The Use of a Novel Container for Secured Transport and Storage of Biological Material for Quantitative Human RNA Analysis
Abstract
1. Introduction
2. Results
2.1. General Properties of the Container
2.2. Study Design
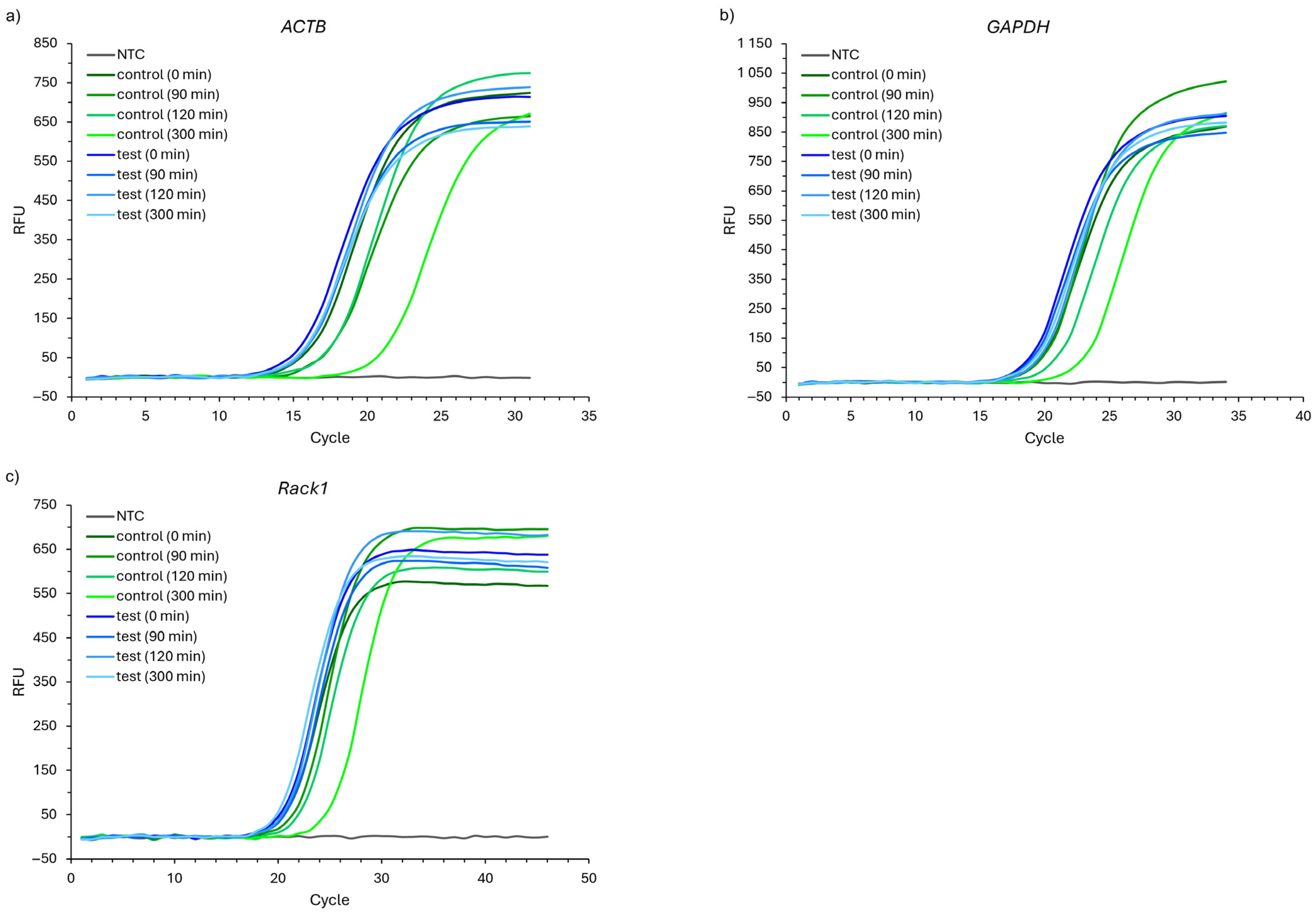
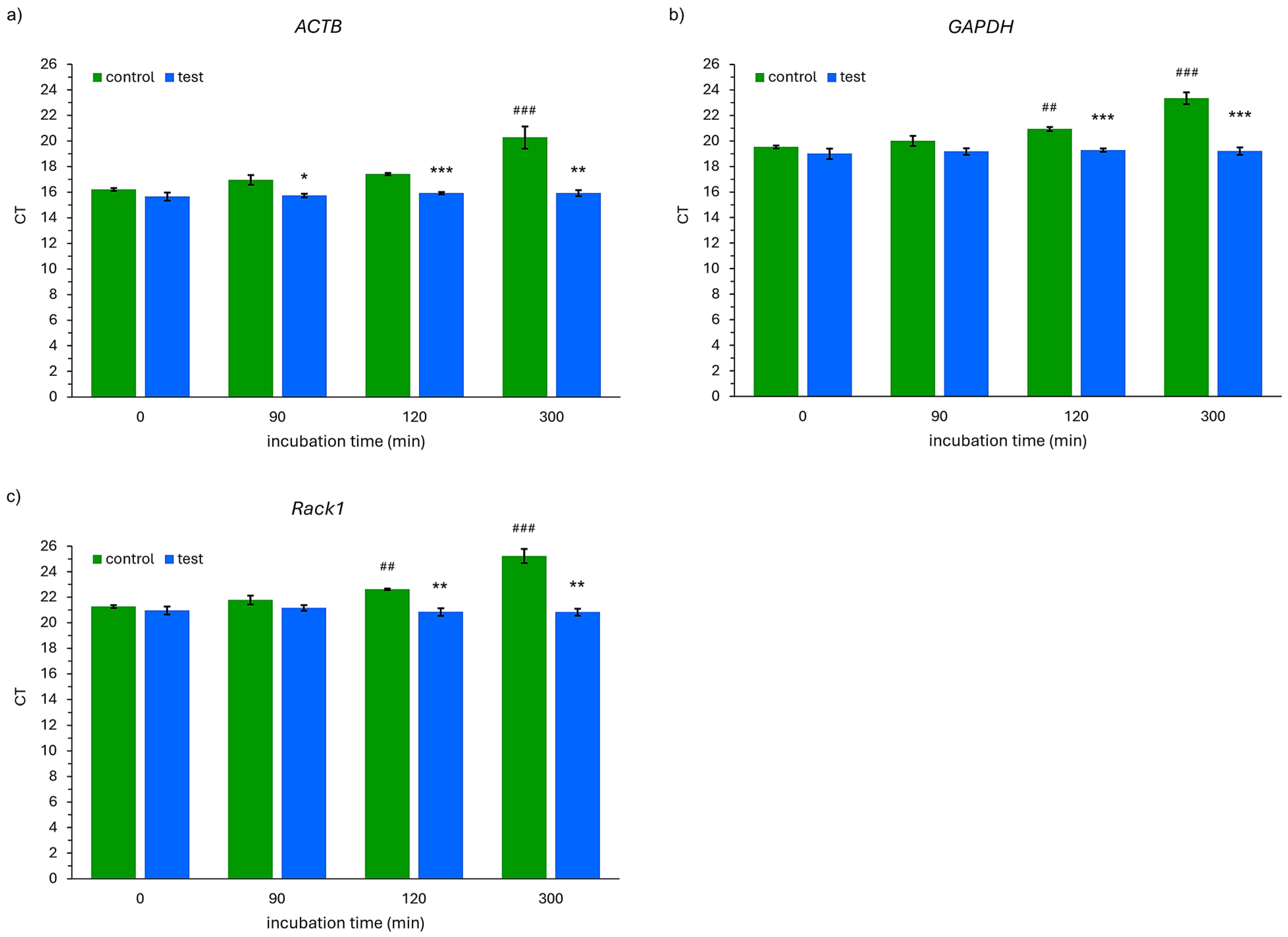
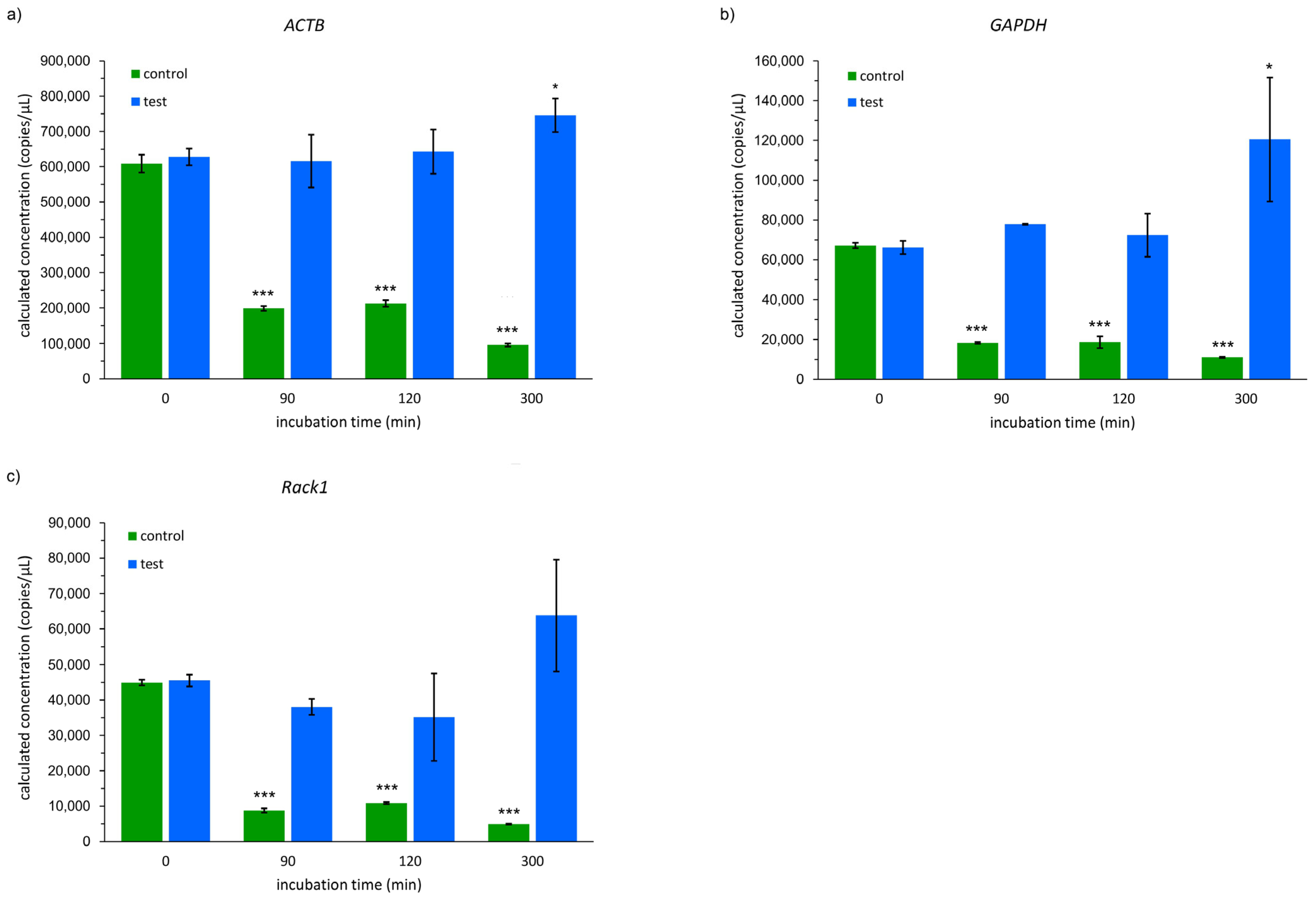
2.3. The Results of RNA Quantitative Analyses
3. Discussion
4. Materials and Methods
4.1. Container Construction
4.2. Analysis of RNA Stability in the Container
4.2.1. Isolation of RNA from Peripheral Nuclear Blood Cells
4.2.2. RNA Integrity Measurement
4.2.3. Reverse Transcription
4.2.4. qPCR Protocol
4.2.5. RT-dPCR Protocol
4.3. Statistical Analysis
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Martins, R.; Queiroz, J.A.; Sousa, F. Ribonucleic Acid Purification. J. Chromatogr. A 2014, 1355, 1–14. [Google Scholar] [CrossRef]
- Cerkovnik, P.; Perhavec, A.; Zgajnar, J.; Novakovic, S. Optimization of an RNA Isolation Procedure from Plasma Samples. Int. J. Mol. Med. 2007, 20, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Fabre, A.L.; Colotte, M.; Luis, A.; Tuffet, S.; Bonnet, J. An Efficient Method for Long-Term Room Temperature Storage of RNA. Eur. J. Hum. Genet. 2014, 22, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Sakanashi, D.; Asai, N.; Nakamura, A.; Miyazaki, N.; Kawamoto, Y.; Ohno, T.; Yamada, A.; Koita, I.; Suematsu, H.; Hagihara, M.; et al. Comparative Evaluation of Nasopharyngeal Swab and Saliva Specimens for the Molecular Detection of SARS-CoV-2 RNA in Japanese Patients with COVID-19. J. Infect. Chemother. 2021, 27, 126–129. [Google Scholar] [CrossRef] [PubMed]
- Khurshid, Z.; Asiri, F.Y.I.; Al Wadaani, H. Human Saliva: Non-Invasive Fluid for Detecting Novel Coronavirus (2019-NCoV). Int. J. Environ. Res. Public Health 2020, 17, 2225. [Google Scholar] [CrossRef] [PubMed]
- Azzi, L.; Carcano, G.; Gianfagna, F.; Grossi, P.; Gasperina, D.D.; Genoni, A.; Fasano, M.; Sessa, F.; Tettamanti, L.; Carinci, F.; et al. Saliva Is a Reliable Tool to Detect SARS-CoV-2. J. Infect. 2020, 81, e45–e50. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.W.; Tsang, O.T.Y.; Yip, C.C.Y.; Chan, K.H.; Wu, T.C.; Chan, J.M.C.; Leung, W.S.; Chik, T.S.H.; Choi, C.Y.C.; Kandamby, D.H.; et al. Consistent Detection of 2019 Novel Coronavirus in Saliva. Clin. Infect. Dis. 2020, 71, 841–843. [Google Scholar] [CrossRef] [PubMed]
- Sri Santosh, T.; Parmar, R.; Anand, H.; Srikanth, K.; Saritha, M. A Review of Salivary Diagnostics and Its Potential Implication in Detection of COVID-19. Cureus 2020, 12, e7708. [Google Scholar] [CrossRef]
- Rodríguez, A.; Duyvejonck, H.; van Belleghem, J.D.; Gryp, T.; van Simaey, L.; Vermeulen, S.; van Mechelen, E.; Vaneechoutte, M. Comparison of Procedures for RNA-Extraction from Peripheral Blood Mononuclear Cells. PloS ONE 2020, 15, e0229423. [Google Scholar] [CrossRef]
- Catts, V.S.; Catts, S.V.; Fernandez, H.R.; Taylor, J.M.; Coulson, E.J.; Lutze-Mann, L.H. A Microarray Study of Post-Mortem MRNA Degradation in Mouse Brain Tissue. Mol. Brain Res. 2005, 138, 164–177. [Google Scholar] [CrossRef]
- Chai, V.; Vassilakos, A.; Lee, Y.; Wright, J.A.; Young, A.H. Optimization of the PAXgeneTM Blood RNA Extraction System for Gene Expression Analysis of Clinical Samples. J. Clin. Lab. Anal. 2005, 19, 182. [Google Scholar] [CrossRef]
- Rollins, B.; Martin, M.V.; Morgan, L.; Vawter, M.P. Analysis Of Whole Genome Biomarker Expression In Blood And Brain. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2010, 153B, 919. [Google Scholar] [CrossRef]
- Weber, D.G.; Casjens, S.; Rozynek, P.; Lehnert, M.; Zilch-Schöneweis, S.; Bryk, O.; Taeger, D.; Gomolka, M.; Kreuzer, M.; Otten, H.; et al. Assessment of MRNA and MicroRNA Stabilization in Peripheral Human Blood for Multicenter Studies and Biobanks. Biomark. Insights 2010, 5, 95. [Google Scholar] [CrossRef]
- Lamot, L.; Niemietz, I.; Brown, K.L. Comparable Type I Interferon Score Determination from PAXgene and Tempus Whole Blood RNA Collection and Isolation Systems. BMC Res. Notes 2019, 12, 511. [Google Scholar] [CrossRef] [PubMed]
- Franken, C.; Remy, S.; Lambrechts, N.; Hollanders, K.; Den Hond, E.; Schoeters, G. Peripheral Blood Collection: The First Step towards Gene Expression Profiling. Biomarkers 2016, 21, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Yip, L.; Fuhlbrigge, R.; Atkinson, M.A.; Fathman, C.G. Impact of Blood Collection and Processing on Peripheral Blood Gene Expression Profiling in Type 1 Diabetes. BMC Genom. 2017, 18, 636. [Google Scholar] [CrossRef] [PubMed]
- Richards, J.; Unger, E.R.; Rajeevan, M.S. Simultaneous Extraction of MRNA and MicroRNA from Whole Blood Stabilized in Tempus Tubes. BMC Res. Notes 2019, 12, 39. [Google Scholar] [CrossRef]
- Guidance on Regulations for the Transport of Infectious Substances. 2021–2022. Available online: https://www.who.int/publications/i/item/9789240019720 (accessed on 17 August 2022).
- Murariu, M.; Dubois, P. PLA Composites: From Production to Properties. Adv. Drug Deliv. Rev. 2016, 107, 17–46. [Google Scholar] [CrossRef] [PubMed]
- Kojabad, A.A.; Farzanehpour, M.; Galeh, H.E.G.; Dorostkar, R.; Jafarpour, A.; Bolandian, M.; Nodooshan, M.M. Droplet Digital PCR of Viral DNA/RNA, Current Progress, Challenges, and Future Perspectives. J. Med. Virol. 2021, 93, 4182–4197. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Bai, R.; Zhao, Z.; Tao, L.; Ma, M.; Ji, Z.; Jian, M.; Ding, Z.; Dai, X.; Bao, F.; et al. Application of Droplet Digital PCR to Detect the Pathogens of Infectious Diseases. Biosci. Rep. 2018, 38, BSR20181170. [Google Scholar] [CrossRef]
- Kozera, B.; Rapacz, M. Reference Genes in Real-Time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef] [PubMed]
- Kloubert, V.; Rink, L. Selection of an Inadequate Housekeeping Gene Leads to Misinterpretation of Target Gene Expression in Zinc Deficiency and Zinc Supplementation Models. J. Trace Elem. Med. Biol. 2019, 56, 192–197. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, G.C.; Treadway, M.B.; Petrone, A.B.; Tennant, C.S.; Lucke-Wold, N.; Chantler, P.D.; Barr, T.L. Leukocyte Dynamics Influence Reference Gene Stability in Whole Blood: Data-Driven QRT-PCR Normalization Is a Robust Alternative for Measurement of Transcriptional Biomarkers. Lab. Med. 2017, 48, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ding, L.; Sandford, A.J. Selection of Reference Genes for Gene Expression Studies in Human Neutrophils by Real-Time PCR. BMC Mol. Biol. 2005, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Plebani, M. Errors in Clinical Laboratories or Errors in Laboratory Medicine? Clin. Chem. Lab. Med. 2006, 44, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Plebani, M. The Detection and Prevention of Errors in Laboratory Medicine. Ann. Clin. Biochem. 2010, 47, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Mrazek, C.; Lippi, G.; Keppel, M.H.; Felder, T.K.; Oberkofler, H.; Haschke-Becher, E.; Cadamuro, J. Errors within the Total Laboratory Testing Process, from Test Selection to Medical Decision-Making—A Review of Causes, Consequences, Surveillance and Solutions. Biochem. Med. 2020, 30, 020502. [Google Scholar] [CrossRef] [PubMed]
- Maddah, H.A. Polypropylene as a Promising Plastic: A Review. Am. J. Polym. Sci. 2016, 6, 1–11. [Google Scholar]
- Sastri, V.R. Commodity Thermoplastics: Polyvinyl Chloride, Polyolefins, Cycloolefins and Polystyrene. In Plastics in Medical Devices. Properties, Requirements, and Applications, 3rd ed.; Sastri, V.R., Ed.; William Andrew Publishing: Oxford, UK, 2022; pp. 113–166. [Google Scholar] [CrossRef]
- Sastri, V.R. Material Requirements for Plastics Used in Medical Devices. In Plastics in Medical Devices. Properties, Requirements, and Applications, 3rd ed.; Sastri, V.R., Ed.; William Andrew Publishing: Oxford, UK, 2022; pp. 65–112. [Google Scholar] [CrossRef]
- Kostrzewa-Nowak, D.; Gos, W. Practical Applications of Target Costing in a Multidisciplinary R&D Project. Sustainability 2022, 15, 124. [Google Scholar] [CrossRef]
- Abdallah, N.M.A.; Zaki, A.M.; Abdel-Salam, S.A. Stability of MERS-CoV RNA on Spin Columns of RNA Extraction Kit at Room Temperature. Diagn. Microbiol. Infect. Dis. 2020, 98, 115182. [Google Scholar] [CrossRef]
- Mathay, C.; Yan, W.; Chuaqui, R.; Skubitz, A.P.N.; Jeon, J.P.; Fall, N.; Betsou, F.; Barnes, M. Short-Term Stability Study of RNA at Room Temperature. Biopreserv. Biobank. 2012, 10, 532–542. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, Q.; Wang, X.; Zhou, X.; He, X.; Liao, Q.; Zhu, F.; Cheng, L.; Zhang, Y. Evaluation of DNA/RNAshells for Room Temperature Nucleic Acids Storage. Biopreserv. Biobank. 2015, 13, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Daum, L.T.; Worthy, S.A.; Yim, K.C.; Nogueras, M.; Schuman, R.F.; Choi, Y.W.; Fischer, G.W. A Clinical Specimen Collection and Transport Medium for Molecular Diagnostic and Genomic Applications. Epidemiol. Infect. 2011, 139, 1764–1773. [Google Scholar] [CrossRef]
- Foss, L.; Reisen, W.K.; Fang, Y.; Kramer, V.; Padgett, K. Evaluation of Nucleic Acid Preservation Cards for West Nile Virus Testing in Dead Birds. PLoS ONE 2016, 11, e0157555. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brisco, M.J.; Morley, A.A. Quantification of RNA Integrity and Its Use for Measurement of Transcript Number. Nucleic Acids Res. 2012, 40, e144. [Google Scholar] [CrossRef]
- Kaukinen, U.; Lyytikäinen, S.; Mikkola, S.; Lönnberg, H. The Reactivity of Phosphodiester Bonds within Linear Single-Stranded Oligoribonucleotides Is Strongly Dependent on the Base Sequence. Nucleic Acids Res. 2002, 30, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Soukup, G.A.; Breaker, R.R. Relationship between Internucleotide Linkage Geometry and the Stability of RNA. RNA 1999, 5, 1308–1325. [Google Scholar] [CrossRef]
- Seyhan, A.A.; Burke, J.M. Mg2+-Independent Hairpin Ribozyme Catalysis in Hydrated RNA Films. RNA 2000, 6, 189–198. [Google Scholar] [CrossRef]
- Li, Y.; Breaker, R.R. Kinetics of RNA Degradation by Specific Base Catalysis of Transesterification Involving the 2’-Hydroxyl Group. J. Am. Chem. Soc. 1999, 121, 5364–5372. [Google Scholar] [CrossRef]
- Imbeaud, S.; Graudens, E.; Boulanger, V.; Barlet, X.; Zaborski, P.; Eveno, E.; Mueller, O.; Schroeder, A.; Auffray, C. Towards Standardization of RNA Quality Assessment Using User-Independent Classifiers of Microcapillary Electrophoresis Traces. Nucleic Acids Res. 2005, 33, e56. [Google Scholar] [CrossRef]
- Vermeulen, J.; De Preter, K.; Lefever, S.; Nuytens, J.; De Vloed, F.; Derveaux, S.; Hellemans, J.; Speleman, F.; Vandesompele, J. Measurable Impact of RNA Quality on Gene Expression Results from Quantitative PCR. Nucleic Acids Res. 2011, 39, e63. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and Quantifying Mammalian Transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Das, M.; Harvey, I.; Chu, L.L.; Sinha, M.; Pelletier, J. Full-Length CDNAs: More than Just Reaching the Ends. Physiol. Genom. 2001, 6, 57–80. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Taylor, S.C.; Laperriere, G.; Germain, H. Droplet Digital PCR versus QPCR for Gene Expression Analysis with Low Abundant Targets: From Variable Nonsense to Publication Quality Data. Sci. Rep. 2017, 7, 2409. [Google Scholar] [CrossRef] [PubMed]
- Pomari, E.; Piubelli, C.; Perandin, F.; Bisoffi, Z. Digital PCR: A New Technology for Diagnosis of Parasitic Infections. Clin. Microbiol. Infect. 2019, 25, 1510–1516. [Google Scholar] [CrossRef] [PubMed]


| T [min] | Control Probe [RIN] | Test Probe [RIN] | pt 1 |
|---|---|---|---|
| 0 | 9.6 ± 0.1 | 9.7 ± 0.1 | 0.600 |
| 15 | 9.5 ± 0.2 | 9.6 ± 0.1 | 0.534 |
| 30 | 9.5 ± 0.2 | 9.7 ± 0.1 | 0.051 |
| 60 | 9.4 ± 0.1 | 9.6 ± 0.1 | 0.003 |
| 90 | 8.3 ± 0.6 *** | 9.4 ± 0.2 *** | 0.001 |
| 120 | 8.3 ± 0.4 *** | 9.5 ± 0.1 | <0.001 |
| 180 | 8.7 ± 0.2 *** | 9.6 ± 0.1 | <0.001 |
| 240 | 7.8 ± 0.7 *** | 9.5 ± 0.1 | <0.001 |
| 300 | 7.9 ± 0.4 *** | 9.6 ± 0.1 | <0.001 |
| Gene | Gene Name | Gene Function | Sequence (5′→3′) | Length | TM 1 (°C) | TA 2 (°C) | Amplicon Size (bp) | GenBank Accession Number | |
|---|---|---|---|---|---|---|---|---|---|
| ACTB | Actin Beta | Cytoskeletal structural protein | Forward | CATGTACGTTGCTATCCAGGC | 21 | 59.1 | 56 | 250 | NM_001101 |
| Reverse | CTCCTTAATGTCACGCACGAT | 21 | 58.4 | ||||||
| Rack1 | Receptor for activated C kinase 1 | Core ribosomal protein of the eukaryotic small (40S) ribosomal subunit | Forward | GATGTGGCCTTCTCCTCTG | 20 | 59.8 | 60 | 224 | NM_006098 |
| Reverse | GCTTGCATTAGCCAGGTTC | 20 | 59.4 | ||||||
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | Glycolysis pathway enzyme | Forward | GAAGGTGAAGGTCGGAGTC | 19 | 57.2 | 60 | 226 | NM_002046 |
| Reverse | GAAGATGGTGATGGGATTTC | 20 | 53.7 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kostrzewa-Nowak, D.; Trzeciak-Ryczek, A.; Lewandowska, K.; van de Wetering, T.; Ciechanowicz, A.; Nowak, R. The Use of a Novel Container for Secured Transport and Storage of Biological Material for Quantitative Human RNA Analysis. Int. J. Mol. Sci. 2025, 26, 228. https://doi.org/10.3390/ijms26010228
Kostrzewa-Nowak D, Trzeciak-Ryczek A, Lewandowska K, van de Wetering T, Ciechanowicz A, Nowak R. The Use of a Novel Container for Secured Transport and Storage of Biological Material for Quantitative Human RNA Analysis. International Journal of Molecular Sciences. 2025; 26(1):228. https://doi.org/10.3390/ijms26010228
Chicago/Turabian StyleKostrzewa-Nowak, Dorota, Alicja Trzeciak-Ryczek, Klaudyna Lewandowska, Thierry van de Wetering, Andrzej Ciechanowicz, and Robert Nowak. 2025. "The Use of a Novel Container for Secured Transport and Storage of Biological Material for Quantitative Human RNA Analysis" International Journal of Molecular Sciences 26, no. 1: 228. https://doi.org/10.3390/ijms26010228
APA StyleKostrzewa-Nowak, D., Trzeciak-Ryczek, A., Lewandowska, K., van de Wetering, T., Ciechanowicz, A., & Nowak, R. (2025). The Use of a Novel Container for Secured Transport and Storage of Biological Material for Quantitative Human RNA Analysis. International Journal of Molecular Sciences, 26(1), 228. https://doi.org/10.3390/ijms26010228









