SARS-CoV-2 Variants: Genetic Insights, Epidemiological Tracking, and Implications for Vaccine Strategies
Abstract
1. Introduction
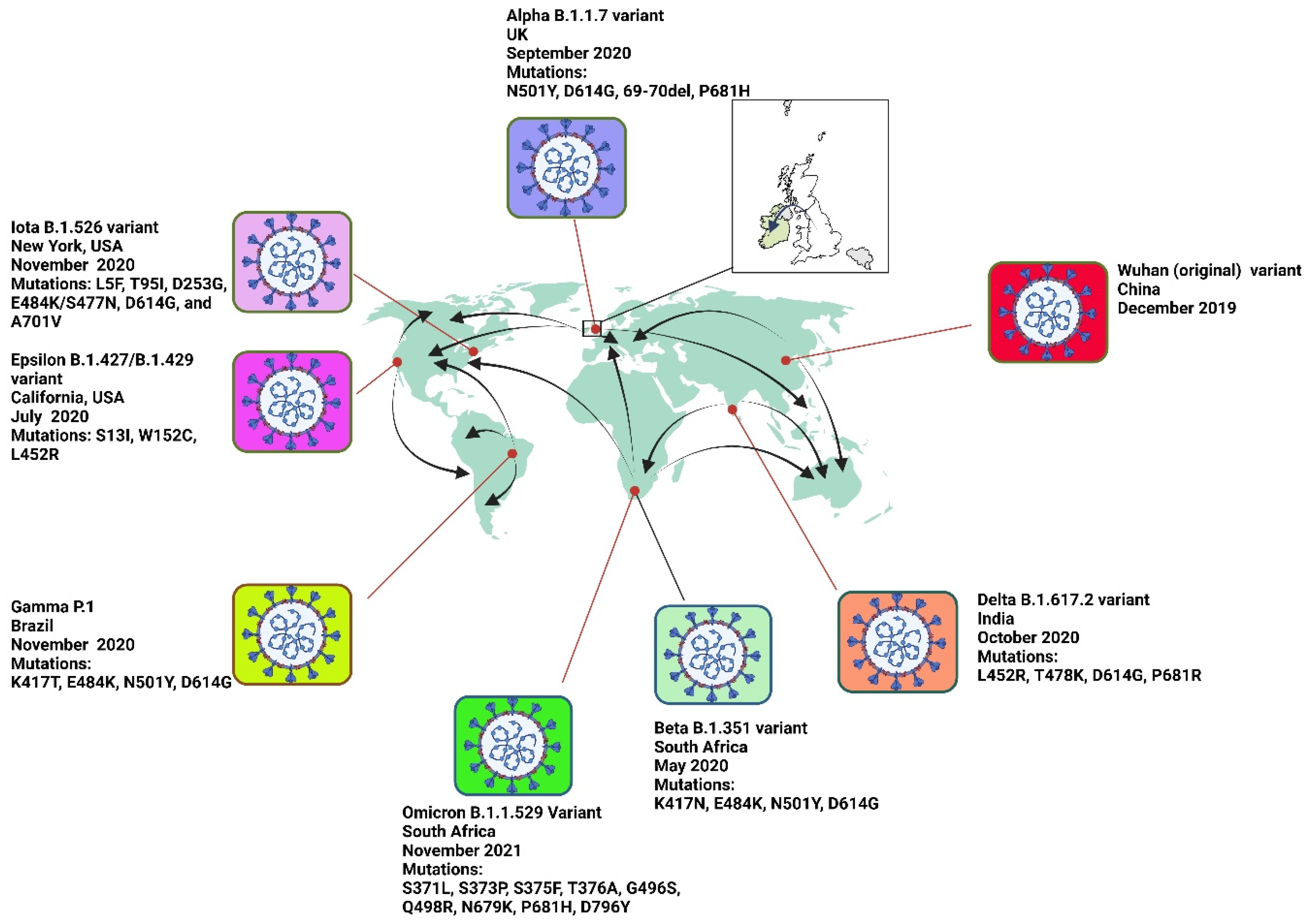
2. SARS-CoV-2 Variants
2.1. Genetic Variability and Evolution
2.1.1. Alpha Variant (B.1.1.7) of SARS-CoV-2
Impact of Mutations on Virulence and Morbidity
2.1.2. Beta Variant (B.1.351) of SARS-CoV-2
Impact of Mutations on Virulence and Morbidity
Beta Variant and Its Impact on Mortality
2.1.3. Gamma Variant (P.1) of SARS-CoV-2
Impact of Mutations on Virulence and Morbidity
Gamma Variant and Its Impact on Mortality
2.1.4. Delta Variant (B.1.617.2) of SARS-CoV-2
Impact of Mutations on Virulence and Morbidity
Delta Variant and Its Impact on Mortality
2.1.5. Omicron Variant (B.1.1.529) of SARS-CoV-2
Mutations and Subvariants
Recombinant XBB Subvariants
Impact of Mutations on Virulence and Morbidity
2.1.6. Non-VOC Variants of SARS-CoV-2
Epsilon Variant (B.1.427/B.1.429)
Iota Variant (B.1.526)
3. Role of SARS-CoV-2 Variants in Viral Fitness
3.1. Increased ACE2 Receptor Affinity
3.2. Enhanced Transmissibility
3.3. Immune Evasion
3.4. Increased Viral Replication
4. Variant Verification and Its Role in Epidemiological Tracking
4.1. Importance of Variant Verification
4.2. Role in Epidemiological Tracking
4.2.1. Monitoring the Spread and Prevalence of Specific Variants
Monitoring the Geographic Distribution
Monitoring Temporal Trends
Population-Specific Variants
4.2.2. Detecting Outbreaks of SARS-CoV-2 Variants
Early Warning Systems
Case Clustering and Hotspots
Rapid Response Strategies
4.2.3. Evaluating Control Measures for SARS-CoV-2 Variants
Effectiveness of Vaccination Campaigns
Impact of Non-Pharmaceutical Interventions (NPIs)
Adjustments to Public Health Policies
4.2.4. Global Surveillance and Collaboration in SARS-CoV-2 Variant Tracking
Data Sharing Platforms
International Cooperation and Coordination
Standardization of Genomic Surveillance Protocols
4.2.5. Public Health Implications of SARS-CoV-2 Variant Verification
Resource Allocation and Planning
Policy Development and Implementation
4.3. Challenges and Limitations of SARS-CoV-2 Variant Verification
4.3.1. Data Quality and Completeness
4.3.2. Ethical and Privacy Considerations
4.3.3. Logistical and Financial Constraints
4.3.4. Methodological Challenges in Genomic and Epidemiological Data Analysis
5. Implications for Vaccine Development and Public Health
5.1. Challenges to Vaccine Effectiveness
5.2. Advancing Vaccine Development
5.3. Vaccine Distribution and Global Health Equity
5.4. Long-Term Pandemic Preparedness
6. Lessons from the Pandemic: Toward a Unified Global Response
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Brant, A.C.; Tian, W.; Majerciak, V.; Yang, W.; Zheng, Z.-M. SARS-CoV-2: From its discovery to genome structure, transcription, and replication. Cell Biosci. 2021, 11, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Cevik, M.; Kuppalli, K.; Kindrachuk, J.; Peiris, M. Virology, transmission, and pathogenesis of SARS-CoV-2. BMJ 2020, 371, m3862. [Google Scholar] [CrossRef]
- Ali, A.; Vijayan, R. Dynamics of the ACE2–SARS-CoV-2/SARS-CoV spike protein interface reveal unique mechanisms. Sci. Rep. 2020, 10, 14214. [Google Scholar] [CrossRef]
- Hoffmann, M.; Arora, P.; Groß, R.; Seidel, A.; Hörnich, B.F.; Hahn, A.S.; Krüger, N.; Graichen, L.; Hofmann-Winkler, H.; Kempf, A.; et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 2021, 184, 2384–2393.e12. [Google Scholar] [CrossRef] [PubMed]
- Pascall, D.J.; Vink, E.; Blacow, R.; Bulteel, N.; Campbell, A.; Campbell, R.; Clifford, S.; Davis, C.; Filipe, A.d.S.; El Sakka, N.; et al. The SARS-CoV-2 Alpha variant was associated with increased clinical severity of COVID-19 in Scotland: A genomics-based retrospective cohort analysis. PLoS ONE 2023, 18, e0284187. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, S.A.J.; Richter, A.; Casey, A.; Osman, H.; Mirza, J.D.; Stockton, J.; Quick, J.; Ratcliffe, L.; Sparks, N.; Cumley, N.; et al. Recurrent SARS-CoV-2 mutations in immunodeficient patients. Virus Evol. 2022, 8, veac050. [Google Scholar] [CrossRef]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Consortium, C.-G.U.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Grint, D.J.; Wing, K.; Houlihan, C.; Gibbs, H.P.; Evans, S.J.W.; Williamson, E.; I McDonald, H.; Bhaskaran, K.; Evans, D.; Walker, A.J.; et al. Severity of Severe Acute Respiratory System Coronavirus 2 (SARS-CoV-2) Alpha Variant (B.1.1.7) in England. Clin. Infect. Dis. 2021, 75, e1120–e1127. [Google Scholar] [CrossRef] [PubMed]
- Volz, E.; Mishra, S.; Chand, M.; Barrett, J.C.; Johnson, R.; Geidelberg, L.; Hinsley, W.R.; Laydon, D.J.; Dabrera, G.; O’Toole, Á.; et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 2021, 593, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Manirambona, E.; Okesanya, O.J.; Olaleke, N.O.; Oso, T.A.; Lucero-Prisno, D.E. Evolution and implications of SARS-CoV-2 variants in the post-pandemic era. Discov. Public Health 2024, 21, 1–12. [Google Scholar] [CrossRef]
- McLean, G.; Kamil, J.; Lee, B.; Moore, P.; Schulz, T.F.; Muik, A.; Sahin, U.; Türeci, Ö.; Pather, S. The Impact of Evolving SARS-CoV-2 Mutations and Variants on COVID-19 Vaccines. mBio 2022, 13, e0297921. [Google Scholar] [CrossRef]
- Lubinski, B.; Fernandes, M.H.; Frazier, L.; Tang, T.; Daniel, S.; Diel, D.G.; Jaimes, J.A.; Whittaker, G.R. Functional evaluation of the P681H mutation on the proteolytic activation of the SARS-CoV-2 variant B.1.1.7 (Alpha) spike. iScience 2021, 25, 103589. [Google Scholar] [CrossRef] [PubMed]
- Zali, A.; Khodadoost, M.; Gholamzadeh, S.; Janbazi, S.; Piri, H.; Taraghikhah, N.; Hannani, K.; Looha, M.A.; Mohammadi, G. Mortality among hospitalized COVID-19 patients during surges of SARS-CoV-2 alpha (B.1.1.7) and delta (B.1.617.2) variants. Sci. Rep. 2022, 12, 18918. [Google Scholar] [CrossRef]
- Lauring, A.S.; Tenforde, M.W.; Chappell, J.D.; Gaglani, M.; A Ginde, A.; McNeal, T.; Ghamande, S.; Douin, D.J.; Talbot, H.K.; Casey, J.D.; et al. Clinical severity of, and effectiveness of mRNA vaccines against, covid-19 from omicron, delta, and alpha SARS-CoV-2 variants in the United States: Prospective observational study. BMJ 2022, 376, e069761. [Google Scholar] [CrossRef]
- Duong, B.V.; Larpruenrudee, P.; Fang, T.; Hossain, S.I.; Saha, S.C.; Gu, Y.; Islam, M.S. Is the SARS CoV-2 Omicron Variant Deadlier and More Transmissible Than Delta Variant? Int. J. Environ. Res. Public Health 2022, 19, 4586. [Google Scholar] [CrossRef]
- Xue, S.; Han, Y.; Wu, F.; Wang, Q. Mutations in the SARS-CoV-2 spike receptor binding domain and their delicate balance between ACE2 affinity and antibody evasion. Protein Cell 2024, 15, 403–418. [Google Scholar] [CrossRef]
- Pantaleo, G.; Correia, B.; Fenwick, C.; Joo, V.S.; Perez, L. Antibodies to combat viral infections: Development strategies and progress. Nat. Rev. Drug Discov. 2022, 21, 676–696. [Google Scholar] [CrossRef]
- Ahmad, L. Implication of SARS-CoV-2 Immune Escape Spike Variants on Secondary and Vaccine Breakthrough Infections. Front. Immunol. 2021, 12, 742167. [Google Scholar] [CrossRef]
- Carabelli, A.M.; Peacock, T.P.; Thorne, L.G.; Harvey, W.T.; Hughes, J.; COVID-19 Genomics UK Consortium; Peacock, S.J.; Barclay, W.S.; de Silva, T.I.; Towers, G.J.; et al. SARS-CoV-2 variant biology: Immune escape, transmission and fitness. Nat. Rev. Microbiol. 2023, 21, 162–177. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Tang, T.; Chen, Y.; Huang, X.; Liang, T. mRNA vaccines in disease prevention and treatment. Signal Transduct. Target. Ther. 2023, 8, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Sturmlechner, I.; Zhang, C.; Farrell, J.; Burow, P.B.; Wickenhagen, A.; Sugrue, E. Exponential growth, high prevalence of SARS-CoV-2, and vaccine effectiveness associated with the Delta variant. Science 2021, 374, 1463. [Google Scholar] [CrossRef]
- Volz, E. Fitness, growth and transmissibility of SARS-CoV-2 genetic variants. Nat. Rev. Genet. 2023, 24, 724–734. [Google Scholar] [CrossRef] [PubMed]
- Banho, C.A.; Sacchetto, L.; Campos, G.R.F.; Bittar, C.; Possebon, F.S.; Ullmann, L.S.; Marques, B.d.C.; da Silva, G.C.D.; Moraes, M.M.; Parra, M.C.P.; et al. Impact of SARS-CoV-2 Gamma lineage introduction and COVID-19 vaccination on the epidemiological landscape of a Brazilian city. Commun. Med. 2022, 2, 1–11. [Google Scholar] [CrossRef]
- Khetran, S.R.; Mustafa, R. Mutations of SARS-CoV-2 Structural Proteins in the Alpha, Beta, Gamma, and Delta Variants: Bioinformatics Analysis. JMIR Bioinform. Biotechnol. 2023, 4, e43906. [Google Scholar] [CrossRef]
- Andeweg, S.P.; Vennema, H.; Veldhuijzen, I.; Smorenburg, N.; Schmitz, D.; Zwagemaker, F.; van Gageldonk-Lafeber, A.B.; Hahné, S.J.M.; Reusken, C.; Knol, M.J.; et al. Elevated risk of infection with SARS-CoV-2 Beta, Gamma, and Delta variants compared with Alpha variant in vaccinated individuals. Sci. Transl. Med. 2022, 15, eabn4338. [Google Scholar] [CrossRef]
- Tao, K.; Tzou, P.L.; Nouhin, J.; Gupta, R.K.; de Oliveira, T.; Kosakovsky Pond, S.L.; Fera, D.; Shafer, R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021, 22, 757–773. [Google Scholar] [CrossRef]
- Giovanetti, M.; Fonseca, V.; Wilkinson, E.; Tegally, H.; San, E.J.; Althaus, C.L.; Xavier, J.; Slavov, S.N.; Viala, V.L.; Lima, A.R.J.; et al. Replacement of the Gamma by the Delta variant in Brazil: Impact of lineage displacement on the ongoing pandemic. Virus Evol. 2022, 8, veac024. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.S.; de Almeida, G.B.; Borges, M.E.; Simon, L.M.; Poloni, S.; Bagattini, Â.M.; da Rosa, M.Q.M.; Filho, J.A.F.D.; Kuchenbecker, R.d.S.; Camey, S.A.; et al. Modelling optimal vaccination strategies against COVID-19 in a context of Gamma variant predominance in Brazil. Vaccine 2022, 40, 6616–6624. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Thambiraja, T.S.; Karuppanan, K.; Subramaniam, G. Omicron and Delta variant of SARS-CoV-2: A comparative computational study of spike protein. J. Med Virol. 2021, 94, 1641–1649. [Google Scholar] [CrossRef] [PubMed]
- Bian, L.; Gao, Q.; Gao, F.; Wang, Q.; He, Q.; Wu, X.; Mao, Q.; Xu, M.; Liang, Z. Impact of the Delta variant on vaccine efficacy and response strategies. Expert Rev. Vaccines 2021, 20, 1201–1209. [Google Scholar] [CrossRef] [PubMed]
- Shiehzadegan, S.; Alaghemand, N.; Fox, M.; Venketaraman, V. Analysis of the Delta Variant B.1.617.2 COVID-19. Clin. Pr. 2021, 11, 778–784. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.; Sun, Y.; Zhou, J.; Ye, Q. The Global Epidemic of the SARS-CoV-2 Delta Variant, Key Spike Mutations and Immune Escape. Front. Immunol. 2021, 12, 751778. [Google Scholar] [CrossRef] [PubMed]
- Saito, A.; Nasser, H.; Uriu, K.; Kosugi, Y.; Irie, T.; Shirakawa, K. SARS-CoV-2 spike P681R mutation enhances and accelerates viral fusion. bioRxiv 2021. [Google Scholar] [CrossRef]
- Fajnzylber, J.; Regan, J.; Coxen, K.; Corry, H.; Wong, C.; Rosenthal, A.; Worrall, D.; Giguel, F.; Piechocka-Trocha, A.; Atyeo, C.; et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat. Commun. 2020, 11, 5493. [Google Scholar] [CrossRef]
- Venkatakrishnan, A.J.; Anand, P.; Lenehan, P.; Ghosh, P.; Suratekar, R.; Siroha, A.; Chowdhury, D.R.; O’Horo, J.C.; Yao, J.D.; Pritt, B.S.; et al. Antigenic minimalism of SARS-CoV-2 is linked to surges in COVID-19 community transmission and vaccine breakthrough infections. medRxiv 2021. [Google Scholar] [CrossRef]
- Khedar, R.S.; Mittal, K.; Ambaliya, H.C.; Mathur, A.; Gupta, J.B.; Sharma, K.K.; Singh, Y.; Sharma, G.; Gupta, A.; Bhargava, V.; et al. Greater Covid-19 Severity and Mortality in Hospitalized Patients in Second (Delta Variant) Wave Compared to the First: Single Centre Prospective Study in India. medRxiv 2021. [Google Scholar] [CrossRef]
- Wang, C.; Liu, B.; Zhang, S.; Huang, N.; Zhao, T.; Lu, Q.; Cui, F. Differences in incidence and fatality of COVID-19 by SARS-CoV-2 Omicron variant versus Delta variant in relation to vaccine coverage: A world-wide review. J. Med Virol. 2022, 95, e28118. [Google Scholar] [CrossRef]
- Kadri, S.S.; Simpson, S.Q. Potential Implications of SARS-CoV-2 Delta Variant Surges for Rural Areas and Hospitals. JAMA 2021, 326, 1003–1004. [Google Scholar] [CrossRef]
- Sezen, Y.I.; Senoglu, S.; Karabela, S.N.; Yesilbag, Z.; Borcak, D.; Unlu, E.C.; Korkusuz, R.; Ozdemir, Y.; Yasar, K.K. Risk factors and the impact of vaccination on mortality in COVID-19 patients. Bratisl. Med J. 2022, 123, 440–443. [Google Scholar] [CrossRef]
- Lorenzo-Redondo, R.; A Ozer, E.; Hultquist, J.F. COVID-19: Is omicron less lethal than delta? BMJ 2022, 378, o1806. [Google Scholar] [CrossRef] [PubMed]
- Karimizadeh, Z.; Dowran, R.; Mokhtari-Azad, T.; Shafiei-Jandaghi, N.-Z. The reproduction rate of severe acute respiratory syndrome coronavirus 2 different variants recently circulated in human: A narrative review. Eur. J. Med Res. 2023, 28, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Abudunaibi, B.; Liu, W.; Guo, Z.; Zhao, Z.; Rui, J.; Song, W.; Wang, Y.; Chen, Q.; Frutos, R.; Su, C.; et al. A comparative study on the three calculation methods for reproduction numbers of COVID-19. Front. Med. 2023, 9, 1079842. [Google Scholar] [CrossRef]
- Ito, K.; Piantham, C.; Nishiura, H. Estimating relative generation times and relative reproduction numbers of Omicron BA.1 and BA.2 with respect to Delta in Denmark. medRxiv 2022. [Google Scholar] [CrossRef]
- Lino, A.; Cardoso, M.A.; Martins-Lopes, P.; Gonçalves, H.M.R. Omicron – The new SARS-CoV-2 challenge? Rev. Med Virol. 2022, 32, e2358. [Google Scholar] [CrossRef] [PubMed]
- Tiecco, G.; Storti, S.; Arsuffi, S.; Degli Antoni, M.; Focà, E.; Castelli, F.; Quiros-Roldan, E. Omicron BA.2 Lineage, the “Stealth” Variant: Is It Truly a Silent Epidemic? A Literature Review. Int. J. Mol. Sci. 2022, 23, 7315. [Google Scholar] [CrossRef]
- Yajima, H.; Nomai, T.; Okumura, K.; Maenaka, K.; The Genotype to Phenotype Japan (G2P-Japan) Consortium; Ito, J.; Hashiguchi, T.; Sato, K.; Matsuno, K.; Nao, N.; et al. Molecular and structural insights into SARS-CoV-2 evolution: From BA.2 to XBB subvariants. mBio 2024, 15, e0322023. [Google Scholar] [CrossRef] [PubMed]
- Garcia, I.; Bråte, J.; Fossum, E.; Rohringer, A.; Moen, L.V.; Hungnes, O.; Fjære, O.; Zaragkoulias, K.; Bragstad, K. Recombinant SARS-CoV-2 Delta/Omicron BA.5 emerging in an immunocompromised long-term infected COVID-19 patient. Sci. Rep. 2024, 14, 25790. [Google Scholar] [CrossRef]
- Ao, D.; He, X.; Hong, W.; Wei, X. The rapid rise of SARS-CoV-2 Omicron subvariants with immune evasion properties: XBB.1.5 and BQ.1.1 subvariants. Medcomm 2023, 4, e239. [Google Scholar] [CrossRef]
- Murdocca, M.; Romeo, I.; Citro, G.; Latini, A.; Centofanti, F.; Bugatti, A.; Caccuri, F.; Caruso, A.; Ortuso, F.; Alcaro, S.; et al. A Dynamic and Effective Peptide-Based Strategy for Promptly Addressing Emerging SARS-CoV-2 Variants of Concern. Pharmaceuticals 2024, 17, 891. [Google Scholar] [CrossRef]
- Chang-Rabley, E.; van Zelm, M.C.; Ricotta, E.E.; Edwards, E.S.J. An Overview of the Strategies to Boost SARS-CoV-2-Specific Immunity in People with Inborn Errors of Immunity. Vaccines 2024, 12, 675. [Google Scholar] [CrossRef] [PubMed]
- Otaki, J.M.; Nakasone, W.; Nakamura, M. Nonself Mutations in the Spike Protein Suggest an Increase in the Antigenicity and a Decrease in the Virulence of the Omicron Variant of SARS-CoV-2. COVID 2022, 2, 407–418. [Google Scholar] [CrossRef]
- Bálint, G.; Vörös-Horváth, B.; Széchenyi, A. Omicron: Increased transmissibility and decreased pathogenicity. Signal Transduct. Target. Ther. 2022, 7, 1–3. [Google Scholar] [CrossRef]
- de Souza, A.S.; Amorim, V.M.d.F.; Guardia, G.D.A.; dos Santos, F.F.; Ulrich, H.; Galante, P.A.F.; de Souza, R.F.; Guzzo, C.R. Severe Acute Respiratory Syndrome Coronavirus 2 Variants of Concern: A Perspective for Emerging More Transmissible and Vaccine-Resistant Strains. Viruses 2022, 14, 827. [Google Scholar] [CrossRef] [PubMed]
- Bager, P.; Wohlfahrt, J.; Bhatt, S.; Stegger, M.; Legarth, R.; Møller, C.H.; Skov, R.L.; Valentiner-Branth, P.; Voldstedlund, M.; Fischer, T.K.; et al. Risk of hospitalisation associated with infection with SARS-CoV-2 omicron variant versus delta variant in Denmark: An observational cohort study. Lancet Infect. Dis. 2022, 22, 967–976. [Google Scholar] [CrossRef] [PubMed]
- Nyberg, T.; Ferguson, N.M.; Nash, S.G.; Webster, H.H.; Flaxman, S.; Andrews, N.; Hinsley, W.; Bernal, J.L.; Kall, M.; Bhatt, S.; et al. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: A cohort study. Lancet 2022, 399, 1303–1312. [Google Scholar] [CrossRef]
- Auvigne, V.; Vaux, S.; Le Strat, Y.; Schaeffer, J.; Fournier, L.; Tamandjou, C.; Montagnat, C.; Coignard, B.; Levy-Bruhl, D.; du Châtelet, I.P. Severe hospital events following symptomatic infection with Sars-CoV-2 Omicron and Delta variants in France, December 2021–January 2022: A retrospective, population-based, matched cohort study. eClinicalMedicine 2022, 48, 101455. [Google Scholar] [CrossRef] [PubMed]
- Carroll, T.; Fox, D.; van Doremalen, N.; Ball, E.; Morris, M.K.; Sotomayor-Gonzalez, A.; Servellita, V.; Rustagi, A.; Yinda, C.K.; Fritts, L.; et al. The B.1.427/1.429 (epsilon) SARS-CoV-2 variants are more virulent than ancestral B.1 (614G) in Syrian hamsters. PLoS Pathog. 2022, 18, e1009914. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Garcia-Knight, M.A.; Khalid, M.M.; Servellita, V.; Wang, C.; Morris, M.K.; Sotomayor-González, A.; Glasner, D.R.; Reyes, K.R.; Gliwa, A.S.; et al. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell 2021, 184, 3426–3437.e8. [Google Scholar] [CrossRef] [PubMed]
- Duerr, R.; Dimartino, D.; Marier, C.; Zappile, P.; Wang, G.; Lighter, J.; Elbel, B.; Troxel, A.B.; Heguy, A. Dominance of Alpha and Iota variants in SARS-CoV-2 vaccine breakthrough infections in New York City. J. Clin. Investig. 2021, 131, e152702. [Google Scholar] [CrossRef] [PubMed]
- Perez-Gomez, R. The Development of SARS-CoV-2 Variants: The Gene Makes the Disease. J. Dev. Biol. 2021, 9, 58. [Google Scholar] [CrossRef] [PubMed]
- Stein, S.C.; Hansen, G.; Ssebyatika, G.; Ströh, L.J.; Ochulor, O.; Herold, E.; Schwarzloh, B.; Mutschall, D.; Zischke, J.; Cordes, A.K.; et al. A human monoclonal antibody neutralizing SARS-CoV-2 Omicron variants containing the L452R mutation. J. Virol. 2024, 98, e0122324. [Google Scholar] [CrossRef] [PubMed]
- Starr, T.N.; Greaney, A.J.; Stewart, C.M.; Walls, A.C.; Hannon, W.W.; Veesler, D.; Bloom, J.D. Deep mutational scans for ACE2 binding, RBD expression, and antibody escape in the SARS-CoV-2 Omicron BA.1 and BA.2 receptor-binding domains. PLoS Pathog. 2022, 18, e1010951. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 2021, 593, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Tada, T.; Zhou, H.; Samanovic, M.I.; Dcosta, B.M.; Cornelius, A.; Herati, R.S.; Mulligan, M.J.; Landau, N.R. Neutralization of SARS-CoV-2 Variants by mRNA and Adenoviral Vector Vaccine-Elicited Antibodies. Front. Immunol. 2022, 13, 797589. [Google Scholar] [CrossRef]
- Thomson, E.C.; Rosen, L.E.; Shepherd, J.G.; Spreafico, R.; Filipe, A.d.S.; Wojcechowskyj, J.A.; Davis, C.; Piccoli, L.; Pascall, D.J.; Dillen, J.; et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell 2021, 184, 1171–1187.e20. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.-L.; Goldsmith, J.A.; Schaub, J.M.; DiVenere, A.M.; Kuo, H.-C.; Javanmardi, K.; Le, K.C.; Wrapp, D.; Lee, A.G.; Liu, Y.; et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 2020, 369, 1501–1505. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.; Logan, N.; Tyson, G.; Orton, R.; Harvey, W.T.; Perkins, J.S.; Mollett, G.; Blacow, R.M.; Peacock, T.P.; Barclay, W.S.; et al. Reduced neutralisation of the Delta (B.1.617.2) SARS-CoV-2 variant of concern following vaccination. PLoS Pathog. 2021, 17, e1010022. [Google Scholar] [CrossRef] [PubMed]
- Newman, J.; Thakur, N.; Peacock, T.P.; Bialy, D.; Elrefaey, A.M.E.; Bogaardt, C.; Horton, D.L.; Ho, S.; Kankeyan, T.; Carr, C.; et al. Neutralizing antibody activity against 21 SARS-CoV-2 variants in older adults vaccinated with BNT162b2. Nat. Microbiol. 2022, 7, 1180–1188. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Beltran, W.F.; Lam, E.C.; St Denis, K.; Nitido, A.D.; Garcia, Z.H.; Hauser, B.M.; Feldman, J.; Pavlovic, M.N.; Gregory, D.J.; Poznansky, M.C.; et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 2021, 184, 2372–2383.e9. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Al-Ahmed, S.H.; Albayat, H.; Alwarthan, S.; Alhajri, M.; Najim, M.A.; AlShehail, B.M.; Al-Adsani, W.; Alghadeer, A.; Abduljabbar, W.A.; et al. Variants of SARS-CoV-2: Influences on the Vaccines’ Effectiveness and Possible Strategies to Overcome Their Consequences. Medicina 2023, 59, 507. [Google Scholar] [CrossRef]
- Willett, B.J. Distinct antigenic properties of the SARS-CoV-2 Omicron lineages BA.4 and BA.5. bioRxiv 2022. [Google Scholar] [CrossRef]
- Tuekprakhon, A.; Nutalai, R.; Dijokaite-Guraliuc, A.; Zhou, D.; Ginn, H.M.; Selvaraj, M.; Liu, C.; Mentzer, A.J.; Supasa, P.; Duyvesteyn, H.M.; et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell 2022, 185, 2422–2433.e13. [Google Scholar] [CrossRef] [PubMed]
- Hirabara, S.M.; Serdan, T.D.A.; Gorjao, R.; Masi, L.N.; Pithon-Curi, T.C.; Covas, D.T.; Curi, R.; Durigon, E.L. SARS-COV-2 Variants: Differences and Potential of Immune Evasion. Front. Cell. Infect. Microbiol. 2022, 11, 781429. [Google Scholar] [CrossRef]
- Gupta, D.; Sharma, P.; Singh, M.; Kumar, M.; Ethayathulla, A.S.; Kaur, P. Structural and functional insights into the spike protein mutations of emerging SARS-CoV-2 variants. Cell. Mol. Life Sci. 2021, 78, 7967–7989. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, J.; Johnson, B.A.; Xia, H.; Ku, Z.; Schindewolf, C.; Widen, S.G.; An, Z.; Weaver, S.C.; Menachery, V.D.; et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. bioRxiv 2021. [Google Scholar] [CrossRef]
- Rahimi, A.; Mirzazadeh, A.; Tavakolpour, S. Genetics and genomics of SARS-CoV-2: A review of the literature with the special focus on genetic diversity and SARS-CoV-2 genome detection. Genomics 2021, 113, 1221–1232. [Google Scholar] [CrossRef] [PubMed]
- Oulas, A.; Zanti, M.; Tomazou, M.; Zachariou, M.; Minadakis, G.; Bourdakou, M.M.; Pavlidis, P.; Spyrou, G.M. Generalized linear models provide a measure of virulence for specific mutations in SARS-CoV-2 strains. PLoS ONE 2021, 16, e0238665. [Google Scholar] [CrossRef]
- Khandia, R.; Singhal, S.; Alqahtani, T.; Kamal, M.A.; El-Shall, N.A.; Nainu, F.; Desingu, P.A.; Dhama, K. Emergence of SARS-CoV-2 Omicron (B.1.1.529) variant, salient features, high global health concerns and strategies to counter it amid ongoing COVID-19 pandemic. Environ. Res. 2022, 209, 112816. [Google Scholar] [CrossRef] [PubMed]
- Sonabend, R.; Whittles, L.K.; Imai, N.; Perez-Guzman, P.N.; Knock, E.S.; Rawson, T.; Gaythorpe, K.A.M.; A Djaafara, B.; Hinsley, W.; FitzJohn, R.G.; et al. Non-pharmaceutical interventions, vaccination, and the SARS-CoV-2 delta variant in England: A mathematical modelling study. Lancet 2021, 398, 1825–1835. [Google Scholar] [CrossRef]
- Malik, J.A.; Ahmed, S.; Mir, A.; Shinde, M.; Bender, O.; Alshammari, F.; Ansari, M.; Anwar, S. The SARS-CoV-2 mutations versus vaccine effectiveness: New opportunities to new challenges. J. Infect. Public Health 2022, 15, 228–240. [Google Scholar] [CrossRef]
- Sheikh, A.; Kerr, S.; Woolhouse, M.; McMenamin, J.; Robertson, C.; Simpson, C.R.; Millington, T.; Shi, T.; Agrawal, U.; Hameed, S.S.; et al. Severity of omicron variant of concern and effectiveness of vaccine boosters against symptomatic disease in Scotland (EAVE II): A national cohort study with nested test-negative design. Lancet Infect. Dis. 2022, 22, 959–966. [Google Scholar] [CrossRef]
- Cunniff, L.; Alyanak, E.; Fix, A.; Novak, M.; Peterson, M.; Mevis, K.; Eiden, A.L.; Bhatti, A. The impact of the COVID-19 pandemic on vaccination uptake in the United States and strategies to recover and improve vaccination rates: A review. Hum. Vaccines Immunother. 2023, 19, 2246502. [Google Scholar] [CrossRef] [PubMed]
- Jang, G.; Kim, J.; Lee, Y.; Son, C.; Ko, K.T.; Lee, H. Analysis of the impact of COVID-19 variants and vaccination on the time-varying reproduction number: Statistical methods. Front. Public Health 2024, 12, 1353441. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, S.M.; O’Shea, K.J.; Chin, K.L.; Strych, U.; Ferguson, M.C.; Bottazzi, M.E.; Wedlock, P.T.; Cox, S.N.; Siegmund, S.S.; Hotez, P.J.; et al. Maintaining face mask use before and after achieving different COVID-19 vaccination coverage levels: A modelling study. Lancet Public Health 2022, 7, e356–e365. [Google Scholar] [CrossRef]
- Günl, F.; Mecate-Zambrano, A.; Rehländer, S.; Hinse, S.; Ludwig, S.; Brunotte, L. Shooting at a Moving Target—Effectiveness and Emerging Challenges for SARS-CoV-2 Vaccine Development. Vaccines 2021, 9, 1052. [Google Scholar] [CrossRef]
- Gorman, M.J.; Patel, N.; Guebre-Xabier, M.; Zhu, A.; Atyeo, C.; Pullen, K.M.; Loos, C.; Goez-Gazi, Y.; Carrion, R., Jr.; Tian, J.-H.; et al. Collaboration between the Fab and Fc contribute to maximal protection against SARS-CoV-2 in nonhuman primates following NVX-CoV2373 subunit vaccine with Matrix-MTM vaccination. bioRxiv 2021, 2021.02.05.429759. [Google Scholar] [CrossRef]
- Silva, A.C.B.; Carvalho, C.A.M. Influence of Mutations on Physicochemical Properties of Spike Proteins from Prototypical SARS-CoV-2 Variants of Concern Detected in Amazonian Countries. Microbiol. Res. 2024, 15, 1334–1345. [Google Scholar] [CrossRef]
- Munnink, B.B.O.; Worp, N.; Nieuwenhuijse, D.F.; Sikkema, R.S.; Haagmans, B.; Fouchier, R.A.M.; Koopmans, M. The next phase of SARS-CoV-2 surveillance: Real-time molecular epidemiology. Nat. Med. 2021, 27, 1518–1524. [Google Scholar] [CrossRef]
- Huang, Q.; Zhang, Q.; Bible, P.W.; Liang, Q.; Zheng, F.; Wang, Y.; Hao, Y.; Liu, Y. A New Way to Trace SARS-CoV-2 Variants Through Weighted Network Analysis of Frequency Trajectories of Mutations. Front. Microbiol. 2022, 13, 859241. [Google Scholar] [CrossRef]
- A Adeyinka, D.; Neudorf, C.; A Camillo, C.; Marks, W.N.; Muhajarine, N. COVID-19 Vaccination and Public Health Countermeasures on Variants of Concern in Canada: Evidence From a Spatial Hierarchical Cluster Analysis. JMIR Public Health Surveill. 2022, 8, e31968. [Google Scholar] [CrossRef]
- Gardy, J.L.; Loman, N.J. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat. Rev. Genet. 2017, 19, 9–20. [Google Scholar] [CrossRef]
- Hale, R.; Crowley, P.; Dervisevic, S.; Coupland, L.; Cliff, P.R.; Ebie, S.; Snell, L.B.; Paul, J.; Williams, C.; Randell, P.; et al. Development of a Multiplex Tandem PCR (MT-PCR) Assay for the Detection of Emerging SARS-CoV-2 Variants. Viruses 2021, 13, 2028. [Google Scholar] [CrossRef] [PubMed]
- Sintchenko, V.; Holmes, E.C. The role of pathogen genomics in assessing disease transmission. BMJ 2015, 350, h1314. [Google Scholar] [CrossRef] [PubMed]
- Barona-Gómez, F.; Delaye, L.; Díaz-Valenzuela, E.; Plisson, F.; Cruz-Pérez, A.; Díaz-Sánchez, M.; García-Sepúlveda, C.A.; Sanchez-Flores, A.; Pérez-Abreu, R.; Valencia-Valdespino, F.J.; et al. Phylogenomics and population genomics of SARS-CoV-2 in Mexico during the pre-vaccination stage reveals variants of interest B.1.1.28.4 and B.1.1.222 or B.1.1.519 and the nucleocapsid mutation S194L associated with symptoms. Microb. Genom. 2021, 7, 000684. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liang, Y.; Yu, D.; Du, B.; Cheng, W.; Li, L.; Yu, Z.; Luo, S.; Zhang, Y.; Wang, H.; et al. A systematic review of Vaccine Breakthrough Infections by SARS-CoV-2 Delta Variant. Int. J. Biol. Sci. 2022, 18, 889–900. [Google Scholar] [CrossRef]
- Luo, C.H.; Morris, C.P.; Sachithanandham, J.; Amadi, A.; Gaston, D.; Li, M.; Swanson, N.J.; Schwartz, M.; Klein, E.Y.; Pekosz, A.; et al. Infection with the SARS-CoV-2 Delta Variant is Associated with Higher Infectious Virus Loads Compared to the Alpha Variant in both Unvaccinated and Vaccinated Individuals. medRxiv 2021. [Google Scholar] [CrossRef]
- Talic, S.; Shah, S.; Wild, H.; Gasevic, D.; Maharaj, A.; Ademi, Z.; Li, X.; Xu, W.; Mesa-Eguiagaray, I.; Rostron, J.; et al. Effectiveness of public health measures in reducing the incidence of covid-19, SARS-CoV-2 transmission, and covid-19 mortality: Systematic review and meta-analysis. BMJ 2021, 375, e068302. [Google Scholar] [CrossRef] [PubMed]
- Anand, U.; Pal, T.; Zanoletti, A.; Sundaramurthy, S.; Varjani, S.; Rajapaksha, A.U.; Barceló, D.; Bontempi, E. The spread of the omicron variant: Identification of knowledge gaps, virus diffusion modelling, and future research needs. Environ. Res. 2023, 225, 115612. [Google Scholar] [CrossRef]
- Arin, K. South Korea Is Keeping Mandatory Masking Indoors. The Korea Herald. Available online: https://www.koreaherald.com/article/2964627 (accessed on 2 January 2025).
- Jefferies, R.; McAdam, J. Locked in: Australia’s COVID-19 Border Closures and the Right to Leave. Aust. Yearb. Int. Law 2023, 41, 185. [Google Scholar] [CrossRef]
- Harrison, C.; Butfield, R.; Yarnoff, B.; Yang, J. Modeling the potential public health and economic impact of different COVID-19 booster dose vaccination strategies with an adapted vaccine in the United Kingdom. Expert Rev. Vaccines 2024, 23, 730–739. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Gonsalves, G.S.; Tan, S.T.; Kelly, J.D.; Rutherford, G.W.; Wachter, R.M.; Schechter, R.; Paltiel, A.D.; Lo, N.C. Comparing frequency of booster vaccination to prevent severe COVID-19 by risk group in the United States. Nat. Commun. 2024, 15, 1883. [Google Scholar] [CrossRef] [PubMed]
- Littlecott, H.; Herd, C.; O’Rourke, J.; Chaparro, L.T.; Keeling, M.; Rubin, G.J.; Fearon, E. Effectiveness of testing, contact tracing and isolation interventions among the general population on reducing transmission of SARS-CoV-2: A systematic review. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2023, 381, 20230131. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Azman, A.S.; Chen, X.; Zou, J.; Tian, Y.; Sun, R.; Xu, X.; Wu, Y.; Lu, W.; Ge, S.; et al. Global landscape of SARS-CoV-2 genomic surveillance and data sharing. Nat. Genet. 2022, 54, 499–507. [Google Scholar] [CrossRef]
- Ziegler, T.; Moen, A.; Zhang, W.; Cox, N.J. Global Influenza Surveillance and Response System: 70 years of responding to the expected and preparing for the unexpected. Lancet 2022, 400, 981–982. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.K.; Stewart, M.A.; Dymond, J.S.; Lewis, S.L. An Implementation Strategy to Develop Sustainable Surveillance Activities Through Adoption of a Target Operating Model. Front. Public Health 2022, 10, 871114. [Google Scholar] [CrossRef]
- Schäfer, M.; Wijaya, K.P.; Rockenfeller, R.; Götz, T. The impact of travelling on the COVID-19 infection cases in Germany. BMC Infect. Dis. 2022, 22, 455. [Google Scholar] [CrossRef] [PubMed]
- The Global Alliance for Genomics and Health Benchmarking Team; Krusche, P.; Trigg, L.; Boutros, P.C.; Mason, C.E.; De La Vega, F.M.; Moore, B.L.; Gonzalez-Porta, M.; Eberle, M.A.; Tezak, Z.; et al. Best practices for benchmarking germline small-variant calls in human genomes. Nat. Biotechnol. 2019, 37, 555–560. [Google Scholar] [CrossRef]
- Struelens, M.J.; Ludden, C.; Werner, G.; Sintchenko, V.; Jokelainen, P.; Ip, M. Real-time genomic surveillance for enhanced control of infectious diseases and antimicrobial resistance. Front. Sci. 2024, 2, 1298248. [Google Scholar] [CrossRef]
- Shringarpure, S.; Xing, E.P. Effects of Sample Selection Bias on the Accuracy of Population Structure and Ancestry Inference. G3 Genes|Genomes|Genetics 2014, 4, 901–911. [Google Scholar] [CrossRef]
- Bannick, M.S.; Gao, F.; Brown, E.R.; E Janes, H. Retrospective, Observational Studies for Estimating Vaccine Effects on the Secondary Attack Rate of SARS-CoV-2. Am. J. Epidemiol. 2023, 192, 1016–1028. [Google Scholar] [CrossRef] [PubMed]
- Bhopal, R.S. Error, bias, and confounding in epidemiology. In Concepts of Epidemiology: Integrating the Ideas, Theories, Principles, and Methods of Epidemiology; Bhopal, R.S., Ed.; Oxford University Press: Oxford, UK, 2016. [Google Scholar] [CrossRef]
- Zabidi, N.Z.; Liew, H.L.; Farouk, I.A.; Puniyamurti, A.; Yip, A.J.W.; Wijesinghe, V.N.; Low, Z.Y.; Tang, J.W.; Chow, V.T.K.; Lal, S.K. Evolution of SARS-CoV-2 Variants: Implications on Immune Escape, Vaccination, Therapeutic and Diagnostic Strategies. Viruses 2023, 15, 944. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.; Govindarajulu, M.; Parise, R.S.; Neel, L.; Shankar, T.; Patel, S.; Lowery, P.; Smith, F.; Dhanasekaran, M.; Moore, T. Emerging SARS-CoV-2 Variants: A Review of Its Mutations, Its Implications and Vaccine Efficacy. Vaccines 2021, 9, 1195. [Google Scholar] [CrossRef] [PubMed]
- Haque, A.; Pant, A.B. Mitigating COVID-19 in the face of emerging virus variants, breakthrough infections and vaccine hesitancy. J. Autoimmun. 2021, 127, 102792. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-L.; Cheng, S.-T.; Shen, C.-F.; Huang, S.-W.; Cheng, C.-M. Impact of the COVID-19 vaccine booster strategy on vaccine protection: A pilot study of a military hospital in Taiwan. Clin. Exp. Vaccine Res. 2023, 12, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Hogan, A.B.; Wu, S.L.; Toor, J.; Mesa, D.O.; Doohan, P.; Watson, O.J.; Winskill, P.; Charles, G.; Barnsley, G.; Riley, E.M.; et al. Long-term vaccination strategies to mitigate the impact of SARS-CoV-2 transmission: A modelling study. PLoS Med. 2023, 20, e1004195. [Google Scholar] [CrossRef] [PubMed]
- Arevalo-Romero, J.A.; Chingaté-López, S.M.; Camacho, B.A.; Alméciga-Díaz, C.J.; Ramirez-Segura, C.A. Next-generation treatments: Immunotherapy and advanced therapies for COVID-19. Heliyon 2024, 10, e26423. [Google Scholar] [CrossRef] [PubMed]
- Modjarrad, K.; Che, Y.; Chen, W.; Wu, H.; Cadima, C.I.; Muik, A.; Maddur, M.S.; Tompkins, K.R.; Martinez, L.T.; Cai, H.; et al. Preclinical characterization of the Omicron XBB.1.5-adapted BNT162b2 COVID-19 vaccine. npj Vaccines 2024, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Jaishwal, P.; Jha, K.; Singh, S.P. Revisiting the dimensions of universal vaccine with special focus on COVID-19: Efficacy versus methods of designing. Int. J. Biol. Macromol. 2024, 277, 134012. [Google Scholar] [CrossRef] [PubMed]
- Al Fayez, N.; Nassar, M.S.; Alshehri, A.A.; Alnefaie, M.K.; Almughem, F.A.; Alshehri, B.Y.; Alawad, A.O.; Tawfik, E.A. Recent Advancement in mRNA Vaccine Development and Applications. Pharmaceutics 2023, 15, 1972. [Google Scholar] [CrossRef] [PubMed]

| Variant | WHO Label | Pango Lineage | Date of Emergence | Key Mutations (Spike Protein) | Transmissibility | Pathogenicity | Immune Escape Potential | Impact on Vaccine Efficacy |
|---|---|---|---|---|---|---|---|---|
| Alpha | B.1.1.7 | United Kingdom | September 2020 | N501Y, D614G, 69-70del, P681H | ~50% more transmissible than the original strain | Slightly increased severity compared to earlier strains | Moderate immune escape (some reduction in neutralization) | Minor reduction in efficacy; vaccines remain effective for severe disease |
| Beta | B.1.351 | South Africa | May 2020 | K417N, E484K, N501Y, D614G | ~50% more transmissible than the original strain | No significant increase in severity observed | High immune escape, especially due to E484K | Significant reduction in neutralization by some vaccines; boosters recommended for enhanced protection |
| Gamma | P.1 | Brazil | November 2020 | K417T, E484K, N501Y, D614G | Increased transmissibility (data not precise) | No significant increase in severity | Moderate immune escape due to E484K | Moderate reduction in efficacy; boosters can improve protection |
| Delta | B.1.617.2 | India | October 2020 | L452R, T478K, D614G, P681R | Highly increased (up to 2x more than Alpha) | Possible increase in severity, particularly unvaccinated | Moderate immune escape | Reduced efficacy, especially after one dose; full vaccination and boosters recommended |
| Omicron | B.1.1.529 | South Africa | November 2021 | S371L, S373P, S375F, T376A, G496S, Q498R, N679K, P681H, D796Y | Extremely high transmissibility; surpasses Delta | Lower severity on average, though risks remain for unvaccinated and high-risk groups | High immune escape due to multiple mutations in RBD (e.g., E484A, Q493K) | Significant reduction in vaccine efficacy for infection; boosters enhance protection against severe disease |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alhamlan, F.S.; Al-Qahtani, A.A. SARS-CoV-2 Variants: Genetic Insights, Epidemiological Tracking, and Implications for Vaccine Strategies. Int. J. Mol. Sci. 2025, 26, 1263. https://doi.org/10.3390/ijms26031263
Alhamlan FS, Al-Qahtani AA. SARS-CoV-2 Variants: Genetic Insights, Epidemiological Tracking, and Implications for Vaccine Strategies. International Journal of Molecular Sciences. 2025; 26(3):1263. https://doi.org/10.3390/ijms26031263
Chicago/Turabian StyleAlhamlan, Fatimah S., and Ahmed A. Al-Qahtani. 2025. "SARS-CoV-2 Variants: Genetic Insights, Epidemiological Tracking, and Implications for Vaccine Strategies" International Journal of Molecular Sciences 26, no. 3: 1263. https://doi.org/10.3390/ijms26031263
APA StyleAlhamlan, F. S., & Al-Qahtani, A. A. (2025). SARS-CoV-2 Variants: Genetic Insights, Epidemiological Tracking, and Implications for Vaccine Strategies. International Journal of Molecular Sciences, 26(3), 1263. https://doi.org/10.3390/ijms26031263






