Comparison of Respiratory Microbiomes in Influenza Versus Other Respiratory Infections: Systematic Review and Analysis
Abstract
:1. Introduction
- (a)
- To compare the differences in human nasal microbiome composition and species between subjects with and without influenza;
- (b)
- To evaluate changes in the human nasal microbiome composition and species as a predictor of influenza severity;
- (c)
- To evaluate the longitudinal changes in the human nasal microbiome in patients with influenza;
- (d)
- To compare differences in the nasal microbiome composition and species in children versus adults with influenza;
- (e)
- To compare differences in the human nasal microbiome in influenza compared to other respiratory infections (such as COVID-19, pneumonia, and chronic rhinosinusitis).
2. Materials and Methods
2.1. Protocol, Registration and Search Strategies
2.2. Study Selection Process and Criteria
2.3. Data Collection and Analysis
- (a)
- Nasal microbiome composition and species in influenza versus healthy subjects;
- (b)
- The relationship between nasal microbiome and influenza severity:
- (i)
- Signature bacterial markers in severe influenza;
- (ii)
- Microbiome diversity across severe influenza and mild to moderate influenza;
- (c)
- Nasal microbiome composition in adult versus pediatric influenza patients;
- (d)
- Nasal microbiome composition and species in influenza versus other respiratory infections (COVID-19, pneumonia, multiple respiratory infections, or chronic rhinosinusitis).
3. Results
3.1. Search Results and Selection of Studies

3.2. Characteristics of Selected Studies
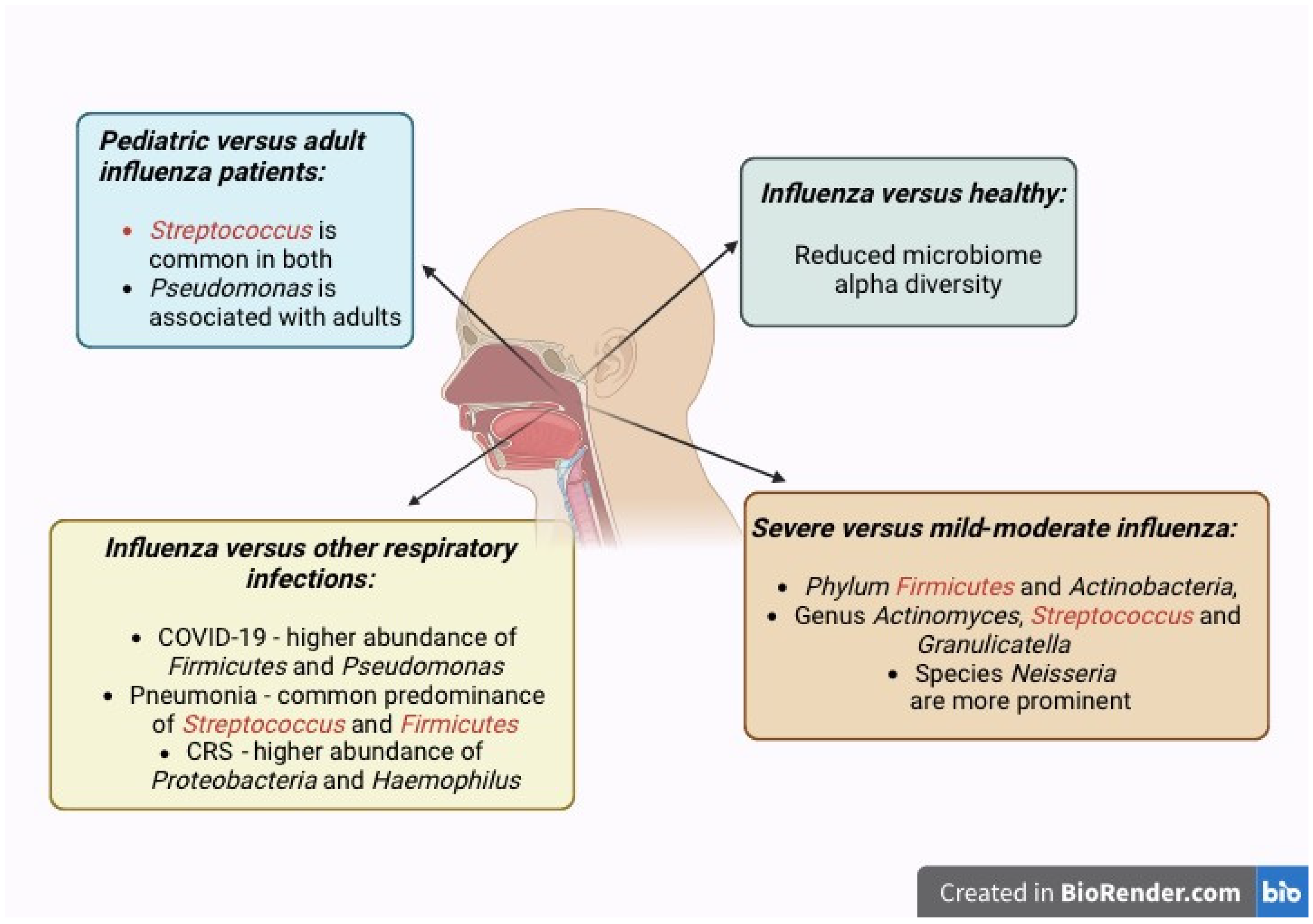
3.3. Differences in Human Nasal Microbiomes Between Individuals with Influenza Versus Without Influenza
3.4. Changes in the Human Nasal Microbiome Composition and Species of Bacteria in Relation to Influenza Severity
3.5. Longitudinal Changes in the Human Nasal Microbiome in Patients with Influenza
3.6. Differences Between Nasal Microbiomes of Children and Adults with Influenza
| Study Subjects | Sample | Gene Sequencing of Variable Region | Signature Microorganisms Associated with Influenza (or Other Respiratory Infections) | References |
|---|---|---|---|---|
| Pediatric patients | ||||
| IAV patients, Healthy controls | OP swab | 16S rRNA V3–V4 | Streptococcus Actinomyces Lactobacillales Veillonellaceae | Hu et al., 2022 [17] |
| IAV patients | NP swab | 16S rRNA | Streptococcus Moraxella Staphylococcus Haemophilus | Langevin et al., 2017 [19] |
| Influenza patients, MP patients, Healthy controls | NP swab OP swab | 16S rRNA V3–V4 | Prevotella | Zhou et al., 2020 [21] |
| Adult patients | ||||
| IAV patients, Healthy controls, Other common virus infections | NP swab | 16S rRNA V1–V3 | Pseudomonas | Kaul et al., 2020 [20] |
| IAV patients, Healthy controls | NP swab OP swab | 16S rRNA V4 | Neisseria | Lee et al., 2019 [24] |
| IAV H7N9 patients, Healthy controls | OP swab Nasal lavage | 16S rRNA | Pseudomonadaceae Fusobacteria Bifidobacteriaceae Bacteroidaceae | Lu et al., 2017 [18] |
| IAV patients, Healthy controls | OP swab | 16S rRNA V1–V3 | Prevotella | Ramos-Sevillano et al., 2019 [22] |
| IAV and IBV patients, COVID-19 patients | NP swab | 16S rRNA V4 | Enterobacteriaceae | Rattanaburi et al., 2022 [25] |
| IAV and IBV patients | NP swab OP swab | 16S rRNA V4 | Prevotella | Tsang et al., 2020 [26] |
| IAV with severe acute respiratory infection | NP aspirate | 16S rRNA V4 | Streptococcus | Borges et al., 2018 [27] |
| Non-IAV with severe acute respiratory infection | ||||
| IAV H1N1 patients | Endotracheal aspirate BAL | 16S rRNA V3–V4 | Proteobacteria Bacteroidetes Firmicutes | Hernández-Terán et al., 2023 [9] |
| IAV H7N9 patients, Healthy controls | OP swab | 16S rRNA V3–V4 | Pseudomonas | Zha et al., 2020 [28] |
| Influenza, parainfluenza, rhinovirus, RSV, COVID-19, adenovirus, metapneumovirus patients, Healthy controls | NP aspirate Sputum OP swab | 16S rRNA V1–V3 | Haemophilus Moraxella | Yi et al., 2014 [23] |
3.7. Differences in the Human Nasal Microbiome in Influenza Compared to Other Respiratory Infections and Conditions (Including COVID-19)
| Study Subjects | Sample | Sequencing Method | Signature Bacteria | References |
|---|---|---|---|---|
| Pneumonia | ||||
| Mycoplasma pneumoniae pneumonia | NP swab OP swab BAL (additional in sick children) | 16S rRNA V3–V4 | Staphylococcus Corynebacterium Mycoplasma | Dai et al., 2018 [30] |
| Healthy controls | ||||
| Pneumonia | NP swab | 16S rRNA V3 | Haemophilus Staphylococcus Streptococcus | Kelly et al., 2017 [31] |
| Healthy controls | ||||
| Pneumonia | NP swab | 16S rRNA V1–V2 | Viral cause: Moraxella lacunata Non-viral cause: Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis | Sakwinska et al., 2014 [29] |
| Healthy controls | ||||
| Community-acquired pneumonia | NP swab OP swab | 16S rRNA V1–V3 | Actinobacteria (Actinomyces), Firmicutes (Streptococcus pneumoniae, Staphylococci) | Weimken et al., 2015 [32] |
| Mycoplasma pneumoniae pneumonia | OP swab Nasal lavage | 16S rRNA | Phylum: Firmicutes Genus: Mycoplasma, Lactobacillus, Ralstonia, Acinetobacter, Actinomyces NP only: Streptococcus OP only: Corynebacterium | Zhou et al., 2020 [21] |
| Influenza | ||||
| Healthy controls | ||||
| COVID-19 | ||||
| IAV and IBV | NP swab | 16S rRNA V4 | Phylum: Firmicutes, Bacteroidetes Genus: Enterobacteriaceae, Staphylococcus, Lautropia, Pseudomonas, Corynebacterium | Rattanaburi et al., 2022 [25] |
| COVID-19 | ||||
| COVID-19-positive respiratory infection | NP swab | 16S rRNA V3–V4 | Phylum: Proteobacteria, Firmicutes, Actinobacteria | Tchoupou Saha et al., 2022 [33] |
| COVID-19 negative respiratory infection | ||||
| Chronic Rhinosinusitis | ||||
| Non-asthmatic CRSwNP patients | Nasal swab | 16S rRNA V1–V2 | Phylum: Proteobacteria Genus: Haemophilus Species: Haemophilus influenzae, Corynebacterium pseudodiphtheriticum | Chalermwatanachai et al., 2018 [34] |
| CRSwNP patients with asthma | ||||
| Healthy controls | ||||
| CRSsNP CRSwNP | Anterior nares, NP, maxillary, and ethmoid sinus swabs (patients) Anterior nares, NP swab (controls) | 16S rRNA V4 | Staphylococcus Corynebacterium Moraxella Haemophilus Streptococcus Prevotella | De Boeck et al., 2019 [35] |
| Healthy controls | ||||
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Man, W.H.; de Steenhuijsen Piters, W.A.A.; Bogaert, D. The microbiota of the respiratory tract: Gatekeeper to respiratory health. Nat. Rev. Microbiol. 2017, 15, 259–270. [Google Scholar] [CrossRef]
- Bassis, C.M.; Tang, A.L.; Young, V.B.; Pynnonen, M.A. The nasal cavity microbiota of healthy adults. Microbiome 2014, 2, 27. [Google Scholar] [CrossRef]
- Chen, M.; He, S.; Miles, P.; Li, C.; Ge, Y.; Yu, X.; Wang, L.; Huang, W.; Kong, X.; Ma, S.; et al. Nasal bacterial microbiome differs between healthy controls and those with asthma and allergic rhinitis. Front. Cell. Infect. Microbiol. 2022, 12, 841995. [Google Scholar] [CrossRef]
- Merenstein, C.; Bushman, F.D.; Collman, R.G. Alterations in the respiratory tract microbiome in COVID-19: Current observations and potential significance. Microbiome 2022, 10, 165. [Google Scholar] [CrossRef]
- Camarinha-Silva, A.; Wos-Oxley, M.L.; Jáuregui, R.; Becker, K.; Pieper, D.H. Validating T-RFLP as a sensitive and high-throughput approach to assess bacterial diversity patterns in human anterior nares. FEMS Microbiol. Ecol. 2012, 79, 98–108. [Google Scholar] [CrossRef]
- Biesbroek, G.; Bosch, A.A.T.M.; Wang, X.; Keijser, B.J.F.; Veenhoven, R.H.; Sanders, E.A.M.; Bogaert, D. The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am. J. Respir. Crit. Care Med. 2014, 190, 298–308. [Google Scholar] [CrossRef]
- Mika, M.; Mack, I.; Korten, I.; Qi, W.; Aebi, S.; Frey, U.; Latzin, P.; Hilty, M. Dynamics of the nasal microbiota in infancy: A prospective cohort study. J. Allergy Clin. Immunol. 2015, 135, 905–912.e11. [Google Scholar] [CrossRef]
- Kumpitsch, C.; Koskinen, K.; Schöpf, V.; Moissl-Eichinger, C. The microbiome of the upper respiratory tract in health and disease. BMC Biol. 2019, 17, 87. [Google Scholar] [CrossRef]
- Hernández-Terán, A.; Vega-Sánchez, A.E.; Mejía-Nepomuceno, F.; Serna-Muñoz, R.; Rodríguez-Llamazares, S.; Salido-Guadarrama, I.; Romero-Espinoza, J.A.; Guadarrama-Perez, C.; Sandoval-Gutierrez, J.L.; Campos, F. Microbiota composition in the lower respiratory tract is associated with severity in patients with acute respiratory distress by influenza. Virol. J. 2023, 20, 19. [Google Scholar] [CrossRef]
- Rhoades, N.S.; Pinski, A.N.; Monsibais, A.N.; Jankeel, A.; Doratt, B.M.; Cinco, I.R.; Ibraim, I.; Messaoudi, I. Acute SARS-CoV-2 infection is associated with an increased abundance of bacterial pathogens, including Pseudomonas aeruginosa in the nose. Cell Rep. 2021, 36, 109637. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Castro-Nallar, E.; Laerte Boechat, J.; Delgado, L.; Azenha Rama, T.; Berrios-Farías, V.; Oliveira, M. The oral bacteriomes of patients with allergic rhinitis and asthma differ from that of healthy controls. Front. Microbiol. 2023, 14, 1197135. [Google Scholar] [CrossRef]
- Wagner Mackenzie, B.; Chang, K.; Zoing, M.; Jain, R.; Hoggard, M.; Biswas, K.; Douglas, R.G.; Taylor, M.W. Longitudinal study of the bacterial and fungal microbiota in the human sinuses reveals seasonal and annual changes in diversity. Sci. Rep. 2019, 9, 17416. [Google Scholar] [CrossRef]
- Yamamoto, S.; Saito, M.; Tamura, A.; Prawisuda, D.; Mizutani, T.; Yotsuyanagi, H. The human microbiome and COVID-19: A systematic review. PLoS ONE 2021, 16, e0253293. [Google Scholar] [CrossRef]
- Paules, C.; Subbarao, K. Influenza. Lancet 2017, 390, 697–708. [Google Scholar] [CrossRef]
- Javanian, M.; Barary, M.; Ghebrehewet, S.; Koppolu, V.; Vasigala, V.; Ebrahimpour, S. A brief review of influenza virus infection. J. Med. Virol. 2021, 93, 4638–4646. [Google Scholar] [CrossRef]
- Peteranderl, C.; Herold, S.; Schmoldt, C. Human influenza virus infections. Semin. Respir. Crit. Care Med. 2016, 37, 487–500. [Google Scholar] [CrossRef]
- Hu, Q.; Liu, B.; Fan, Y.; Zheng, Y.; Wen, F.; Yu, U.; Wang, W. Multi-omics association analysis reveals interactions between the oropharyngeal microbiome and the metabolome in pediatric patients with influenza A virus pneumonia. Front. Cell. Infect. Microbiol. 2022, 12, 1011254. [Google Scholar] [CrossRef]
- Lu, H.F.; Ang, L.; Zhang, T.; Ren, Z.G.; He, K.X.; Zhang, H.; Yang, J.Z.; Luo, Q.X.; Zhou, K.; Chen, C.L.; et al. Disordered oropharyngeal microbial communities in H7N9 patients with or without secondary bacterial lung infection. Emerg. Microbes Infect. 2017, 6, e112. [Google Scholar] [CrossRef]
- Langevin, S.; Pichon, M.; Smith, E.; Morrison, J.; Bent, Z.; Green, R.; Barker, K.; Solberg, O.; Gillet, Y.; Javouhey, E.; et al. Early nasopharyngeal microbial signature associated with severe influenza in children: A retrospective pilot study. J. Gen. Virol. 2017, 98, 2425–2437. [Google Scholar] [CrossRef]
- Kaul, D.; Rathnasinghe, R.; Ferres, M.; Tan, G.S.; Barrera, A.; Pickett, B.E.; Methe, B.A.; Das, S.R.; Budnik, I.; Halpin, R.A.; et al. Microbiome disturbance and resilience dynamics of the upper respiratory tract during influenza A virus infection. Nat. Commun. 2020, 11, 2537. [Google Scholar] [CrossRef]
- Zhou, Q.; Xie, G.; Liu, Y.; Wang, H.; Yang, Y.; Shen, K.; Dai, W.; Li, S.; Zheng, Y. Different nasopharynx and oropharynx microbiota imbalance in children with Mycoplasma pneumoniae or influenza virus infection. Microb. Pathog. 2020, 144, 104189. [Google Scholar] [CrossRef]
- Ramos-Sevillano, E.; Wade, W.G.; Mann, A.; Gilbert, A.; Lambkin-Williams, R.; Killingley, B.; Nguyen-Van-Tam, J.S.; Tang, C.M. The effect of influenza virus on the human oropharyngeal microbiome. Clin. Infect. Dis. 2019, 68, 1993–2002. [Google Scholar] [CrossRef]
- Yi, H.; Yong, D.; Lee, K.; Cho, Y.J.; Chun, J. Profiling bacterial community in upper respiratory tracts. BMC Infect. Dis. 2014, 14, 583. [Google Scholar] [CrossRef]
- Lee, K.H.; Foxman, B.; Kuan, G.; López, R.; Shedden, K.; Ng, S.; Balmaseda, A.; Gordon, A. The respiratory microbiota: Associations with influenza symptomatology and viral shedding. Ann. Epidemiol. 2019, 37, 51–56.e6. [Google Scholar] [CrossRef]
- Rattanaburi, S.; Sawaswong, V.; Chitcharoen, S.; Sivapornnukul, P.; Nimsamer, P.; Suntronwong, N.; Puenpa, J.; Poovorawan, Y.; Payungporn, S. Bacterial microbiota in upper respiratory tract of COVID-19 and influenza patients. Exp. Biol. Med. 2022, 247, 409–415. [Google Scholar] [CrossRef]
- Tsang, T.K.; Lee, K.H.; Foxman, B.; Balmaseda, A.; Gresh, L.; Sanchez, N.; Ojeda, S.; Lopez, R.; Yang, Y.; Kuan, G.; et al. Association between the respiratory microbiome and susceptibility to influenza virus infection. Clin. Infect. Dis. 2020, 71, 1195–1203. [Google Scholar] [CrossRef]
- Borges, L.G.D.A.; Giongo, A.; Pereira, L.D.M.; Trindade, F.J.; Gregianini, T.S.; Campos, F.S.; Ghedin, E.; da Veiga, A.B.G. Comparison of the nasopharynx microbiome between influenza and non-influenza cases of severe acute respiratory infections: A pilot study. Health Sci. Rep. 2018, 1, e47. [Google Scholar] [CrossRef]
- Zha, H.; Lu, H.; Wu, J.; Chang, K.; Wang, Q.; Zhang, H.; Li, J.; Luo, Q.; Lu, Y.; Li, L. Vital members in the more dysbiotic oropharyngeal microbiotas in H7N9-infected patients. Front. Med. 2020, 7, 396. [Google Scholar] [CrossRef]
- Sakwinska, O.; Bastic Schmid, V.; Berger, B.; Bruttin, A.; Keitel, K.; Lepage, M.; Moine, D.; Ngom Bru, C.; Brussow, H.; Gervaix, A. Nasopharyngeal microbiota in healthy children and pneumonia patients. J. Clin. Microbiol. 2014, 52, 1590–1594. [Google Scholar] [CrossRef]
- Dai, W.; Wang, H.; Zhou, Q.; Feng, X.; Lu, Z.; Li, D.; Yang, Z.; Liu, Y.; Li, Y.; Xie, G.; et al. The concordance between upper and lower respiratory microbiota in children with Mycoplasma pneumoniae pneumonia. Emerg. Microbes Infect. 2018, 7, 92. [Google Scholar] [CrossRef]
- Kelly, M.S.; Surette, M.G.; Smieja, M.; Pernica, J.M.; Rossi, L.; Luinstra, K.; Steenhoff, A.P.; Feemster, K.A.; Goldfarb, D.M.; Arscott-Mills, T.; et al. The nasopharyngeal microbiota of children with respiratory infections in Botswana. Pediatr. Infect. Dis. J. 2017, 36, e211–e218. [Google Scholar] [CrossRef]
- Wiemken, T.L.; Jala, V.R.; Kelley, R.R.; Peyrani, P.; Mattingly, W.A.; Arnold, F.W.; Cabral, P.W.; Cavallazzi, R.; Haribabu, B.; Ramirez, J.A. The upper respiratory tract microbiome of hospitalised patients with community-acquired pneumonia of unknown aetiology: A pilot study. Pneumonia 2015, 6, 83–89. [Google Scholar] [CrossRef]
- Tchoupou Saha, O.F.; Dubourg, G.; Yacouba, A.; Bossi, V.; Raoult, D.; Lagier, J.C. Profile of the nasopharyngeal microbiota affecting the clinical course in COVID-19 patients. Front. Microbiol. 2022, 13, 871627. [Google Scholar] [CrossRef]
- Chalermwatanachai, T.; Vilchez-Vargas, R.; Holtappels, G.; Lacoere, T.; Jáuregui, R.; Kerckhof, F.M.; Pieper, D.H.; Van der Wiele, T.; Vaneechoutte, M.; Van Zele, T.; et al. Chronic rhinosinusitis with nasal polyps is characterized by dysbacteriosis of the nasal microbiota. Sci. Rep. 2018, 8, 7926. [Google Scholar] [CrossRef]
- De Boeck, I.; Wittouck, S.; Martens, K.; Claes, J.; Jorissen, M.; Steelant, B.; van den Broek, M.F.L.; Seys, S.F.; Hellings, P.W.; Vanderveken, O.M.; et al. Anterior nares diversity and pathobionts represent sinus microbiome in chronic rhinosinusitis. mSphere 2019, 4, e00532-19. [Google Scholar] [CrossRef]
- Galiana, A.; Aguirre, E.; Rodriguez, J.C.; Mira, A.; Santibañez, M.; Candela, I.; Llavero, J.; Garcinuno, P.; Lopez, F.; Ruiz, M.; et al. Sputum microbiota in moderate versus severe patients with COPD. Eur. Respir. J. 2014, 43, 1787–1790. [Google Scholar] [CrossRef]
- Garcia-Nuñez, M.; Millares, L.; Pomares, X.; Ferrari, R.; Pérez-Brocal, V.; Gallego, M.; Espasa, M.; Moya, A.; Monso, E. Severity-related changes of bronchial microbiome in chronic obstructive pulmonary disease. J. Clin. Microbiol. 2014, 52, 4217–4223. [Google Scholar] [CrossRef]
- Bomar, L.; Brugger, S.D.; Lemon, K.P. Bacterial microbiota of the nasal passages across the span of human life. Curr. Opin. Microbiol. 2018, 41, 8–14. [Google Scholar] [CrossRef]
- Gupta, A.; Karyakarte, R.; Joshi, S.; Das, R.; Jani, K.; Shouche, Y.; Sharma, A. Nasopharyngeal microbiome reveals the prevalence of opportunistic pathogens in SARS-CoV-2 infected individuals and their association with host types. Microbes Infect. 2022, 24, 104880. [Google Scholar] [CrossRef]
- Mac Aogáin, M. Unsung heroes? Decoding the protective effects of airway microbiota in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2024, 210, 136–138. [Google Scholar] [CrossRef]
- Li, L.; Mac Aogáin, M.; Xu, T.; Jaggi, T.K.; Chan, L.L.Y.; Qu, J.; Wei, L.; Liao, S.; Cheng, H.S.; Keir, H.R.; et al. Neisseria species as pathobionts in bronchiectasis. Cell Host Microbe 2022, 30, 1311–1327.e8. [Google Scholar] [CrossRef]
- Rudd, J.M.; Ashar, H.K.; Chow, V.T.; Teluguakula, N. Lethal synergism between influenza and Streptococcus pneumoniae. J. Infect. Pulm. Dis. 2016, 2. [Google Scholar] [CrossRef]
- Narayana Moorthy, A.; Narasaraju, T.; Rai, P.; Perumalsamy, R.; Tan, K.B.; Wang, S.; Engelward, B.; Chow, V.T. In vivo and in vitro studies on the roles of neutrophil extracellular traps during secondary pneumococcal pneumonia after primary pulmonary influenza infection. Front. Immunol. 2013, 4, 56. [Google Scholar] [CrossRef]
- Jochems, S.P.; Marcon, F.; Carniel, B.F.; Holloway, M.; Mitsi, E.; Smith, E.; Gritzfeld, J.F.; Solórzano, C.; Reiné, J.; Pojar, S.; et al. Inflammation induced by influenza virus impairs human innate immune control of pneumococcus. Nat. Immunol. 2018, 19, 1299–1308. [Google Scholar] [CrossRef]
- Wang, J.; Li, F.; Sun, R.; Gao, X.; Wei, H.; Li, L.J.; Tian, Z. Bacterial colonization dampens influenza-mediated acute lung injury via induction of M2 alveolar macrophages. Nat. Commun. 2013, 4, 2106. [Google Scholar] [CrossRef]
- Avalos-Fernandez, M.; Alin, T.; Métayer, C.; Thiébaut, R.; Enaud, R.; Delhaes, L. The respiratory microbiota alpha-diversity in chronic lung diseases: First systematic review and meta-analysis. Respir. Res. 2022, 23, 214. [Google Scholar] [CrossRef]
- Chen, J.; Li, T.; Ye, C.; Zhong, J.; Huang, J.D.; Ke, Y.; Sun, H. The lung microbiome: A new frontier for lung and brain disease. Int. J. Mol. Sci. 2023, 24, 2170. [Google Scholar] [CrossRef]
- Lira-Lucio, J.A.; Falfán-Valencia, R.; Ramírez-Venegas, A.; Buendía-Roldán, I.; Rojas-Serrano, J.; Mejía, M.; Pérez-Rubio, G. Lung microbiome participation in local immune response regulation in respiratory diseases. Microorganisms 2020, 8, 1059. [Google Scholar] [CrossRef]
- Keir, H.R.; Chalmers, J.D. COUNTERPOINT: Is chronic bacterial infection clinically relevant in COPD? No. Chest 2022, 162, 972–976. [Google Scholar] [CrossRef]
- Yin, T.; Wagner Mackenzie, B.; Radcliff, F.; Broderick, D.; Biswas, K.; Douglas, R. Sinus microbial communities in patients with chronic rhinosinusitis are unpredictable over time. Int. Forum Allergy Rhinol. 2024; in press. [Google Scholar] [CrossRef]

| References | Type of Sample | Disease Severity (n) | Alpha Diversity (Shannon Index) | Alpha Diversity (Simpson Index) | Alpha Diversity (Chao Index) |
|---|---|---|---|---|---|
| Langevin et al., 2017 [19] | NP swab | 14 severe influenza | Severe vs. mild: Influenza outcome = 12.44 vs. 0.0014 | Severe vs. mild: Influenza outcome = 14.66 vs. 0.0006 | Severe vs. mild: Influenza outcome = 10.72 vs. 0.0027 |
| 22 mild influenza | Days since symptom onset = 0.9 vs. 0.3519 | Days since symptom onset = 2.49 vs. 0.1258 | Days since symptom onset = 0.24 vs. 0.6314 | ||
| Lee et al., 2019 [24] | NP swab, OP swab | 124 index cases | Shedding duration: 25th and 75th quartiles of Shannon index = 3.1 and 3.6 days | - | Serial interval: Chao index AF 0.992; 25th and 75th quartiles of Chao index = 3.8 and 3.0 days |
| Serial interval: Shannon index AF 0.72; 25th and 75th quartiles of Shannon index = 3.7 and 3.2 days | Time to shedding onset: AF 0.995; 25th and 75th quartiles of Chao index = 5.8 and 5.2 days | ||||
| Lu et al., 2017 [18] | OP swab, nasal lavage | 21 severe | Graphical representation only | ||
| 30 mild-moderate | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, Y.; Lee, Y.-J.; Yap, K.; Samuel, M.; Chow, V.T. Comparison of Respiratory Microbiomes in Influenza Versus Other Respiratory Infections: Systematic Review and Analysis. Int. J. Mol. Sci. 2025, 26, 778. https://doi.org/10.3390/ijms26020778
Hao Y, Lee Y-J, Yap K, Samuel M, Chow VT. Comparison of Respiratory Microbiomes in Influenza Versus Other Respiratory Infections: Systematic Review and Analysis. International Journal of Molecular Sciences. 2025; 26(2):778. https://doi.org/10.3390/ijms26020778
Chicago/Turabian StyleHao, Yunrui, Ying-Jou Lee, Kihan Yap, Miny Samuel, and Vincent T. Chow. 2025. "Comparison of Respiratory Microbiomes in Influenza Versus Other Respiratory Infections: Systematic Review and Analysis" International Journal of Molecular Sciences 26, no. 2: 778. https://doi.org/10.3390/ijms26020778
APA StyleHao, Y., Lee, Y.-J., Yap, K., Samuel, M., & Chow, V. T. (2025). Comparison of Respiratory Microbiomes in Influenza Versus Other Respiratory Infections: Systematic Review and Analysis. International Journal of Molecular Sciences, 26(2), 778. https://doi.org/10.3390/ijms26020778






