellow Eye Pain During Sequential Cataract Surgery by Regulating Granulocyte Colony Stimulating Factor CSF3. Front. Cell. Neurosci. 2022, 16, 841733. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.R.; Mahdi, R.R.; Oh, H.M.; Amadi-Obi, A.; Levy-Clarke, G.; Burton, J.; Eseonu, A.; Lee, Y.; Chan, C.C.; Egwuagu, C.E. Suppressor of cytokine signaling-1 (SOCS1) inhibits lymphocyte recruitment into the retina and protects SOCS1 transgenic rats and mice from ocular inflammation. Investig. Ophthalmol. Vis. Sci. 2011, 52, 6978–6986. [Google Scholar] [CrossRef] [PubMed]
- Akira, M.; Takahiro, Y.; Masaki, T.; Nobuhisa, M. Contribution of HLA-A and HLA-B genes to genetic predisposition in ocular Behçet’s disease. Investig. Ophthalmol. Vis. Sci. 2018, 59, 6049. [Google Scholar]
- Chen, G.; Yan, F.; Wei, W.; Wang, F.; Wang, Z.; Nie, J.; Jin, M.; Pang, Y.; Qin, M.; Wang, L.; et al. CD38 deficiency protects the retina from ischaemia/reperfusion injury partly via suppression of TLR4/MyD88/NF-κB signaling. Exp. Eye Res. 2022, 219, 109058. [Google Scholar] [CrossRef] [PubMed]
- Syc-Mazurek, S.B.; Fernandes, K.A.; Libby, R.T. JUN is important for ocular hypertension-induced retinal ganglion cell degeneration. Cell Death Dis. 2017, 8, e2945. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, H.L.; Woods, A.; Clowers, J.S.; Planck, S.R.; Rosenbaum, J.T. The NLRP3 inflammasome is active but not essential in endotoxin-induced uveitis. Inflamm. Res. 2012, 61, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Burgaletto, C.; Platania, C.B.M.; Di Benedetto, G.; Munafo, A.; Giurdanella, G.; Federico, C.; Caltabiano, R.; Saccone, S.; Conti, F.; Bernardini, R.; et al. Targeting the miRNA-155/TNFSF10 network restrains inflammatory response in the retina in a mouse model of Alzheimer’s disease. Cell Death Dis. 2021, 12, 905. [Google Scholar] [CrossRef]
- Ennis, S.; Jomary, C.; Mullins, R.; Cree, A.; Chen, X.; Macleod, A.; Jones, S.; Collins, A.; Stone, E.; Lotery, A. Association between the SERPING1 gene and age-related macular degeneration: A two-stage case-control study. Lancet 2008, 372, 1828–1834. [Google Scholar] [CrossRef]
- Russo, R.C.; Garcia, C.C.; Teixeira, M.M.; Amaral, F.A. The CXCL8/IL-8 chemokine family and its receptors in inflammatory diseases. Expert. Rev. Clin. Immunol. 2014, 10, 593–619. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, H. Roles of IL-6 in Ocular Inflammation: A Review. Ocul. Immunol. Inflamm. 2018, 26, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Walker, G.B.; Kurji, K.; Fang, E.; Law, G.; Prasad, S.S.; Kojic, L.; Cao, S.; White, V.; Cui, J.Z.; et al. Parainflammation associated with advanced glycation endproduct stimulation of RPE in vitro: Implications for age-related degenerative diseases of the eye. Cytokine 2013, 62, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Saada, J.; McAuley, R.J.; Marcatti, M.; Tang, T.Z.; Motamedi, M.; Szczesny, B. Oxidative stress induces Z-DNA-binding protein 1-dependent activation of microglia via mtDNA released from retinal pigment epithelial cells. J. Biol. Chem. 2022, 298, 101523. [Google Scholar] [CrossRef] [PubMed]
- Ming-Xuan, W.; Jing, Z.; Hong, Z.; Ke, L.; Ling-Zhi, N.; Yuan-Ping, W.; Ya-Juan, Z. Potential Protective and Therapeutic Roles of the Nrf2 Pathway in Ocular Diseases: An Update. Oxidative Med. Cell. Longev. 2020, 2020, 9410952. [Google Scholar] [CrossRef] [PubMed]
- Dikoglu, E.; Alfaiz, A.; Gorna, M.; Bertola, D.; Chae, J.H.; Cho, T.J.; Derbent, M.; Alanay, Y.; Guran, T.; Kim, O.H.; et al. Mutations in LONP1, a mitochondrial matrix protease, cause CODAS syndrome. Am. J. Med. Genet. A 2015, 167, 1501–1509. [Google Scholar] [CrossRef]
- Kroeger, H.; Chiang, W.C.; Felden, J.; Nguyen, A.; Lin, J.H. ER stress and unfolded protein response in ocular health and disease. FEBS J. 2019, 286, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.E.; DeWeerd, A.J.; Ildefonso, C.J.; Lewin, A.S.; Ash, J.D. Mitochondrial oxidative stress in the retinal pigment epithelium (RPE) led to metabolic dysfunction in both the RPE and retinal photoreceptors. Redox Biol. 2019, 24, 101201. [Google Scholar] [CrossRef]
- Thomas, V.; Claus, C.; Maria, N. Autophagy in the limbal epithelium during UV-induced stem cell damage. Investig. Ophthalmol. Vis. Sci. 2021, 62, 870. [Google Scholar]
- Christensen, N.J.; Demharter, S.; Machado, M.; Pederson, L.; Salvatore, M.; Stentoft-Hansen, V.; Iglesias, M.T. Identifying interactions in omics data for clinical biomarker discovery using symbolic regression. Bioinformatics 2022, 38, 3749–3758. [Google Scholar] [CrossRef]
- Seifert, U.; Bialy, L.P.; Ebstein, F.; Bech-Otschir, D.; Voigt, A.; Schröter, F.; Krüger, E. Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell 2010, 142, 613–624. [Google Scholar] [CrossRef]
- Wie, S.H.; Du, P.; Luong, T.Q.; Rought, S.E.; Beliakova-Bethell, N.; Lozach, J.; Woelk, C.H. HIV downregulates interferon-stimulated genes in primary macrophages. J. Interferon Cytokine Res. 2013, 33, 90–95. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular Signatures Database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Li, P.; Li, S.H.; Wu, J.; Zang, W.F.; Dhingra, S.; Sun, L.; Li, R.K. Interleukin-6 downregulation with mesenchymal stem cell differentiation results in loss of immunoprivilege. J. Cell. Mol. Med. 2013, 17, 1136–1145. [Google Scholar] [CrossRef]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016295. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, J.J.; Ma, A.; Lipsky, P. Cytokines and autoimmunity. Nat. Rev. Immunol. 2002, 2, 37–45. [Google Scholar] [CrossRef] [PubMed]
- West, A.P.; Shadel, G.S.; Ghosh, S. Mitochondria in innate immune responses. Nat. Rev. Immunol. 2011, 11, 389–402. [Google Scholar] [CrossRef]
- Grootjans, J.; Kaser, A.; Kaufman, R.J.; Blumberg, R.S. The unfolded protein response in immunity and inflammation. Nat. Rev. Immunol. 2016, 16, 469–484. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Kaufman, R.J. The impact of the unfolded protein response on human disease. J. Cell Biol. 2012, 197, 857–867. [Google Scholar] [CrossRef]
- Ekholm, M.; Kahan, T. The impact of the renin-angiotensin-aldosterone system on inflammation, coagulation, and atherothrombotic complications, and to aggravated COVID-19. Front. Pharmacol. 2021, 12, 640185. [Google Scholar] [CrossRef]
- Ransy, C.; Vaz, C.; Lombès, A.; Bouillaud, F. Use of H2O2 to Cause Oxidative Stress, the Catalase Issue. Int. J. Mol. Sci. 2020, 21, 9149. [Google Scholar] [CrossRef] [PubMed]
- Denning, T.L.; Takaishi, H.; Crowe, S.E.; Boldogh, I.; Jevnikar, A.; Ernst, P.B. Oxidative stress induces the expression of Fas and Fas ligand and apoptosis in murine intestinal epithelial cells. Free Radic. Biol. Med. 2002, 33, 1641–1650. [Google Scholar] [CrossRef]
- Pei, J.; Pan, X.; Wei, G.; Hua, Y. Research progress of glutathione peroxidase family (GPX) in redoxidation. Front. Pharmacol. 2023, 14, 1147414. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Wang, Q.; Xu, J.; Xu, X.; Padilla, M.T.; Ren, G.; Lin, Y. Attenuation of TNFSF10/TRAIL-induced apoptosis by an autophagic survival pathway involving TRAF2-and RIPK1/RIP1-mediated MAPK8/JNK activation. Autophagy 2012, 8, 1811–1821. [Google Scholar] [CrossRef] [PubMed]
- Oshima, T.; Sonoda, K.H.; Nakao, S.; Hijioka, K.; Taniguchi, M.; Ishibashi, T. Protective role for CD1d-reactive invariant natural killer T cells in cauterization-induced corneal inflammation. Investig. Ophthalmol. Vis. Sci. 2008, 49, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Payton, R.; Dai, W.; Lu, L. Hyperosmotic stress-induced ATF-2 activation through Polo-like kinase 3 in human corneal epithelial cells. J. Biol. Chem. 2011, 286, 1951–1958. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, T.; Falkowski, M.; Park, J.W.; Keegan, S.; Elliott, M.; Wang, J.J.; Zhang, S.X. Loss of XBP1 accelerates age-related decline in retinal function and neurodegeneration. Mol. Neurodegener. 2018, 13, 16. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Wang, H.; Wang, Q.; Deng, H. Overexpression of CD38 decreases cellular NAD levels and alters the expression of proteins involved in energy metabolism and antioxidant defense. J. Proteome Res. 2014, 13, 786–795. [Google Scholar] [CrossRef] [PubMed]
- Brunk, U.T.; Svensson, I. Oxidative stress, growth factor starvation and Fas activation may all cause apoptosis through lysosomal leak. Redox Rep. 1999, 4, 3–11. [Google Scholar] [CrossRef]
- Zhao, T.; Singhal, S.S.; Piper, J.T.; Cheng, J.; Pandya, U.; Clark-Wronski, J.; Awasthi, Y.C. The role of human glutathione S-transferases hGSTA1-1 and hGSTA2-2 in protection against oxidative stress. Arch. Biochem. Biophys. 1999, 367, 216–224. [Google Scholar] [CrossRef]
- Hollander, M.C.; Fornace Jr, A.J. Induction of fos RNA by DNA-damaging agents. Cancer Res. 1989, 49, 1687–1692. [Google Scholar] [PubMed]
- Fessler, E.; Eckl, E.M.; Schmitt, S.; Mancilla, I.A.; Meyer-Bender, M.F.; Hanf, M.; Jae, L.T. A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol. Nature 2020, 579, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Adachi, M.; Zhao, S.; Hareyama, M.; Koong, A.C.; Luo, D.; Shinomura, Y. Preventing oxidative stress: A new role for XBP1. Cell Death Differ. 2009, 16, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Liu, Y.; Meng, J.; Wang, X.; Liu, X.; Li, W.; Hou, S. Lgals3bp in Microglia Promotes Retinal Angiogenesis through Pi3k/Akt Pathway During Hypoxia. Investig. Ophthalmol. Vis. Sci. 2022, 63, 25. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Lauer, T.W.; Sick, A.; Hackett, S.F.; Campochiaro, P.A. Oxidative stress modulates complement factor H expression in retinal pigmented epithelial cells by acetylation of FOXO3. J. Biol. Chem. 2007, 282, 22414–22425. [Google Scholar] [CrossRef] [PubMed]
- Sandbach, J.M.; Coscun, P.E.; Grossniklaus, H.E.; Kokoszka, J.E.; Newman, N.J.; Wallace, D.C. Ocular pathology in mitochondrial superoxide dismutase (Sod2)–deficient mice. Investig. Ophthalmol. Vis. Sci. 2001, 42, 2173–2178. [Google Scholar]
- Hou, A.; Lan, W.; Law, K.P.; Khoo, S.C.; Tin, M.Q.; Lim, Y.P.; Tong, L. Evaluation of global differential gene and protein expression in primary Pterygium: S100A8 and S100A9 as possible drivers of a signaling network. PLoS ONE 2014, 9, e97402. [Google Scholar] [CrossRef] [PubMed]
- Maranzana, E.; Barbero, G.; Falasca, A.I.; Lenaz, G.; Genova, M.L. Mitochondrial respiratory supercomplex association limits production of reactive oxygen species from complex I. Antioxid. Redox Signal. 2013, 19, 1469–1480. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, V.S.; Torres, F.F.; Da Silva, D.G.H. FoxO3 and oxidative stress: A multifaceted role in cellular adaptation. J. Mol. Med. 2023, 101, 83–99. [Google Scholar] [CrossRef]
- Petrera, A.; Kern, U.; Linz, D.; Gomez-Auli, A.; Hohl, M.; Gassenhuber, J.; Schilling, O. Proteomic profiling of cardiomyocyte-specific cathepsin A overexpression links cathepsin A to the oxidative stress response. J. Proteome Res. 2016, 15, 3188–3195. [Google Scholar] [CrossRef]
- Le, S.; Fu, X.; Pang, M.; Zhou, Y.; Yin, G.; Zhang, J.; Fan, D. The Antioxidative Role of Chaperone-Mediated Autophagy as a Downstream Regulator of Oxidative Stress in Human Diseases. Technol. Cancer Res. Treat. 2022, 21, 15330338221114178. [Google Scholar] [CrossRef]
- Bang, M.S.; Rho, C.R.; Kang, B.H.; Cho, K.J.; Oh, C.H. Overexpression of CFH gene in pterygiumv patients. Trop. J. Pharm. Res. 2017, 16, 633–639. [Google Scholar] [CrossRef]
- Tong, L.; Zhou, L.; Beuerman, R.; Simonyi, S.; Hollander, D.A.; Stern, M.E. Effects of punctal occlusion on global tear proteins in patients with dry eye. Ocul. Surf. 2017, 15, 736–741. [Google Scholar] [CrossRef]
- Sun, N.; Zhang, H. Pyroptosis in pterygium pathogenesis. Biosci. Rep. 2018, 38, BSR20180282. [Google Scholar] [CrossRef]
- Krishnamoorthy, R.R.; Crawford, M.J.; Chaturvedi, M.M.; Jain, S.K.; Aggarwal, B.B.; Al-Ubaidi, M.R.; Agarwal, N. Photo-oxidative stress down-modulates the activity of nuclear factor-κB via involvement of caspase-1, leading to apoptosis of photoreceptor cells. J. Biol. Chem. 1999, 274, 3734–3743. [Google Scholar] [CrossRef]
- Ivanenkov, Y.A.; Balakin, K.V.; Lavrovsky, Y. Small Molecule Inhibitors of NF- B and JAK/STAT Signal Transduction Pathways as Promising Anti-Inflammatory Therapeutics. Mini Rev. Med. Chem. 2011, 11, 55–78. [Google Scholar] [CrossRef]
- Coskun, M.; Salem, M.; Pedersen, J.; Nielsen, O.H. Involvement of JAK/STAT signaling in the pathogenesis of inflammatory bowel disease. Pharmacol. Res. 2013, 76, 1–8. [Google Scholar] [CrossRef]
- Fenwick, P.S.; Macedo, P.; Kilty, I.C.; Barnes, P.J.; Donnelly, L.E. Effect of JAK inhibitors on release of CXCL9, CXCL10 and CXCL11 from human airway epithelial cells. PLoS ONE 2015, 10, e0128757. [Google Scholar] [CrossRef]
- John, L.; Samuel, C.E. Induction of stress granules by interferon and down-regulation by the cellular RNA adenosine deaminase ADAR1. Virology 2014, 454, 299–310. [Google Scholar] [CrossRef]
- Sun, Y.; Qiao, L.; Xia, H.H.X.; Lin, M.C.; Zou, B.; Yuan, Y.; Wong, B.C. Regulation of XAF1 expression in human colon cancer cell by interferon β: Activation by the transcription regulator STAT1. Cancer Lett. 2008, 260, 62–71. [Google Scholar] [CrossRef]
- Yang, D.; Liang, Y.; Zhao, S.; Ding, Y.; Zhuang, Q.; Shi, Q.; Han, J. ZBP1 mediates interferon-induced necroptosis. Cell. Mol. Immunol. 2020, 17, 356–368. [Google Scholar] [CrossRef]
- Mathieu, N.A.; Paparisto, E.; Barr, S.D.; Spratt, D.E. HERC5 and the ISGylation pathway: Critical modulators of the antiviral immune response. Viruses 2021, 13, 1102. [Google Scholar] [CrossRef]
- Tur, J.; Farrera, C.; Sánchez-Tilló, E.; Vico, T.; Guerrero-Gonzalez, P.; Fernandez-Elorduy, A.; Celada, A. Induction of CIITA by IFN-γ in macrophages involves STAT1 activation by JAK and JNK. Immunobiology 2021, 226, 152114. [Google Scholar] [CrossRef]
- Heinrich, P.C.; Behrmann, I.; Müller-Newen, G.; Schaper, F.; Graeve, L. Interleukin-6-type cytokine signaling through the gp130/Jak/STAT pathway. Biochem. J. 1998, 334, 297–314. [Google Scholar] [CrossRef]
- Riley, J.K.; Takeda, K.; Akira, S.; Schreiber, R.D. Interleukin-10 receptor signaling through the JAK-STAT pathway: Requirement for two distinct receptor-derived signals for anti-inflammatory action. J. Biol. Chem. 1999, 274, 16513–16521. [Google Scholar] [CrossRef]
- O’Shea, J.J.; Plenge, R. JAK and STAT Signaling Molecules in Immunoregulation and Immune-Mediated Disease. Immunity 2012, 36, 542–550. [Google Scholar] [CrossRef]
- Mogensen, T.H. IRF and STAT Transcription Factors—From Basic Biology to Roles in Infection, Protective Immunity, and Primary Immunodeficiencies. Front. Immunol. 2019, 9, 3047. [Google Scholar] [CrossRef]
- Lu, G.F.; Chen, S.C.; Xia, Y.P.; Ye, Z.M.; Cao, F.; Hu, B. Synergistic inflammatory signaling by cGAS may be involved in the development of atherosclerosis. Aging 2021, 13, 5650–5673. [Google Scholar] [CrossRef]
- Hurt, E.M.; Thomas, S.B.; Peng, B.; Farrar, W.L. Integrated molecular profiling of SOD2 expression in multiple myeloma. Blood 2007, 109, 3953–3962. [Google Scholar] [CrossRef]
- Park, M.C.; Jeong, H.; Son, S.H.; Kim, Y.; Han, D.; Goughnour, P.C.; Kim, S. Novel morphologic and genetic analysis of cancer cells in a 3D microenvironment identifies STAT3 as a regulator of tumor permeability barrier function. Cancer Res. 2016, 76, 1044–1054. [Google Scholar] [CrossRef] [PubMed]
- Kanda, N.; Watanabe, S. Prolactin enhances interferon-γ-induced production of CXC ligand 9 (CXCL9), CXCL10, and CXCL11 in human keratinocytes. Endocrinology 2007, 148, 2317–2325. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, M.; Shiromoto, Y.; Ota, H.; Song, C.; Kossenkov, A.V.; Wickramasinghe, J.; Nishikura, K. ADAR1 controls apoptosis of stressed cells by inhibiting Staufen1-mediated mRNA decay. Nat. Struct. Mol. Biol. 2017, 24, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Denzer, L.; Muranyi, W.; Schroten, H.; Schwerk, C. The role of PLVAP in endothelial cells. Cell Tissue Res. 2023, 392, 393–412. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Wei, B.; Liu, Y.; Liu, T.; Zeng, S.; Gan, J.; Qi, G. Depletion of PARP10 inhibits the growth and metastatic potential of oral squamous cell carcinoma. Front. Genet. 2022, 13, 1035638. [Google Scholar] [CrossRef]
- Yu, L.F.; Wang, J.; Zou, B.; Lin, M.C.; Wu, Y.L.; Xia, H.H.; Wong, B.C. XAF1 mediates apoptosis through an extracellular signal-regulated kinase pathway in colon cancer. Cancer Interdiscip. Int. J. Am. Cancer Soc. 2007, 109, 1996–2003. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Hua, H.; Xiao, G.; Yang, X.; Yang, Q.; Jin, L. ZC3HAV1 promotes the proliferation and metastasis via regulating KRAS in pancreatic cancer. Aging 2021, 13, 18482. [Google Scholar] [CrossRef]
- Muendlein, H.I.; Connolly, W.M.; Magri, Z.; Smirnova, I.; Ilyukha, V.; Gautam, A.; Poltorak, A. ZBP1 promotes LPS-induced cell death and IL-1β release via RHIM-mediated interactions with RIPK1. Nat. Commun. 2021, 12, 86. [Google Scholar] [CrossRef]
- Gordon, E.M.; Venkatesan, N.; Salazar, R.; Tang, H.; Schmeidler-Sapiro, K.; Buckley, S.; Warburton, D.; Hall, F.L. Factor XII-induced mitogenesis is mediated via a distinct signal transduction pathway that activates a mitogen-activated protein kinase. Proc. Natl. Acad. Sci. USA 1996, 93, 2174–2179. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Xu, Q.; Kwon, M.J.; Matta, R.; Liu, Y.; Hong, S.C.; Chang, C.H. ERK and p38 MAPK signaling pathways negatively regulate CIITA gene expression in dendritic cells and macrophages. J. Immunol. 2006, 177, 70–76. [Google Scholar] [CrossRef]
- Ueda, T.; Bruchovsky, N.; Sadar, M.D. Activation of the androgen receptor N-terminal domain by interleukin-6 via MAPK and STAT3 signal transduction pathways. J. Biol. Chem. 2002, 277, 7076–7085. [Google Scholar] [CrossRef]
- Raza, A.; Crothers, J.W.; McGill, M.M.; Mawe, G.M.; Teuscher, C.; Krementsov, D.N. Anti-inflammatory roles of p38α MAPK in macrophages are context dependent and require IL-10. J. Leukoc. Biol. 2017, 102, 1219–1227. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, M.; Chen, L.; Yang, K.; Shan, Y.; Zhu, L.; Wang, C. The TAK1-JNK cascade is required for IRF3 function in the innate immune response. Cell Res. 2009, 19, 412–428. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; Du, C.; Wang, J. The cGAS-STING pathway in hematopoiesis and its physiopathological significance. Front. Immunol. 2020, 11, 573915. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.W.; Kuo, C.Y.; Fan, C.C.; Fang, W.C.; Jiang, S.S.; Lo, Y.K.; Lee, A.Y. Overexpression of Lon contributes to survival and aggressive phenotype of cancer cells through mitochondrial complex I-mediated generation of reactive oxygen species. Cell Death Dis. 2013, 4, e681. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Puerto, M.C.; Verhagen, L.P.; Braat, A.K.; Lam, E.F.; Coffer, P.J.; Lorenowicz, M.J. Activation of autophagy by FOXO3 regulates redox homeostasis during osteogenic differentiation. Autophagy 2016, 12, 1804–1816. [Google Scholar] [CrossRef]
- Hempel, N.; Carrico, P.M.; Melendez, J.A. Manganese superoxide dismutase (Sod2) and redox-control of signaling events that drive metastasis. Anti-Cancer Agents Med. Chem. 2011, 11, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Kanda, N.; Shimizu, T.; Tada, Y.; Watanabe, S. IL-18 enhances IFN-γ-induced production of CXCL9, CXCL10, and CXCL11 in human keratinocytes. Eur. J. Immunol. 2007, 37, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shoshan, S.O.; Kagan, P.; Sultan, M.; Barabash, Z.; Dor, C.; Jacob-Hirsch, J.; Safran, M. ADAR1 deletion induces NF κ B and interferon signaling dependent liver inflammation and fibrosis. RNA Biol. 2017, 14, 587–602. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.S.; Park, M.R.; Sun, E.G.; Choi, W.; Hwang, J.E.; Bae, W.K.; Rhee, J.H.; Cho, S.H.; Chung, I.J. Gal-3BP negatively regulates NF-κB signaling by inhibiting the activation of TAK1. Front. Immunol. 2019, 10, 1760. [Google Scholar] [CrossRef]
- Forlani, G.; Shallak, M.; Gatta, A.; Shaik, A.K.; Accolla, R.S. The NLR member CIITA: Master controller of adaptive and intrinsic immunity and unexpected tool in cancer immunotherapy. Biomed. J. 2023, 46, 100631. [Google Scholar] [CrossRef]
- Matsusaka, T.; Fujikawa, K.; Nishio, Y.; Mukaida, N.; Matsushima, K.; Kishimoto, T.; Akira, S. Transcription factors NF-IL6 and NF-kappa B synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc. Natl. Acad. Sci. USA 1993, 90, 10193–10197. [Google Scholar] [CrossRef] [PubMed]
- Tamassia, N.; Castellucci, M.; Rossato, M.; Gasperini, S.; Bosisio, D.; Giacomelli, M.; Bazzoni, F. Uncovering an IL-10-dependent NF-KB recruitment to the IL-1ra promoter that is impaired in STAT3 functionally defective patients. FASEB J. 2010, 24, 1365–1375. [Google Scholar] [CrossRef] [PubMed]
- Thanos, D.; Maniatis, T. NF-κB: A lesson in family values. Cell 1995, 80, 529–532. [Google Scholar] [CrossRef] [PubMed]
- Alpay, M.; Wanju, K. The Importance of Protein Expression SOD2 in Response to Oxidative Stress for Different Cancer Cells. Kafkas Üniversitesi Vet. Fakültesi Derg. 2014, 20, 507–511. [Google Scholar]
- Di Bona, D.; Cippitelli, M.; Fionda, C.; Cammà, C.; Licata, A.; Santoni, A.; Craxì, A. Oxidative stress inhibits IFN-α-induced antiviral gene expression by blocking the JAK–STAT pathway. J. Hepatol. 2006, 45, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Wada, T.; Penninger, J.M. Mitogen-activated protein kinases in apoptosis regulation. Oncogene 2004, 23, 2838–2849. [Google Scholar] [CrossRef] [PubMed]
- Kerstin, N.S.; Paul, A.; Peter, C.; Patrick, A.B. The roles of hydrogen peroxide and superoxide as messengers in activation of transcription factor NF-kB. Chem. Biol. 1995, 2, 13–22. [Google Scholar]
- Gestal-Mato, U.; Herhaus, L. Autophagy-dependent regulation of MHC-I molecule presentation. J. Cell. Biochem. 2023. early view. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Liao, Y.; Xiao, L.; Wu, R.; Zhao, S.; Chen, H.; Yuan, Z. Autophagy regulates MAVS signaling activation in a phosphorylation-dependent manner in microglia. Cell Death Differ. 2017, 24, 276–287. [Google Scholar] [CrossRef]
- He, Q.; Cai, Y.; Huang, J.; He, X.; Han, W.; Chen, W. Impairment of autophagy promotes human conjunctival fibrosis and pterygium occurrence via enhancing SQSTM1–NF-κB signaling pathway. J. Mol. Cell Biol. 2023, 15, mjad009. [Google Scholar] [CrossRef]
- Li, Y.; Jin, R.; Li, L.; Choi, J.S.; Kim, J.; Yoon, H.J.; Yoon, K.C. Blue light induces impaired autophagy through nucleotide-binding oligomerization domain 2 activation on the mouse ocular surface. Int. J. Mol. Sci. 2021, 22, 2015. [Google Scholar] [CrossRef] [PubMed]
- Lindqvist, L.M.; Vaux, D.L. BCL2 and related prosurvival proteins require BAK1 and BAX to affect autophagy. Autophagy 2014, 10, 1474–1475. [Google Scholar] [CrossRef] [PubMed]
- Petrovski, G.; Zahuczky, G.; Májai, G.; Fésüs, L. Phagocytosis of cells dying through autophagy evokes a pro-inflammatory response in macrophages. Autophagy 2007, 3, 508–510. [Google Scholar] [CrossRef] [PubMed]
- Hou, A.; Tin, M.; Tong, L. Profiling expression of NF-κB signaling target genes in IKK-2 inhibitor treated pterygium and conjunctiva fibroblast cells. Investig. Ophthalmol. Vis. Sci. 2014, 55, 6306. [Google Scholar]
- Shayegan, M.R.; Khakzad, M.R.; Gharaee, H.; Varasteh, A.R.; Sankian, M. Evaluation of transforming growth factor-beta1 gene expression in pterygium tissue of atopic patients. J. Chin. Med. Assoc. 2016, 79, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Wahl, S.M. Transforming growth factor beta (TGF-β) in inflammation: A cause and a cure. J. Clin. Immunol. 1992, 12, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Larrayoz, I.M.; Rúa, O.; Velilla, S.; Martínez, A. Transcriptomic profiling explains racial disparities in pterygium patients treated with doxycycline. Investig. Ophthalmol. Vis. Sci. 2014, 55, 7553–7561. [Google Scholar] [CrossRef] [PubMed]
- Wolf, J.; Hajdu, R.I.; Boneva, S.; Schlecht, A.; Lapp, T.; Wacker, K.; Agostini, H.; Reinhard, T.; Auw-Hädrich, C.; Schlunck, G.; et al. Characterization of the Cellular Microenvironment and Novel Specific Biomarkers in Pterygia Using RNA Sequencing. Front. Med. 2022, 8, 714458. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef] [PubMed]
- Kevin, B.; Sharmila, R.; Myles, L. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. 2018. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 13 August 2023).
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Merico, D.; Isserlin, R.; Stueker, O.; Emili, A.; Bader, G.D. Enrichment Map: A Network-Based Method for Gene-Set Enrichment Visualization and Interpretation. PLoS ONE 2010, 5, e13984. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Korotkevich, G.; Sukhov, V.; Budin, N.; Shpak, B.; Artyomov, M.N.; Sergushichev, A. Fast Gene Set Enrichment Analysis. bioRxiv 2021. [Google Scholar] [CrossRef]
- Rath, S.; Sharma, R.; Gupta, R.; Ast, T.; Chan, C.; Durham, T.J.; Goodman, R.P.; Grabarek, Z.; Haas, M.E.; Hung, W.H.; et al. MitoCarta3.0: An updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 2021, 49, D1541–D1547. [Google Scholar] [CrossRef]
- Guarnieri, J.W.; Dybas, J.M.; Fazelinia, H.; Kim, M.S.; Frere, J.; Zhang, Y.; Soto Albrecht, Y.; Murdock, D.G.; Angelin, A.; Singh, L.N.; et al. Core mitochondrial genes are down-regulated during SARS-CoV-2 infection of rodent and human hosts. Sci. Transl. Med. 2023, 15, eabq1533. [Google Scholar] [CrossRef]



| Asian (Korean) | European (Germany) | Inflammation | Eye Disease or Pterygium Pathogenesis | Oxidative Stress or DNA Damage |
|---|---|---|---|---|
| up | up | EI; FYN, CD1D, CD38, CXCR6, CD8A, FAS ISR; GSTA1, FOS, ATF2, DELE1 UPR; PDIA4, XBP1, EDEM3 | FYN [44], CD1D [45], ATF2 [46], XBP1 [47] | CD38 [48], FAS [49], GSTA1 [50], FOS [51], DELE1 [52], XBP1 [53] |
| down | down | II; IFITM3, IFIT3, LGALS3BP, ZC3HAV1, ZBP1 ISR; SLC6A9 MII; ZBP1 UPR; LONP1, SCAF1, FOXO3, SOD2 RAAS; CTSA, CMA1, ZBP1 | LGALS3BP [54], FOXO3 [55], SOD2 [56], CMA1 [57] | ZBP1 [24], SCAF1 [58], FOXO3 [59], SOD2 [28], CTSA [60], CMA1 [61] |
| up | down | MII; MAVS RAAS; C1QA, SERPING1, HYAL1 | C1QA [62], SERPING1 [63] | |
| down | up | II; CASP1, NFKB1 MII; CASP1, IL1R1 ISR; ADARB1, EIF2B2 RAAS; SDC2 | CASP1 [64] | CASP1 [65] |
| Case NO | Age Mean (std) | Sex | Tissue (Diagnosis) | Race |
|---|---|---|---|---|
| 1 | 61 | F | Primary pterygium/healthy conjunctiva | Asian (Korea) |
| 2 | 65 | F | Primary pterygium/healthy conjunctiva | Asian (Korea) |
| 3 | 61 | M | Primary pterygium/healthy conjunctiva | Asian (Korea) |
| 4 | 72 | M | Primary pterygium/healthy conjunctiva | Asian (Korea) |
| 5 | 72 | M | Primary pterygium/healthy conjunctiva | Asian (Korea) |
| 6 | 64 | F | Primary pterygium/healthy conjunctiva | Asian (Korea) |
| 7–14 | 57.6 (8.5) | 6/2 (M/F) | Primary pterygium | European (Germany) |
| 15–22 | 55.8 (7.9) | 6/2 (M/F) | Healthy conjunctiva | European (Germany) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.-A.; Choi, Y.; Kim, T.G.; Jeong, J.; Yu, S.; Kim, T.; Sheen, K.; Lee, Y.; Choi, T.; Park, Y.H.; et al. Multi-System-Level Analysis with RNA-Seq on Pterygium Inflammation Discovers Association between Inflammatory Responses, Oxidative Stress, and Oxidative Phosphorylation. Int. J. Mol. Sci. 2024, 25, 4789. https://doi.org/10.3390/ijms25094789
Kim Y-A, Choi Y, Kim TG, Jeong J, Yu S, Kim T, Sheen K, Lee Y, Choi T, Park YH, et al. Multi-System-Level Analysis with RNA-Seq on Pterygium Inflammation Discovers Association between Inflammatory Responses, Oxidative Stress, and Oxidative Phosphorylation. International Journal of Molecular Sciences. 2024; 25(9):4789. https://doi.org/10.3390/ijms25094789
Chicago/Turabian StyleKim, Ye-Ah, Yueun Choi, Tae Gi Kim, Jisu Jeong, Sanghyeon Yu, Taeyoon Kim, Kisung Sheen, Yoonsung Lee, Taesoo Choi, Yong Hwan Park, and et al. 2024. "Multi-System-Level Analysis with RNA-Seq on Pterygium Inflammation Discovers Association between Inflammatory Responses, Oxidative Stress, and Oxidative Phosphorylation" International Journal of Molecular Sciences 25, no. 9: 4789. https://doi.org/10.3390/ijms25094789
APA StyleKim, Y.-A., Choi, Y., Kim, T. G., Jeong, J., Yu, S., Kim, T., Sheen, K., Lee, Y., Choi, T., Park, Y. H., Kang, M. S., & Kim, M. S. (2024). Multi-System-Level Analysis with RNA-Seq on Pterygium Inflammation Discovers Association between Inflammatory Responses, Oxidative Stress, and Oxidative Phosphorylation. International Journal of Molecular Sciences, 25(9), 4789. https://doi.org/10.3390/ijms25094789






