Identification of Ovarian High-Grade Serous Carcinoma with Mitochondrial Gene Variation
Abstract
1. Introduction
2. Results
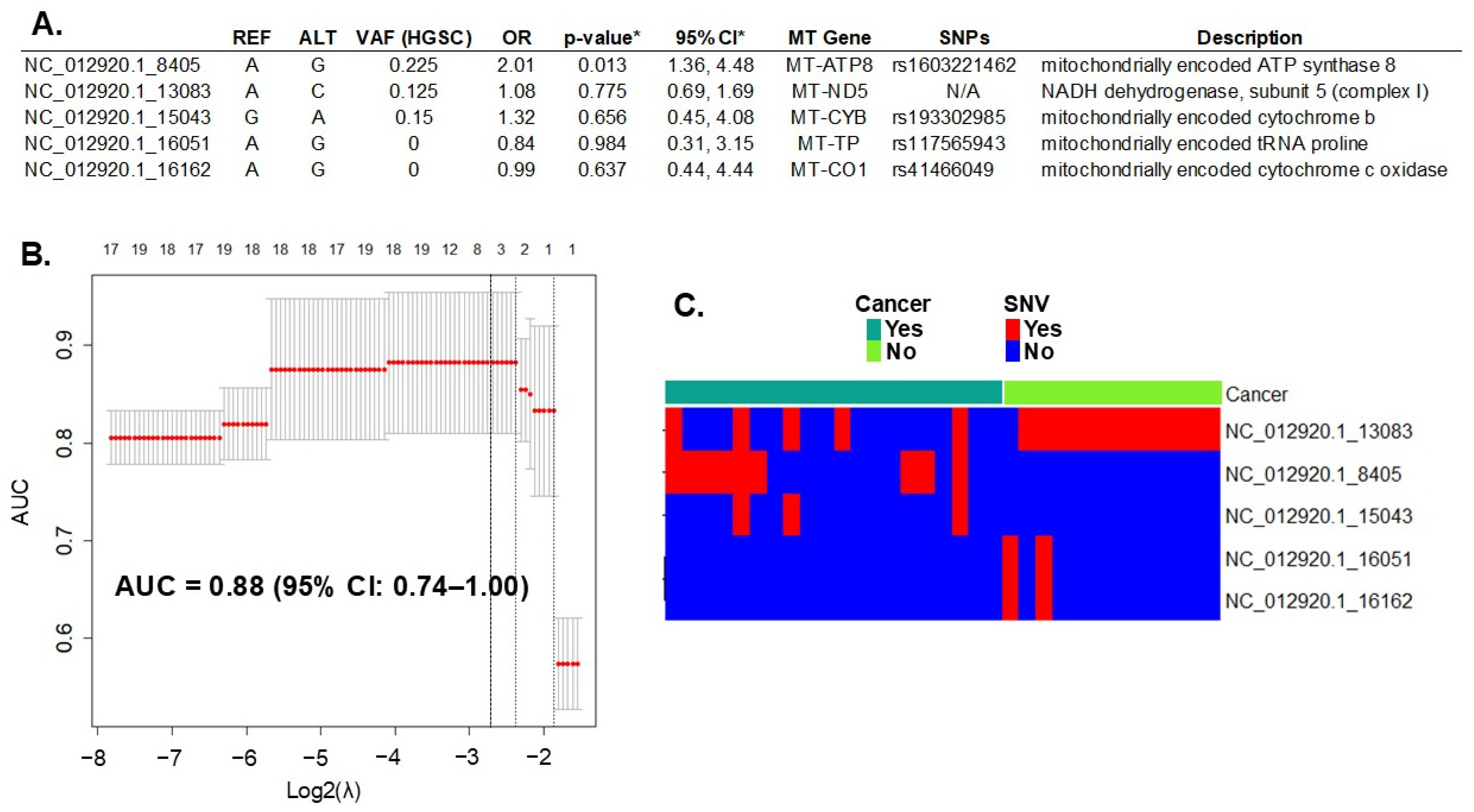
2.1. mtDNA Single Nucleotide Variation (SNV)
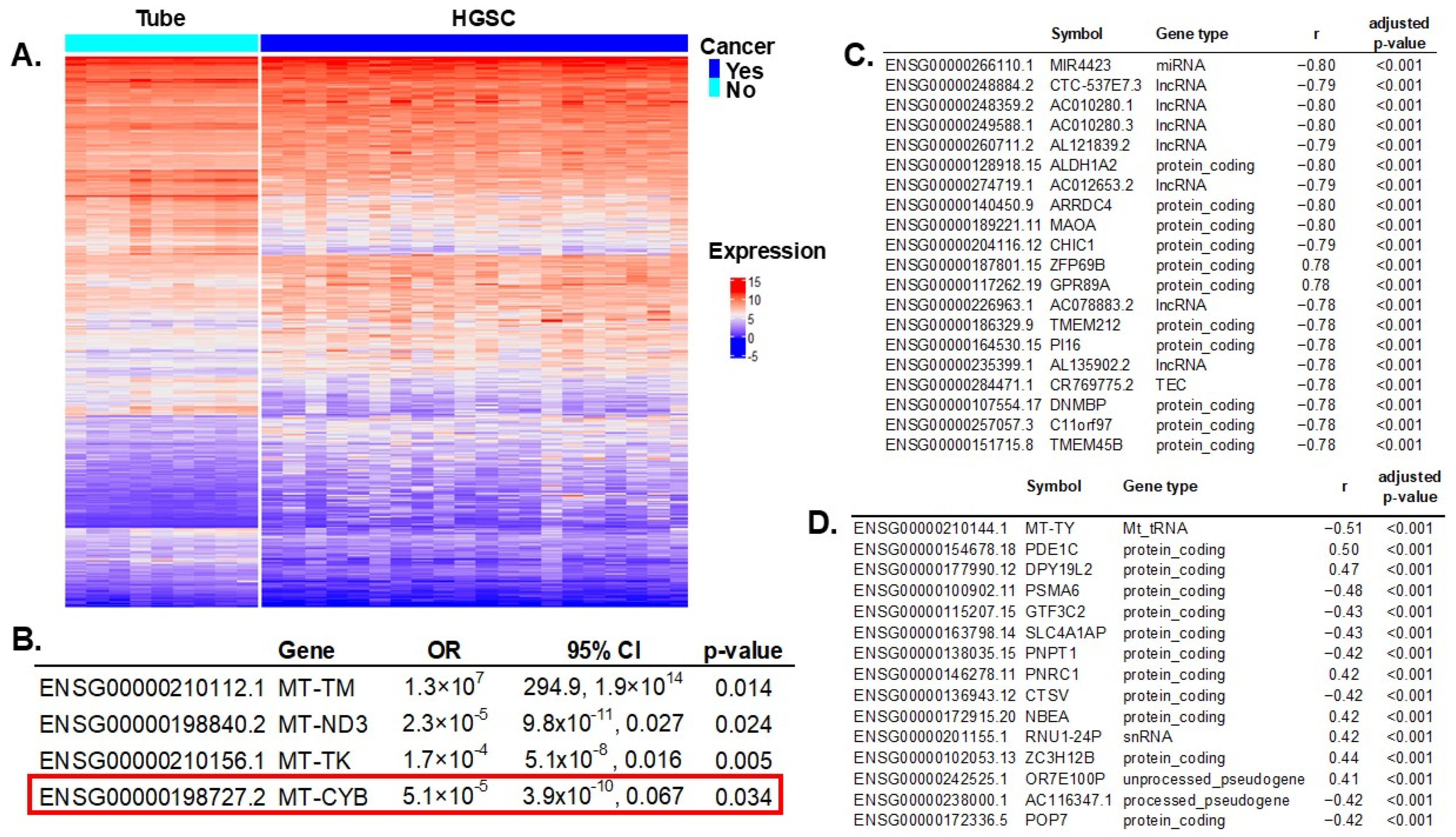
2.2. Correlation of Significant mtDNA Variants with Gene Expression
2.3. Pathway Enrichment Analysis of Correlated Genes
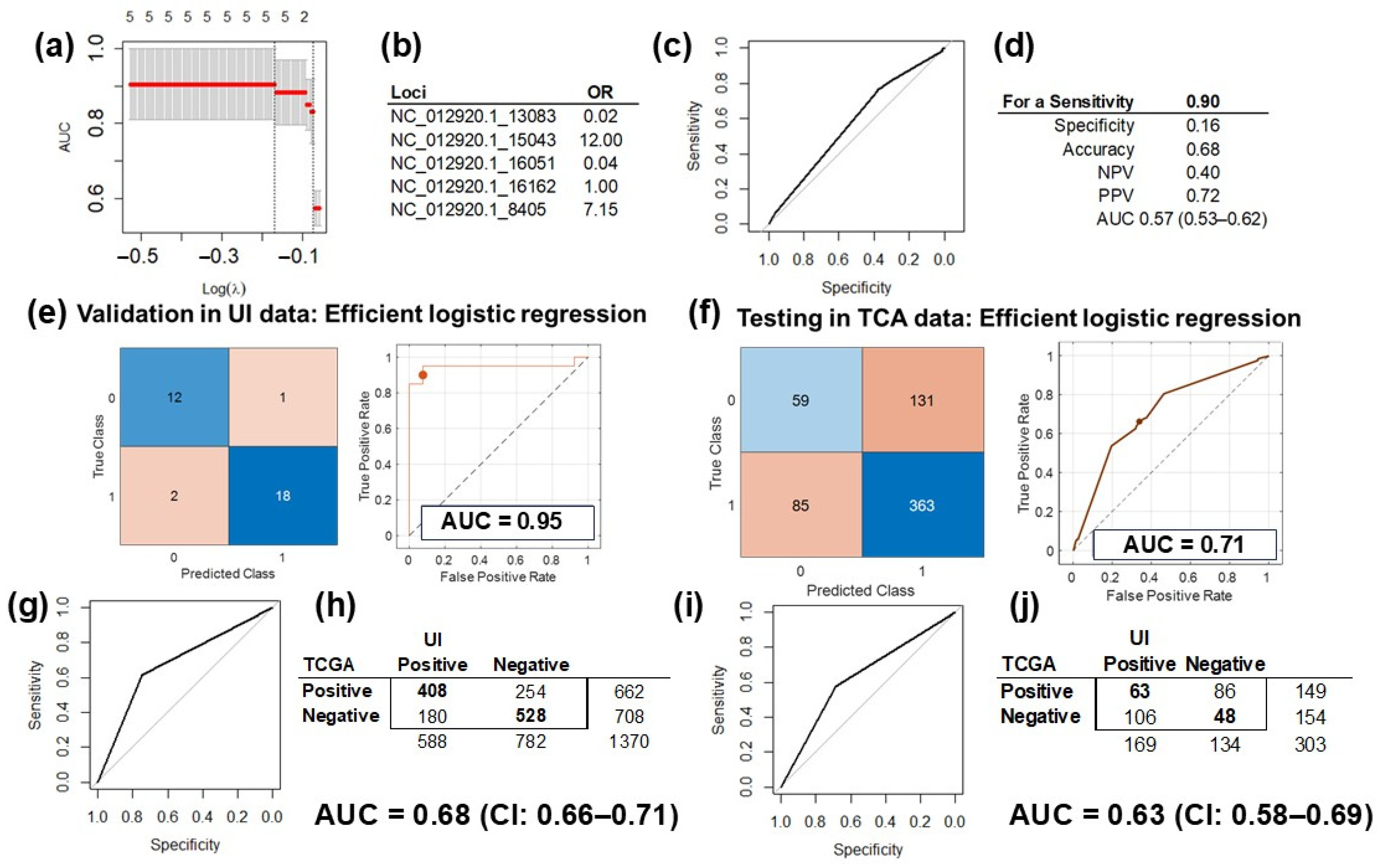
2.4. Prediction Model Training, Validation and Testing
2.5. Correlation Analysis Validation
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Specimen Acquisition
5.2. DNA Sequencing
5.3. RNA Sequencing
5.4. Single Nucleotide Variation (SNV) Analysis
5.5. Correlation of mtDNA Variables with Gene Expression and Pathway Analysis
5.6. Prediction Model Testing
5.6.1. Model Testing with the TCGA Database
5.6.2. Testing of Prediction Model of HGSC in Independent Analytical Platform
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Elias, K.M.; Guo, J.; Bast, R.C., Jr. Early Detection of Ovarian Cancer. Hematol./Oncol. Clin. N. Am. 2018, 32, 903–914. [Google Scholar] [CrossRef]
- Gonzalez Bosquet, J.; Newtson, A.M.; Chung, R.K.; Thiel, K.W.; Ginader, T.; Goodheart, M.J.; Leslie, K.K.; Smith, B.J. Prediction of chemo-response in serous ovarian cancer. Mol. Cancer 2016, 15, 66. [Google Scholar] [CrossRef]
- US Preventive Services Task Force; Grossman, D.C.; Curry, S.J.; Owens, D.K.; Barry, M.J.; Davidson, K.W.; Doubeni, C.A.; Epling, J.W., Jr.; Kemper, A.R.; Krist, A.H.; et al. Screening for Ovarian Cancer: US Preventive Services Task Force Recommendation Statement. JAMA 2018, 319, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Lam, E.T.; Bracci, P.M.; Holly, E.A.; Chu, C.; Poon, A.; Wan, E.; White, K.; Kwok, P.Y.; Pawlikowska, L.; Tranah, G.J. Mitochondrial DNA sequence variation and risk of pancreatic cancer. Cancer Res. 2012, 72, 686–695. [Google Scholar] [CrossRef]
- Kabekkodu, S.P.; Bhat, S.; Mascarenhas, R.; Mallya, S.; Bhat, M.; Pandey, D.; Kushtagi, P.; Thangaraj, K.; Gopinath, P.M.; Satyamoorthy, K. Mitochondrial DNA variation analysis in cervical cancer. Mitochondrion 2014, 16, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Nakai, T.; Sakurada, A.; Endo, T.; Kobayashi, H.; Masuda, S.; Makishima, M.; Esumi, M. Caution for simple sequence repeat number variation in the mitochondrial DNA D-loop to determine cancer-specific variants. Oncol. Lett. 2019, 17, 1883–1888. [Google Scholar] [CrossRef] [PubMed]
- Gentiluomo, M.; Katzke, V.A.; Kaaks, R.; Tjonneland, A.; Severi, G.; Perduca, V.; Boutron-Ruault, M.C.; Weiderpass, E.; Ferrari, P.; Johnson, T.; et al. Mitochondrial DNA Copy-Number Variation and Pancreatic Cancer Risk in the Prospective EPIC Cohort. Cancer Epidemiol. Biomark. Prev. 2020, 29, 681–686. [Google Scholar] [CrossRef] [PubMed]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef]
- Phallen, J.; Sausen, M.; Adleff, V.; Leal, A.; Hruban, C.; White, J.; Anagnostou, V.; Fiksel, J.; Cristiano, S.; Papp, E.; et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 2017, 9, eaan2415. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef]
- Hou, J.M.; Krebs, M.G.; Lancashire, L.; Sloane, R.; Backen, A.; Swain, R.K.; Priest, L.J.; Greystoke, A.; Zhou, C.; Morris, K.; et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J. Clin. Oncol. 2012, 30, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Casagrande, G.M.S.; Silva, M.O.; Reis, R.M.; Leal, L.F. Liquid Biopsy for Lung Cancer: Up-to-Date and Perspectives for Screening Programs. Int. J. Mol. Sci. 2023, 24, 2505. [Google Scholar] [CrossRef]
- Zhu, G.; Pei, L.; Xia, H.; Tang, Q.; Bi, F. Role of oncogenic KRAS in the prognosis, diagnosis and treatment of colorectal cancer. Mol. Cancer 2021, 20, 143. [Google Scholar] [CrossRef]
- Mazzeo, R.; Sears, J.; Palmero, L.; Bolzonello, S.; Davis, A.A.; Gerratana, L.; Puglisi, F. Liquid biopsy in triple-negative breast cancer: Unlocking the potential of precision oncology. ESMO Open 2024, 9, 103700. [Google Scholar] [CrossRef] [PubMed]
- Nahm, F.S. Receiver operating characteristic curve: Overview and practical use for clinicians. Korean J. Anesth. 2022, 75, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Bosquet, J.; Cardillo, N.D.; Reyes, H.D.; Smith, B.J.; Leslie, K.K.; Bender, D.P.; Goodheart, M.J.; Devor, E.J. Using Genomic Variation to Distinguish Ovarian High-Grade Serous Carcinoma from Benign Fallopian Tubes. Int. J. Mol. Sci. 2022, 23, 14814. [Google Scholar] [CrossRef]
- Roy, T.; Oliveira, S.; Gonzalez Bosquet, J.; Wu, X. 3D Supervised Contrastive-Learning Network for Classification of Ovarian Neoplasms. In Proceedings of the Medical Imaging with Deep Learning 2023, Nashville, TN, USA, 10 July 2023. [Google Scholar]
- Linder, K.; Watson, R.; Ulmer, K.; Bender, D.; Goodheart, M.J.; Devor, E.; Gonzalez Bosquet, J. Prediction of Ovarian Cancer with Deep Machine Learning and Alternative Splicing. Med. Res. Arch. 2023, 11, 1–16. [Google Scholar] [CrossRef]
- Miller, M.D.; Devor, E.J.; Salinas, E.A.; Newtson, A.M.; Goodheart, M.J.; Leslie, K.K.; Gonzalez-Bosquet, J. Population Substructure Has Implications in Validating Next-Generation Cancer Genomics Studies with TCGA. Int. J. Mol. Sci. 2019, 20, 1192. [Google Scholar] [CrossRef]
- Esposti, M.D.; De Vries, S.; Crimi, M.; Ghelli, A.; Patarnello, T.; Meyer, A. Mitochondrial cytochrome b: Evolution and structure of the protein. Biochim. Biophys. Acta 1993, 1143, 243–271. [Google Scholar] [CrossRef]
- Weiss, H.; Linke, P.; Haiker, H.; Leonard, K. Structure and function of the mitochondrial ubiquinol: Cytochrome c reductase and NADH: Ubiquinone reductase. Biochem. Soc. Trans. 1987, 15, 100–102. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Guo, W.; Wang, X.; Zhou, K.; Guo, S.; Liu, Y.; Sun, T.; Li, S.; Xu, Z.; Yuan, Q.; et al. Mutational profiling of mitochondrial DNA reveals an epithelial ovarian cancer-specific evolutionary pattern contributing to high oxidative metabolism. Clin. Transl. Med. 2024, 14, e1523. [Google Scholar] [CrossRef] [PubMed]
- Hahn, A.; Zuryn, S. Mitochondrial Genome (mtDNA) Mutations that Generate Reactive Oxygen Species. Antioxidants 2019, 8, 392. [Google Scholar] [CrossRef]
- Klimova, T.; Chandel, N.S. Mitochondrial complex III regulates hypoxic activation of HIF. Cell Death Differ. 2008, 15, 660–666. [Google Scholar] [CrossRef]
- Lou, C.; Ma, X.; Chen, Z.; Zhao, Y.; Yao, Q.; Zhou, C.; Zhao, X.; Meng, X. The mtDNA fragments within exosomes might be novel diagnostic biomarkers of non-small cell lung cancer. Pathol. Res. Pract. 2023, 249, 154718. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Morales, S.; Perez-Amado, C.J.; Langley, E.; Hidalgo-Miranda, A. Overview of mitochondrial germline variants and mutations in human disease: Focus on breast cancer. Int. J. Oncol. 2018, 53, 923–936. [Google Scholar] [CrossRef] [PubMed]
- Larman, T.C.; DePalma, S.R.; Hadjipanayis, A.G.; Cancer Genome Atlas Research, N.; Protopopov, A.; Zhang, J.; Gabriel, S.B.; Chin, L.; Seidman, C.E.; Kucherlapati, R.; et al. Spectrum of somatic mitochondrial mutations in five cancers. Proc. Natl. Acad. Sci. USA 2012, 109, 14087–14091. [Google Scholar] [CrossRef] [PubMed]
- Kopinski, P.K.; Singh, L.N.; Zhang, S.; Lott, M.T.; Wallace, D.C. Mitochondrial DNA variation and cancer. Nat. Rev. Cancer 2021, 21, 431–445. [Google Scholar] [CrossRef]
- Wallace, D.C. Mitochondria and cancer. Nat. Rev. Cancer 2012, 12, 685–698. [Google Scholar] [CrossRef]
- Kalsbeek, A.M.; Chan, E.F.; Grogan, J.; Petersen, D.C.; Jaratlerdsiri, W.; Gupta, R.; Lyons, R.J.; Haynes, A.M.; Horvath, L.G.; Kench, J.G.; et al. Mutational load of the mitochondrial genome predicts pathological features and biochemical recurrence in prostate cancer. Aging 2016, 8, 2702–2712. [Google Scholar] [CrossRef] [PubMed]
- Permuth-Wey, J.; Chen, Y.A.; Tsai, Y.Y.; Chen, Z.; Qu, X.; Lancaster, J.M.; Stockwell, H.; Dagne, G.; Iversen, E.; Risch, H.; et al. Inherited variants in mitochondrial biogenesis genes may influence epithelial ovarian cancer risk. Cancer Epidemiol. Biomark. Prev. 2011, 20, 1131–1145. [Google Scholar] [CrossRef]
- Yuan, Y.; Ju, Y.S.; Kim, Y.; Li, J.; Wang, Y.; Yoon, C.J.; Yang, Y.; Martincorena, I.; Creighton, C.J.; Weinstein, J.N.; et al. Comprehensive molecular characterization of mitochondrial genomes in human cancers. Nat. Genet. 2020, 52, 342–352. [Google Scholar] [CrossRef]
- Chen, K.; Holschneider, D.P.; Wu, W.; Rebrin, I.; Shih, J.C. A spontaneous point mutation produces monoamine oxidase A/B knock-out mice with greatly elevated monoamines and anxiety-like behavior. J. Biol. Chem. 2004, 279, 39645–39652. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Shi, C.T.; Chen, H.; Zhou, Q.; Ding, W.; Chen, F.; Liang, Z.W.; Teng, Y.J.; Shao, Q.X.; Dong, X.Q. Role of LncRNA MIR99AHG in breast cancer: Bioinformatic analysis and preliminary verification. Heliyon 2023, 9, e19805. [Google Scholar] [CrossRef]
- Maeda, Y.; Ide, T.; Koike, M.; Uchiyama, Y.; Kinoshita, T. GPHR is a novel anion channel critical for acidification and functions of the Golgi apparatus. Nat. Cell Biol. 2008, 10, 1135–1145. [Google Scholar] [CrossRef] [PubMed]
- Marquez, R.T.; Baggerly, K.A.; Patterson, A.P.; Liu, J.; Broaddus, R.; Frumovitz, M.; Atkinson, E.N.; Smith, D.I.; Hartmann, L.; Fishman, D.; et al. Patterns of gene expression in different histotypes of epithelial ovarian cancer correlate with those in normal fallopian tube, endometrium, and colon. Clin. Cancer Res. 2005, 11, 6116–6126. [Google Scholar] [CrossRef] [PubMed]
- Cogliati, S.; Lorenzi, I.; Rigoni, G.; Caicci, F.; Soriano, M.E. Regulation of Mitochondrial Electron Transport Chain Assembly. J. Mol. Biol. 2018, 430, 4849–4873. [Google Scholar] [CrossRef] [PubMed]
- Berner, M.J.; Wall, S.W.; Echeverria, G.V. Deregulation of mitochondrial gene expression in cancer: Mechanisms and therapeutic opportunities. Br. J. Cancer 2024, 131, 1415–1424. [Google Scholar] [CrossRef]
- Yi, C.; Zhang, X.; Chen, X.; Huang, B.; Song, J.; Ma, M.; Yuan, X.; Zhang, C. A novel 8-genome instability-associated lncRNAs signature predicting prognosis and drug sensitivity in gastric cancer. Int. J. Immunopathol. Pharmacol. 2022, 36, 3946320221103195. [Google Scholar] [CrossRef] [PubMed]
- Jing, Z.; Guo, S.; Zhang, P.; Liang, Z. LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma. Technol. Cancer Res. Treat. 2020, 19, 1533033820985787. [Google Scholar] [CrossRef]
- Ferrasi, A.C.; Fernandez, G.J.; Grotto, R.M.T.; Silva, G.F.; Goncalves, J.; Costa, M.C.; Enguita, F.J.; Pardini, M. New LncRNAs in Chronic Hepatitis C progression: From fibrosis to hepatocellular carcinoma. Sci. Rep. 2020, 10, 9886. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Pei, Y.; Yu, J.; Ding, W.; Yang, Y.; Liu, F.; Liu, L.; Huang, J.; Yuan, S.; Wang, Z.; et al. High-throughput sequencing approach for the identification of lncRNA biomarkers in hepatocellular carcinoma and revealing the effect of ZFAS1/miR-150-5p on hepatocellular carcinoma progression. PeerJ 2023, 11, e14891. [Google Scholar] [CrossRef] [PubMed]
- Perdomo, C.; Campbell, J.D.; Gerrein, J.; Tellez, C.S.; Garrison, C.B.; Walser, T.C.; Drizik, E.; Si, H.; Gower, A.C.; Vick, J.; et al. MicroRNA 4423 is a primate-specific regulator of airway epithelial cell differentiation and lung carcinogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 18946–18951. [Google Scholar] [CrossRef] [PubMed]
- Srinivasainagendra, V.; Sandel, M.W.; Singh, B.; Sundaresan, A.; Mooga, V.P.; Bajpai, P.; Tiwari, H.K.; Singh, K.K. Migration of mitochondrial DNA in the nuclear genome of colorectal adenocarcinoma. Genome Med. 2017, 9, 31. [Google Scholar] [CrossRef] [PubMed]
- Martignetti, J.A.; Camacho-Vanegas, O.; Priedigkeit, N.; Camacho, C.; Pereira, E.; Lin, L.; Garnar-Wortzel, L.; Miller, D.; Losic, B.; Shah, H.; et al. Personalized ovarian cancer disease surveillance and detection of candidate therapeutic drug target in circulating tumor DNA. Neoplasia 2014, 16, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Oikkonen, J.; Zhang, K.; Salminen, L.; Schulman, I.; Lavikka, K.; Andersson, N.; Ojanpera, E.; Hietanen, S.; Grenman, S.; Lehtonen, R.; et al. Prospective Longitudinal ctDNA Workflow Reveals Clinically Actionable Alterations in Ovarian Cancer. JCO Precis. Oncol. 2019, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Vanderstichele, A.; Busschaert, P.; Smeets, D.; Landolfo, C.; Van Nieuwenhuysen, E.; Leunen, K.; Neven, P.; Amant, F.; Mahner, S.; Braicu, E.I.; et al. Chromosomal Instability in Cell-Free DNA as a Highly Specific Biomarker for Detection of Ovarian Cancer in Women with Adnexal Masses. Clin. Cancer Res. 2017, 23, 2223–2231. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, M.; Kawashima, A.; Yasuhara, R.; Hayakawa, Y.; Miyamoto, S.; Iizuka, C.; Sekizawa, A. Massively parallel sequencing of cell-free DNA in plasma for detecting gynaecological tumour-associated copy number alteration. Sci. Rep. 2018, 8, 11205. [Google Scholar] [CrossRef]
- Gupta, R.; Othman, T.; Chen, C.; Sandhu, J.; Ouyang, C.; Fakih, M. Guardant360 Circulating Tumor DNA Assay Is Concordant with FoundationOne Next-Generation Sequencing in Detecting Actionable Driver Mutations in Anti-EGFR Naive Metastatic Colorectal Cancer. Oncologist 2020, 25, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Rolfo, C.; Mack, P.; Scagliotti, G.V.; Aggarwal, C.; Arcila, M.E.; Barlesi, F.; Bivona, T.; Diehn, M.; Dive, C.; Dziadziuszko, R.; et al. Liquid Biopsy for Advanced NSCLC: A Consensus Statement From the International Association for the Study of Lung Cancer. J. Thorac. Oncol. 2021, 16, 1647–1662. [Google Scholar] [CrossRef]
- Gonzalez Bosquet, J.; Devor, E.J.; Newtson, A.M.; Smith, B.J.; Bender, D.P.; Goodheart, M.J.; McDonald, M.E.; Braun, T.A.; Thiel, K.W.; Leslie, K.K. Creation and validation of models to predict response to primary treatment in serous ovarian cancer. Sci. Rep. 2021, 11, 5957. [Google Scholar] [CrossRef]
- Gonzalez Bosquet, J.; Marchion, D.C.; Chon, H.; Lancaster, J.M.; Chanock, S. Analysis of chemotherapeutic response in ovarian cancers using publically available high-throughput data. Cancer Res. 2014, 74, 3902–3912. [Google Scholar] [CrossRef] [PubMed]
- Erickson, B.K.; Conner, M.G.; Landen, C.N., Jr. The role of the fallopian tube in the origin of ovarian cancer. Am. J. Obs. Gynecol. 2013, 209, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Shih, I.M.; Wang, Y.; Wang, T.L. The Origin of Ovarian Cancer Species and Precancerous Landscape. Am. J. Pathol. 2021, 191, 26–39. [Google Scholar] [CrossRef]
- Ploner, A. Heatplus: Heatmaps with Row and/or Column Covariates and Colored Clusters. Available online: https://github.com/alexploner/Heatplus (accessed on 1 February 2025).
- Reyes, H.D.; Devor, E.J.; Warrier, A.; Newtson, A.M.; Mattson, J.; Wagner, V.; Duncan, G.N.; Leslie, K.K.; Gonzalez-Bosquet, J. Differential DNA methylation in high-grade serous ovarian cancer (HGSOC) is associated with tumor behavior. Sci. Rep. 2019, 9, 17996. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Miller, M.D.; Salinas, E.A.; Newtson, A.M.; Sharma, D.; Keeney, M.E.; Warrier, A.; Smith, B.J.; Bender, D.P.; Goodheart, M.J.; Thiel, K.W.; et al. An integrated prediction model of recurrence in endometrial endometrioid cancers. Cancer Manag. Res. 2019, 11, 5301–5315. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Alganmi, N.; Abusamra, H. Evaluation of an optimized germline exomes pipeline using BWA-MEM2 and Dragen-GATK tools. PLoS ONE 2023, 18, e0288371. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef] [PubMed]
- Riley, R.D.; Snell, K.I.E.; Martin, G.P.; Whittle, R.; Archer, L.; Sperrin, M.; Collins, G.S. Penalization and shrinkage methods produced unreliable clinical prediction models especially when sample size was small. J. Clin. Epidemiol. 2021, 132, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
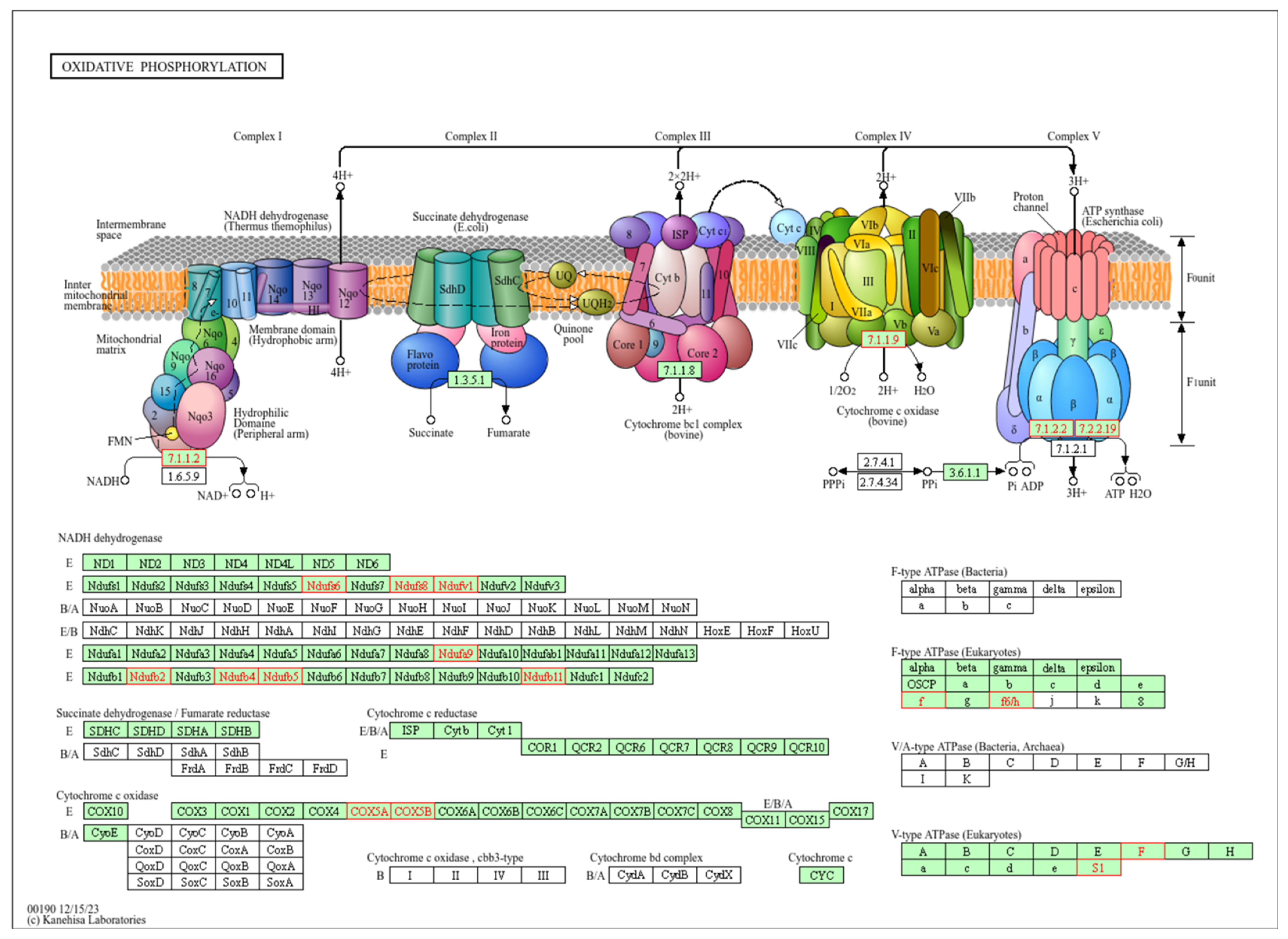
| ID | Category | Description | Fold | Adjusted p-Value |
|---|---|---|---|---|
| hsa00190 | Energy metabolism | Oxidative phosphorylation | 2.84 | <0.001 |
| hsa05415 | Cardiovascular disease | Diabetic cardiomyopathy | 2.29 | <0.001 |
| hsa04714 | Environmental adaptation | Thermogenesis | 2.17 | <0.001 |
| hsa05016 | Neurodegenerative disease | Huntington disease | 1.89 | 0.001 |
| hsa05014 | Neurodegenerative disease | Amyotrophic lateral sclerosis | 1.80 | 0.001 |
| hsa05012 | Neurodegenerative disease | Parkinson disease | 1.88 | 0.001 |
| hsa04932 | Endocrine and metabolic disease | Non-alcoholic fatty liver disease | 2.12 | 0.003 |
| hsa05020 | Neurodegenerative disease | Prion disease | 1.76 | 0.004 |
| hsa05208 | Cancer: overview | Chemical carcinogenesis - reactive oxygen species | 1.81 | 0.006 |
| hsa05010 | Neurodegenerative disease | Alzheimer disease | 1.55 | 0.009 |
| hsa05022 | Neurodegenerative disease | Pathways of neurodegeneration - multiple diseases | 1.46 | 0.014 |
| hsa00510 | Glycan biosynthesis and metabolism | N-Glycan biosynthesis | 2.59 | 0.017 |
| hsa00600 | Lipid metabolism | Sphingolipid metabolism | 2.54 | 0.019 |
| hsa04723 | Nervous system | Retrograde endocannabinoid signaling | 1.84 | 0.020 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzalez Bosquet, J.; Wagner, V.; Polio, A.; Linder, K.E.; Bender, D.P.; Goodheart, M.J.; Schickling, B.M. Identification of Ovarian High-Grade Serous Carcinoma with Mitochondrial Gene Variation. Int. J. Mol. Sci. 2025, 26, 1347. https://doi.org/10.3390/ijms26031347
Gonzalez Bosquet J, Wagner V, Polio A, Linder KE, Bender DP, Goodheart MJ, Schickling BM. Identification of Ovarian High-Grade Serous Carcinoma with Mitochondrial Gene Variation. International Journal of Molecular Sciences. 2025; 26(3):1347. https://doi.org/10.3390/ijms26031347
Chicago/Turabian StyleGonzalez Bosquet, Jesus, Vincent Wagner, Andrew Polio, Katharine E. Linder, David P. Bender, Michael J. Goodheart, and Brandon M. Schickling. 2025. "Identification of Ovarian High-Grade Serous Carcinoma with Mitochondrial Gene Variation" International Journal of Molecular Sciences 26, no. 3: 1347. https://doi.org/10.3390/ijms26031347
APA StyleGonzalez Bosquet, J., Wagner, V., Polio, A., Linder, K. E., Bender, D. P., Goodheart, M. J., & Schickling, B. M. (2025). Identification of Ovarian High-Grade Serous Carcinoma with Mitochondrial Gene Variation. International Journal of Molecular Sciences, 26(3), 1347. https://doi.org/10.3390/ijms26031347






