Decoding PHR-Orchestrated Stress Adaptation: A Genome-Wide Integrative Analysis of Transcriptional Regulation Under Abiotic Stress in Eucalyptus grandis
Abstract
1. Introduction
2. Results
2.1. Genome-Wide Characterization of PHRs in Eucalyptus grandis
2.2. Phylogenetic Analysis of Eucalyptus PHR Proteins
2.3. Distinct Motifs Were Present or Absent Among Different Groups of PHRs
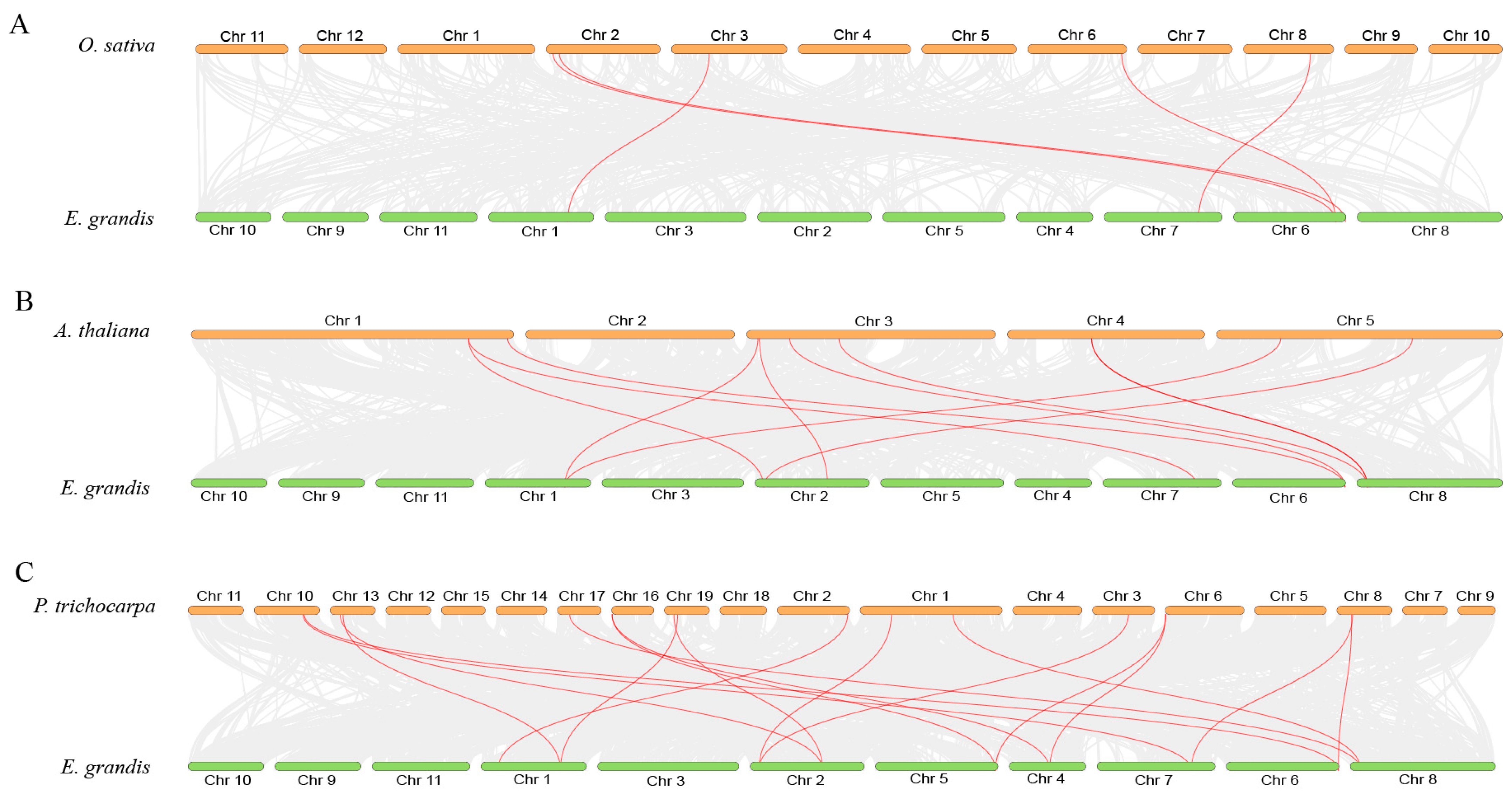
2.4. Gene Duplication and Syntenic Analyses of EgPHRs
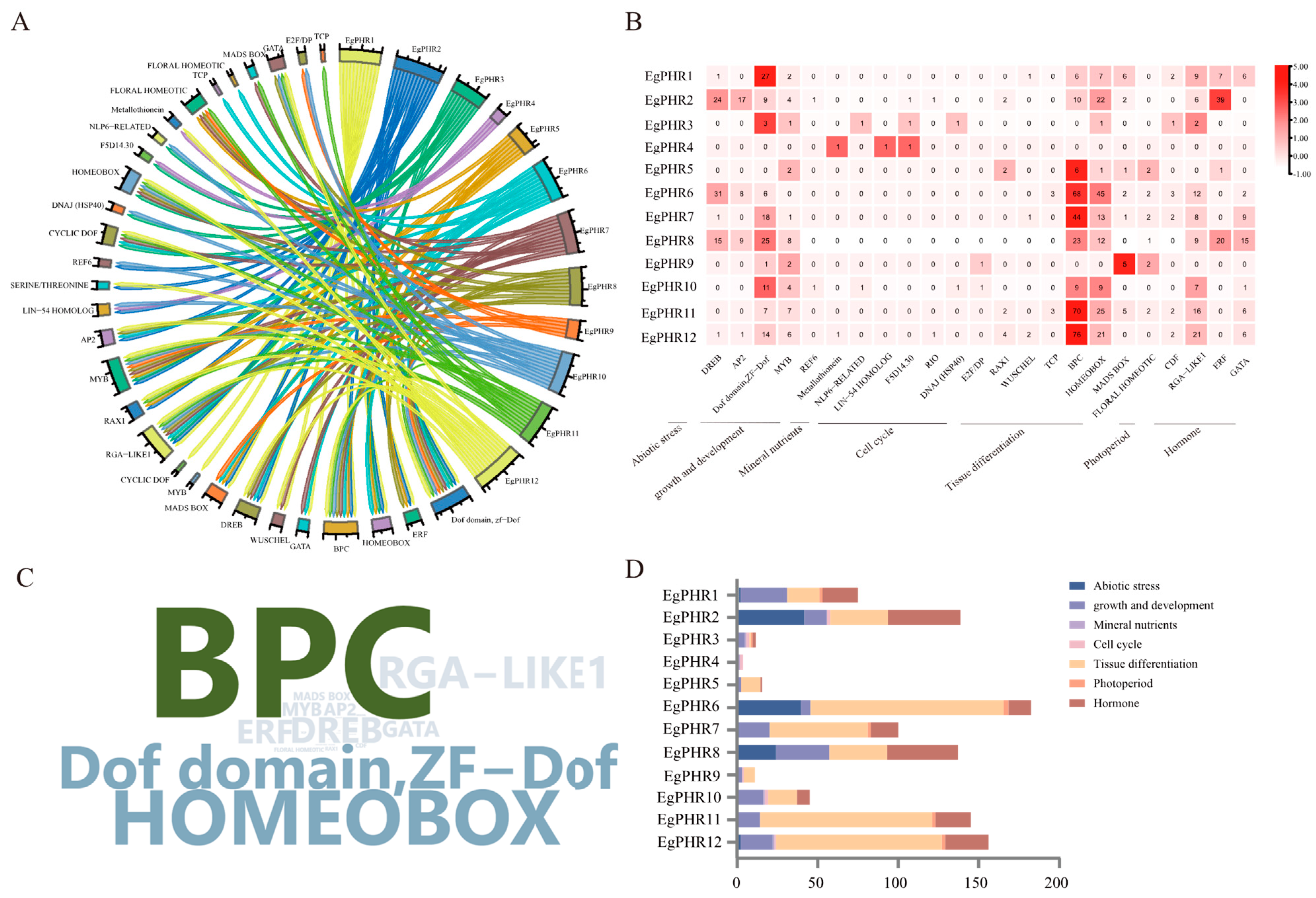
2.5. The Potential Upstream Regulatory Transcription Factors of EgPHRs
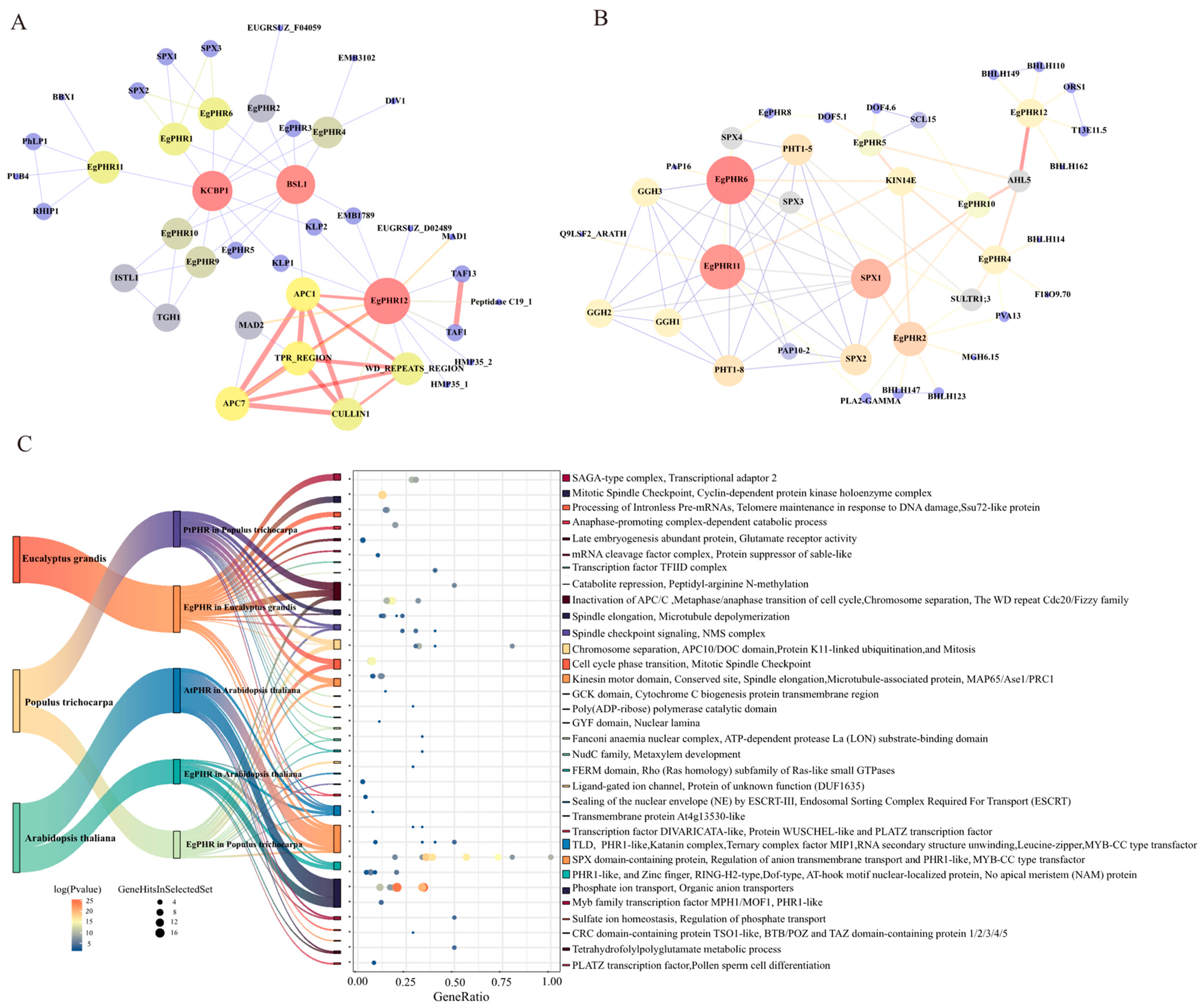
2.6. Protein–Protein Interaction Network of EgPHRs
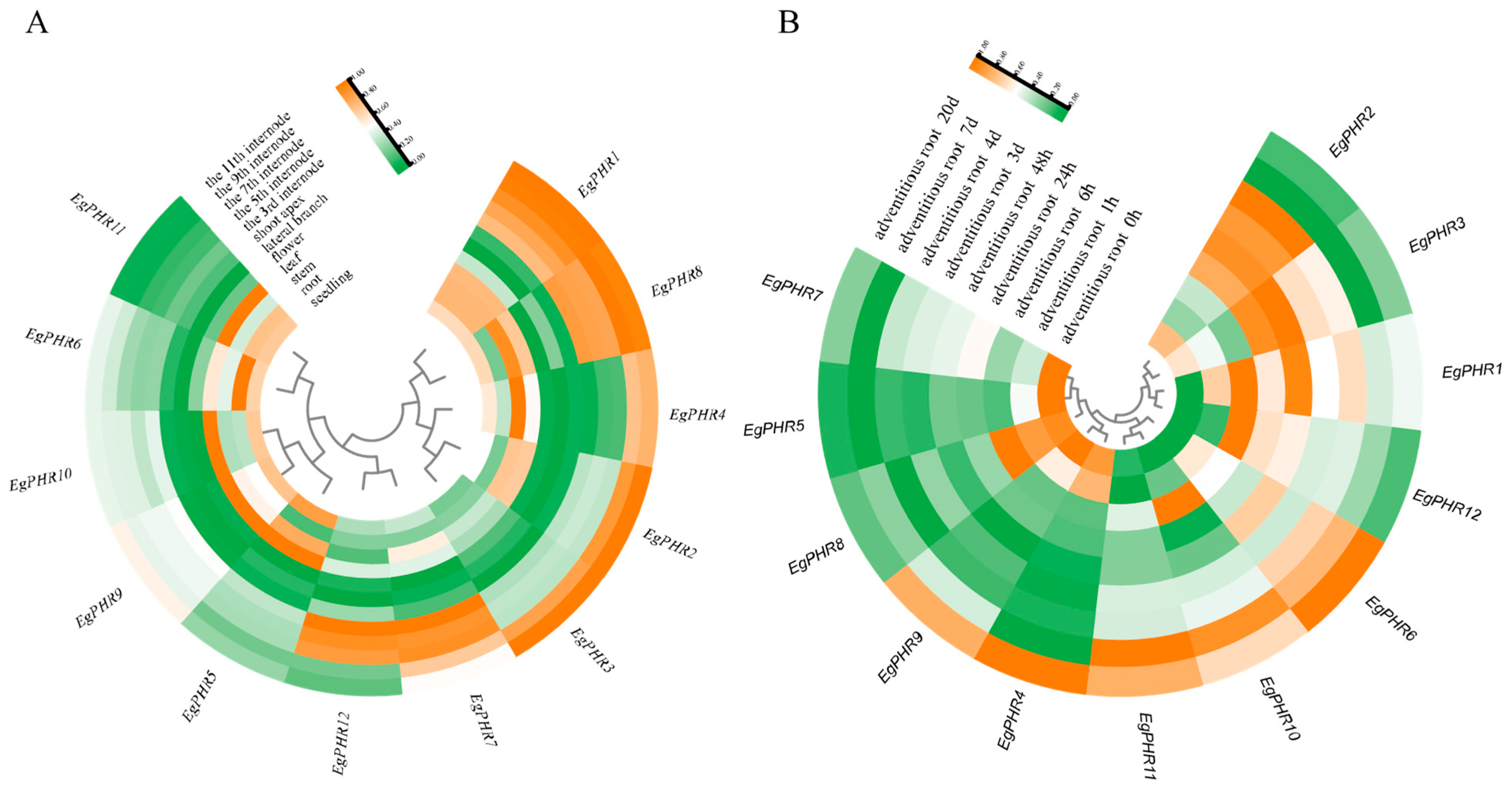
2.7. Gene Expression Patterns of EgPHRs in the Development of Different Tissues
2.8. Gene Expression Patterns of EgPHRs During the Adventitious Root Development
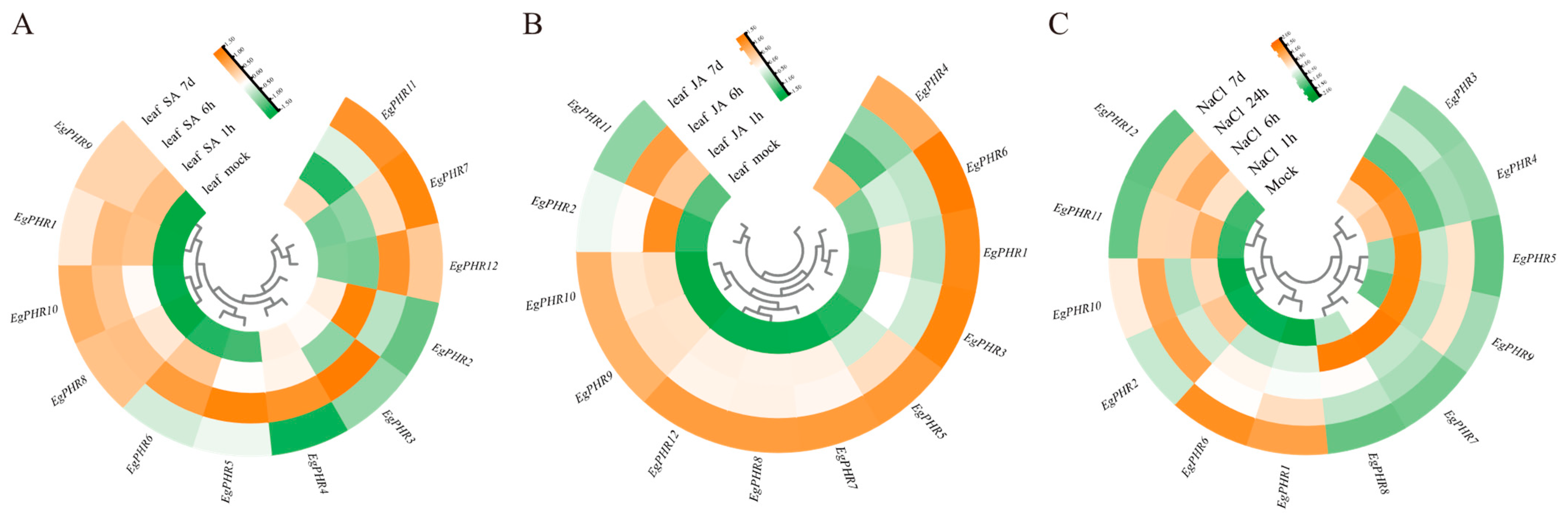
2.9. Gene Expression Pattern of EgPHRs Under SA and JA Treatment
2.10. Gene Expression Pattern of EgPHRs Under Salt Stress
2.11. Gene Expression Pattern of EgPHRs Under Cold Stress
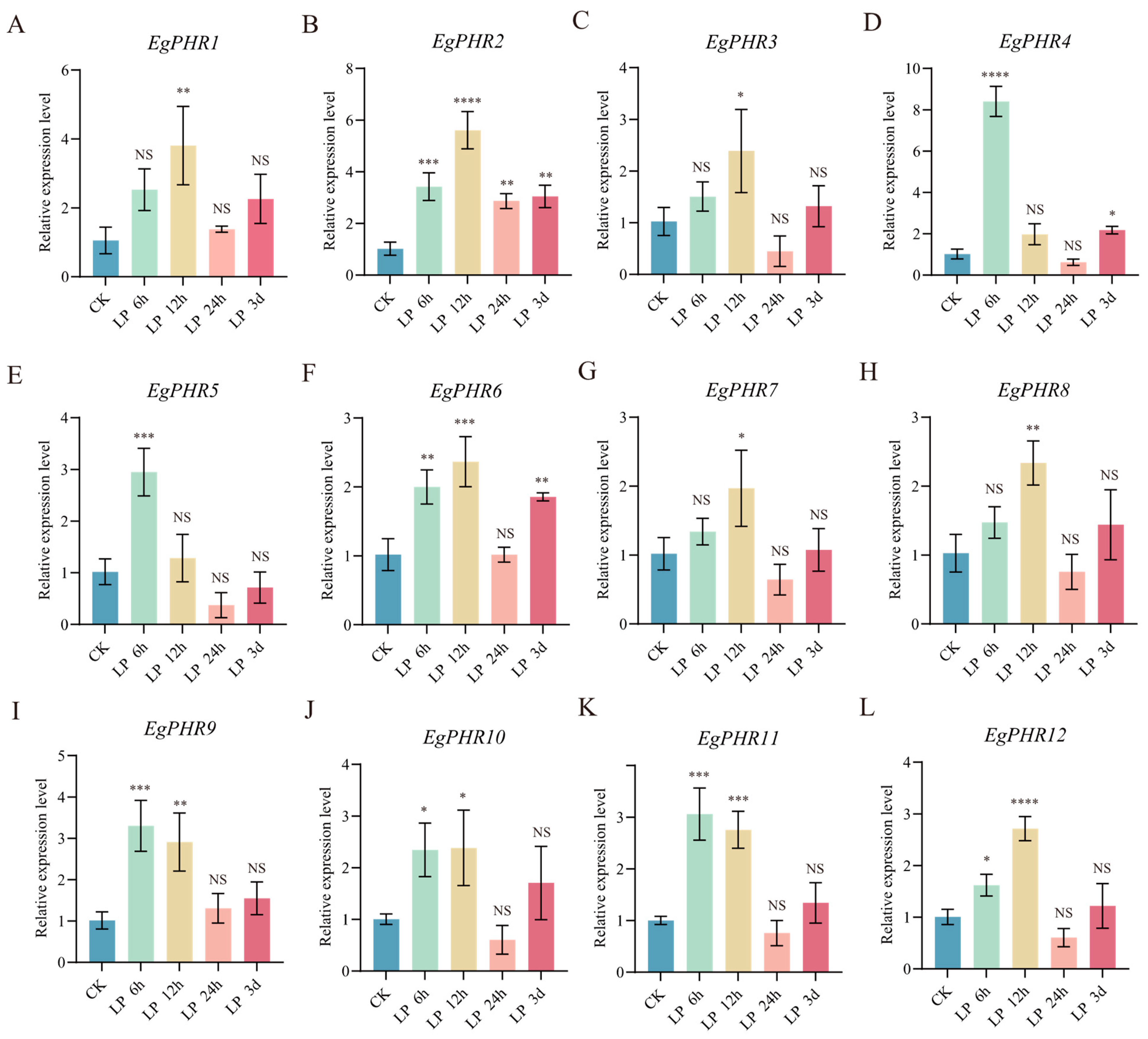
2.12. Gene Expression Pattern of EgPHRs Under Low-Phosphate Starvation
2.13. Gene Expression Pattern of EgPHRs Under Nitrogen Starvation
2.14. Gene Expression Pattern of EgPHRs Under Boron Deficiency
3. Discussion
4. Materials and Methods
4.1. Characterization of EgPHRs in Eucalyptus grandis
4.2. Phylogenetic Analysis
4.3. Conserved Domains, Motifs, and Gene Structure Analysis
4.4. Duplication Event Analysis and Multi-Species Analysis of Covariance
4.5. Prediction and Analysis of Potential Upstream Regulatory Transcription Factors for EgPHRs
4.6. Protein–Protein Interaction Analysis
4.7. Abiotic Stress Treatments and RT-qPCR Analysis
4.8. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| EgPHRs | Eucalyptus grandis phosphate starvation responses |
| Eg | Eucalyptus grandis |
| At | Arabidopsis thaliana |
| Potri | Populus trichocarpa |
| Os | Oryza sativa |
| Chr | Chromosome |
| FPKM | Fragments per kilobase of transcript per million mapped reads |
| HMM | Hidden Markov Model |
| RT-qPCR | Quantitative reverse transcription PCR |
| WGS | Whole-genome sequencing |
| PUE | Phosphorus use efficiency |
| PPI | Protein–protein interaction |
| SPX | SYG1, Pho81, XPR1 |
| PAP | Purple acid phosphatase |
| WUS | WUSCHEL |
| KNOX1 | Knotted1-like homeobox 1 |
| WOX13 | WUSCHEL-related homeobox 13 |
| SIP1 | Smad-interacting protein 1 |
| BSL1 | BSU1-like family of Ser/Thr protein phosphatases 1 |
| KCBP1 | Kinesin-like calmodulin binding protein 1 |
References
- Fang, X.; Yang, D.; Deng, L.; Zhang, Y.; Lin, Z.; Zhou, J.; Chen, Z.; Ma, X.; Guo, M.; Lu, Z.; et al. Phosphorus uptake, transport, and signaling in woody and model plants. For. Res. 2024, 4, e017. [Google Scholar] [CrossRef]
- Hong, L.; Wang, Q.; Zhang, J.; Chen, X.; Liu, Y.; Asiegbu, F.O.; Wu, P.; Ma, X.; Wang, K. Advances in the beneficial endophytic fungi for the growth and health of woody plants. For. Res. 2024, 4, e028. [Google Scholar] [CrossRef]
- Paz-Ares, J.; Puga, M.I.; Rojas-Triana, M.; Martinez-Hevia, I.; Diaz, S.; Poza-Carrion, C.; Minambres, M.; Leyva, A. Plant adaptation to low phosphorus availability: Core signaling, crosstalks, and applied implications. Mol. Plant 2022, 15, 104–124. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, Y.F.; Wu, W.H. Potassium and phosphorus transport and signaling in plants. J. Integr. Plant Biol. 2021, 63, 34–52. [Google Scholar] [CrossRef]
- Meharg, A.A. Marschner’s Mineral Nutrition of Higher Plants, 3rd ed.; Marschner, P., Ed.; Elsevier: Amsterdam, The Netherlands; Academic Press: Amsterdam, The Netherlands, 2011; p. 684. ISBN 978-0-12-384905-2. [Google Scholar]
- Malhotra, H.; Vandana; Sharma, S.; Pandey, R. Phosphorus Nutrition: Plant Growth in Response to Deficiency and Excess. In Plant Nutrients and Abiotic Stress Tolerance; Springer: Singapore, 2018; pp. 171–190. [Google Scholar]
- Bayu, D.; Dejene, A.; Alemayehu, R.; Gezahegn, B. Improving available phosphorus in acidic soil using biochar. JSSEM 2017, 8, 87–94. [Google Scholar] [CrossRef]
- Lei, J.; Yin, J.; Chen, S.; Fenton, O.; Liu, R.; Chen, Q.; Fan, B.; Zhang, S. Understanding phosphorus mobilization mechanisms in acidic soil amended with calcium-silicon-magnesium-potassium fertilizer. Sci. Total Environ. 2024, 916, 170294. [Google Scholar] [CrossRef] [PubMed]
- Liu, D. Root developmental responses to phosphorus nutrition. J. Integr. Plant Biol. 2021, 63, 1065–1090. [Google Scholar] [CrossRef]
- Sega, P.; Pacak, A. Plant PHR Transcription Factors: Put on A Map. Genes 2019, 10, 1018. [Google Scholar] [CrossRef]
- Zhu, J.; Lau, K.; Puschmann, R.; Harmel, R.K.; Zhang, Y.; Pries, V.; Gaugler, P.; Broger, L.; Dutta, A.K.; Jessen, H.J.; et al. Two bifunctional inositol pyrophosphate kinases/phosphatases control plant phosphate homeostasis. eLife 2019, 8, e43582. [Google Scholar] [CrossRef]
- Zhou, J.; Hu, Q.; Xiao, X.; Yao, D.; Ge, S.; Ye, J.; Li, H.; Cai, R.; Liu, R.; Meng, F.; et al. Mechanism of phosphate sensing and signaling revealed by rice SPX1-PHR2 complex structure. Nat. Commun. 2021, 12, 7040. [Google Scholar] [CrossRef]
- Guan, Z.; Zhang, Q.; Zhang, Z.; Zuo, J.; Chen, J.; Liu, R.; Savarin, J.; Broger, L.; Cheng, P.; Wang, Q.; et al. Mechanistic insights into the regulation of plant phosphate homeostasis by the rice SPX2–PHR2 complex. Nat. Commun. 2022, 13, 1581. [Google Scholar] [CrossRef]
- Bustos, R.; Castrillo, G.; Linhares, F.; Puga, M.I.; Rubio, V.; Perez-Perez, J.; Solano, R.; Leyva, A.; Paz-Ares, J. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010, 6, e1001102. [Google Scholar] [CrossRef]
- Lin, W.Y.; Huang, T.K.; Chiou, T.J. Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell 2013, 25, 4061–4074. [Google Scholar] [CrossRef]
- Ayadi, A.; David, P.; Arrighi, J.F.; Chiarenza, S.; Thibaud, M.C.; Nussaume, L.; Marin, E. Reducing the genetic redundancy of Arabidopsis PHOSPHATE TRANSPORTER1 transporters to study phosphate uptake and signaling. Physiol. Plant 2015, 167, 1511–1526. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Liu, F.; Han, G.; Cheng, B. Genome-wide identification and comparative analysis of phosphate starvation-responsive transcription factors in maize and three other gramineous plants. Plant Cell Rep. 2018, 37, 711–726. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, P.; Wang, H.; Feng, J.; Li, Y.; Wang, S.; Li, Y.; Guo, Y.; Li, L.; Su, Y.; et al. Genome-wide investigation and expression pattern of PHR family genes in cotton under low phosphorus stress. PeerJ 2022, 10, e14584. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zheng, Z.; Zhu, Y.; Kong, S.; Liu, D. PHOSPHATE RESPONSE 1 family members act distinctly to regulate transcriptional responses to phosphate starvation. Plant Physiol. 2023, 191, 1324–1343. [Google Scholar] [CrossRef]
- Yue, C.; Chen, Q.; Hu, J.; Li, C.; Luo, L.; Zeng, L. Genome-Wide Identification and Characterization of GARP Transcription Factor Gene Family Members Reveal Their Diverse Functions in Tea Plant (Camellia sinensis). Front. Plant Sci. 2022, 13, 947072. [Google Scholar] [CrossRef]
- Li, R.; An, J.-P.; You, C.-X.; Wang, X.-F.; Hao, Y.-J. Overexpression of MdPHR1 Enhanced Tolerance to Phosphorus Deficiency by Increasing MdPAP10 Transcription in Apple (Malus × Domestica). J. Plant Growth Regul. 2020, 40, 1753–1763. [Google Scholar] [CrossRef]
- Chen, N.; Tong, S.; Yang, J.; Qin, J.; Wang, W.; Chen, K.; Shi, W.; Li, J.; Liu, J.; Jiang, Y. PtoWRKY40 interacts with PtoPHR1-LIKE3 while regulating the phosphate starvation response in poplar. Plant Physiol. 2022, 190, 2688–2705. [Google Scholar] [CrossRef]
- Liu, T.; Xie, Y. Analysis and Prospects of the Drivers of the Rapid Development of Eucalyptus Plantation Forests in China. Eucalyptus Technol. 2020, 37, 38–47. [Google Scholar] [CrossRef]
- Tang, X.; Lei, P.; You, Q.; Liu, Y.; Jiang, S.; Ding, J.; Chen, J.; You, H. Monitoring Seasonal Growth of Eucalyptus Plantation under Different Forest Age and Slopes Based on Multi-Temporal UAV Stereo Images. Forests 2023, 14, 2231. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Khan, A.; Hashem, A.; Abd_Allah, E.F.; Al-Harrasi, A. The molecular mass and isoelectric point of plant proteomes. BMC Genom. 2019, 20, 631. [Google Scholar] [CrossRef]
- Guruprasad, K.; Reddy, B.V.; Pandit, M.W. Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 1990, 4, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Ninkuu, V.; Yan, J.; Fu, Z.; Yang, T.; Zhang, L.; Ren, J.; Li, G.; Zeng, H. Genome-wide identification, phylogenomics, and expression analysis of benzoxazinoids gene family in rice (Oryza sativa). Plant Stress 2023, 10, 100214. [Google Scholar] [CrossRef]
- Zhou, J.; Jiao, F.; Wu, Z.; Li, Y.; Wang, X.; He, X.; Zhong, W.; Wu, P. OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol. 2008, 146, 1673–1686. [Google Scholar] [CrossRef]
- Guo, M.; Ruan, W.; Li, C.; Huang, F.; Zeng, M.; Liu, Y.; Yu, Y.; Ding, X.; Wu, Y.; Wu, Z.; et al. Integrative Comparison of the Role of the PHOSPHATE RESPONSE1 Subfamily in Phosphate Signaling and Homeostasis in Rice. Plant Physiol. 2015, 168, 1762–1776. [Google Scholar] [CrossRef]
- Shi, J.; Zhao, B.; Zheng, S.; Zhang, X.; Wang, X.; Dong, W.; Xie, Q.; Wang, G.; Xiao, Y.; Chen, F.; et al. A phosphate starvation response-centered network regulates mycorrhizal symbiosis. Cell 2021, 184, 5527–5540.e5518. [Google Scholar] [CrossRef]
- Naidoo, R.; Ferreira, L.; Berger, D.K.; Myburg, A.A.; Naidoo, S. The identification and differential expression of Eucalyptus grandis pathogenesis-related genes in response to salicylic acid and methyl jasmonate. Front. Plant Sci. 2013, 4, 42592. [Google Scholar]
- Ye, W.; Wang, T.; Wei, W.; Lou, S.; Lan, F.; Zhu, S.; Li, Q.; Ji, G.; Lin, C.; Wu, X.; et al. The Full-Length Transcriptome of Spartina alterniflora Reveals the Complexity of High Salt Tolerance in Monocotyledonous Halophyte. Plant Cell Physiol. 2020, 61, 882–896. [Google Scholar] [CrossRef]
- Chen, B.; Liu, T.; Yang, Z.; Yang, S.; Chen, J. PacBio Full-Length Transcriptome Sequencing Reveals the Mechanism of Salt Stress Response in Sonneratia apetala. Plants 2023, 12, 3849. [Google Scholar] [CrossRef] [PubMed]
- Oberschelp, G.P.J.; Guarnaschelli, A.B.; Teson, N.; Harrand, L.; Podestá, F.E.; Margarit, E. Cold acclimation and freezing tolerance in three Eucalyptus species: A metabolomic and proteomic approach. Plant Physiol. Biochem. 2020, 154, 316–327. [Google Scholar] [CrossRef] [PubMed]
- Medici, A.; Szponarski, W.; Dangeville, P.; Safi, A.; Dissanayake, I.M.; Saenchai, C.; Emanuel, A.; Rubio, V.; Lacombe, B.; Ruffel, S.; et al. Identification of Molecular Integrators Shows that Nitrogen Actively Controls the Phosphate Starvation Response in Plants. Plant Cell 2019, 31, 1171–1184. [Google Scholar] [CrossRef]
- Luo, J.; Liang, Z.; Wu, M.; Mei, L. Genome-wide identification of BOR genes in poplar and their roles in response to various environmental stimuli. Environ. Exp. Bot. 2019, 164, 101–113. [Google Scholar] [CrossRef]
- Wang, T.; Ye, W.; Zhang, J.; Li, H.; Zeng, W.; Zhu, S.; Ji, G.; Wu, X.; Ma, L. Alternative 3′-untranslated regions regulate high-salt tolerance of Spartina alterniflora. Plant Physiol. 2023, 191, 2570–2587. [Google Scholar] [CrossRef]
- Lin, Z.; Guo, C.; Lou, S.; Jin, S.; Zeng, W.; Guo, Y.; Fang, J.; Xu, Z.; Zuo, Z.; Ma, L. Functional analyses unveil the involvement of moso bamboo (Phyllostachys edulis) group I and II NIN-LIKE PROTEINS in nitrate signaling regulation. Plant Sci. 2021, 306, 110862. [Google Scholar] [CrossRef]
- Fan, C.; Lyu, M.; Zeng, B.; He, Q.; Wang, X.; Lu, M.Z.; Liu, B.; Liu, J.; Esteban, E.; Pasha, A.; et al. Profiling of the gene expression and alternative splicing landscapes of Eucalyptus grandis. Plant Cell Environ. 2024, 47, 1363–1378. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Naito, Y.; Nakano, T.; Ohtsuki, N.; Suzuki, K. NSR1/MYR2 is a negative regulator of ASN1 expression and its possible involvement in regulation of nitrogen reutilization in Arabidopsis. Plant Sci. 2017, 263, 219–225. [Google Scholar] [CrossRef]
- Zhao, C.; Hanada, A.; Yamaguchi, S.; Kamiya, Y.; Beers, E.P. The Arabidopsis Myb genes MYR1 and MYR2 are redundant negative regulators of flowering time under decreased light intensity. Plant J. 2011, 66, 502–515. [Google Scholar] [CrossRef]
- Hu, B.; Chu, C. Nitrogen–phosphorus interplay: Old story with molecular tale. New Phytol. 2020, 225, 1455–1460. [Google Scholar] [CrossRef]
- Hu, B.; Jiang, Z.; Wang, W.; Qiu, Y.; Zhang, Z.; Liu, Y.; Li, A.; Gao, X.; Liu, L.; Qian, Y.; et al. Nitrate–NRT1.1B–SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat. Plants 2019, 5, 401–413. [Google Scholar] [CrossRef]
- Long, Y.; Peng, J. Interaction between Boron and Other Elements in Plants. Genes 2023, 14, 130. [Google Scholar] [CrossRef]
- Kaya, C.; Tuna, A.L.; Dikilitas, M.; Ashraf, M.; Koskeroglu, S.; Guneri, M. Supplementary phosphorus can alleviate boron toxicity in tomato. Sci. Hortic. 2009, 121, 284–288. [Google Scholar] [CrossRef]
- Gunes, A.; Alpaslan, M. Boron uptake and toxicity in maize genotypes in relation to boron and phosphorus supply. J. Plant Nutr. 2000, 23, 541–550. [Google Scholar] [CrossRef]
- Masood, S.; Zhao, X.Q.; Shen, R.F. Bacillus pumilus increases boron uptake and inhibits rapeseed growth under boron supply irrespective of phosphorus fertilization. AoB Plants 2019, 11, plz036. [Google Scholar] [CrossRef]
- He, K.; Du, J.; Han, X.; Li, H.; Kui, M.; Zhang, J.; Huang, Z.; Fu, Q.; Jiang, Y.; Hu, Y. PHOSPHATE STARVATION RESPONSE1 (PHR1) interacts with JASMONATE ZIM-DOMAIN (JAZ) and MYC2 to modulate phosphate deficiency-induced jasmonate signaling in Arabidopsis. Plant Cell 2023, 35, 2132–2156. [Google Scholar] [CrossRef]
- Li, L.; Zhang, X.; Li, D.; Su, H.; He, Y.; Xu, Z.; Zhao, Y.; Hong, Y.; Li, Q.; Xu, P.; et al. CsPHRs-CsJAZ3 incorporates phosphate signaling and jasmonate pathway to regulate catechin biosynthesis in Camellia sinensis. Hortic. Res. 2024, 11, uhae178. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Deng, L.; Zhou, X.; Xing, Y.; Li, G.; Chen, Y.; Huang, Y.; Ma, X.; Liu, Z.-J.; Li, M.; et al. Unveiling the PHR-centered regulatory network orchestrating the phosphate starvation signaling in Chinese fir (Cunninghamia lanceolata). bioRxiv 2024. [Google Scholar] [CrossRef]
- Wang, T.; Jin, Y.; Deng, L.; Li, F.; Wang, Z.; Zhu, Y.; Wu, Y.; Qu, H.; Zhang, S.; Liu, Y.; et al. The transcription factor MYB110 regulates plant height, lodging resistance, and grain yield in rice. Plant Cell 2024, 36, 298–323. [Google Scholar] [CrossRef] [PubMed]
- Kavka, M.; Polle, A. Phosphate uptake kinetics and tissue-specific transporter expression profiles in poplar (Populus × canescens) at different phosphorus availabilities. BMC Plant Biol. 2016, 16, 206. [Google Scholar] [CrossRef]
- Yadav, R.K.; Perales, M.; Gruel, J.; Girke, T.; Jönsson, H.; Reddy, G.V. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 2011, 25, 2025–2030. [Google Scholar] [PubMed]
- Long, X.; Zhang, J.; Wang, D.; Weng, Y.; Liu, S.; Li, M.; Hao, Z.; Cheng, T.; Shi, J.; Chen, J. Expression dynamics of WOX homeodomain transcription factors during somatic embryogenesis in Liriodendron hybrids. For. Res. 2023, 3, 15. [Google Scholar] [CrossRef]
- Furumizu, C.; Alvarez, J.P.; Sakakibara, K.; Bowman, J.L. Antagonistic Roles for KNOX1 and KNOX2 Genes in Patterning the Land Plant Body Plan Following an Ancient Gene Duplication. PLoS Genet. 2015, 11, e1004980. [Google Scholar] [CrossRef]
- Ogura, N.; Sasagawa, Y.; Ito, T.; Tameshige, T.; Kawai, S.; Sano, M.; Doll, Y.; Iwase, A.; Kawamura, A.; Suzuki, T.; et al. WUSCHEL-RELATED HOMEOBOX 13 suppresses de novo shoot regeneration via cell fate control of pluripotent callus. Sci. Adv. 2023, 9, eadg6983. [Google Scholar] [CrossRef]
- Kojc, N.; Zidar, N.; Gale, N.; Poljak, M.; Fujs Komloš, K.; Cardesa, A.; Höfler, H.; Becker, K.-F. Transcription factors Snail, Slug, Twist, and SIP1 in spindle cell carcinoma of the head and neck. Virchows Arch. 2009, 454, 549–555. [Google Scholar] [CrossRef]
- Fujita, A.; Lord, M.; Hiroko, T.; Hiroko, F.; Chen, T.; Oka, C.; Misumi, Y.; Chant, J. Rax1, a protein required for the establishment of the bipolar budding pattern in yeast. Gene 2004, 327, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Park, C.H.; Wang, Z.Y.; Nickels, B.E.; Dong, J. A spatiotemporal molecular switch governs plant asymmetric cell division. Nat. Plants 2021, 7, 667–680. [Google Scholar] [CrossRef]
- Smirnova, E.A.; Reddy, A.S.; Bowser, J.; Bajer, A.S. Minus end-directed kinesin-like motor protein, Kcbp, localizes to anaphase spindle poles in Haemanthus endosperm. Cell Motil. Cytoskelet. 1998, 41, 271–280. [Google Scholar] [CrossRef]
- Wang, T.; Yang, Y.; Lou, S.; Wei, W.; Zhao, Z.; Ren, Y.; Lin, C.; Ma, L. Genome-Wide Characterization and Gene Expression Analyses of GATA Transcription Factors in Moso Bamboo (Phyllostachys edulis). Int. J. Mol. Sci. 2019, 21, 14. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Wang, T.; Li, Q.; Lou, S.; Yang, Y.; Peng, L.; Lin, Z.; Hu, Q.; Ma, L. GSK3/shaggy-like kinase 1 ubiquitously regulates cell growth from Arabidopsis to Moso bamboo (Phyllostachys edulis). Plant Sci. 2019, 283, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Wu, J.; Zhang, D.; Cai, X.; Du, L.; Lu, L.; Liu, C.; Chen, S.; Yao, Q.; Xie, S.; et al. The GRAS gene family and its roles in pineapple (Ananas comosus L.) developmental regulation and cold tolerance. BMC Plant Biol. 2024, 24, 1204. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Ewels, P.; Magnusson, M.; Lundin, S.; Kaller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]




| Name | Sequence ID | Size (AA) | MW (KDa) | PI | Instability Index | GRAVY | Subcellular Localization | Group |
|---|---|---|---|---|---|---|---|---|
| EgPHR1 | LOC104436960 | 581 | 63.21 | 5.98 | 67.24 | −0.80 | Nucleus | I |
| EgPHR2 | LOC104450718 | 498 | 55.00 | 5.68 | 71.40 | −0.72 | Nucleus | I |
| EgPHR3 | LOC104425247 | 414 | 45.41 | 6.79 | 42.30 | −0.63 | Nucleus | II |
| EgPHR4 | LOC104445780 | 384 | 43.14 | 7.62 | 71.83 | −0.76 | Nucleus | I |
| EgPHR5 | LOC104442716 | 378 | 41.88 | 6.82 | 46.29 | −0.73 | Nucleus | II |
| EgPHR6 | LOC104432550 | 371 | 40.93 | 5.20 | 53.12 | −0.76 | Nucleus | I |
| EgPHR7 | LOC104453942 | 353 | 39.24 | 7.14 | 59.45 | −0.75 | Nucleus | II |
| EgPHR8 | LOC104451042 | 344 | 37.97 | 7.90 | 46.69 | −0.75 | Nucleus | III |
| EgPHR9 | LOC104447696 | 330 | 36.09 | 5.69 | 53.79 | −0.78 | Nucleus | I |
| EgPHR10 | LOC104441194 | 330 | 35.67 | 6.39 | 44.07 | −0.70 | Nucleus | I |
| EgPHR11 | LOC104456029 | 316 | 34.12 | 6.08 | 45.25 | −0.34 | Nucleus | III |
| EgPHR12 | LOC104442369 | 294 | 32.13 | 9.04 | 50.46 | −0.69 | Nucleus | III |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, H.; Xing, Y.; Li, G.; Wang, X.; Zhou, X.; Lu, Z.; Ma, L.; Yang, D. Decoding PHR-Orchestrated Stress Adaptation: A Genome-Wide Integrative Analysis of Transcriptional Regulation Under Abiotic Stress in Eucalyptus grandis. Int. J. Mol. Sci. 2025, 26, 2958. https://doi.org/10.3390/ijms26072958
Xu H, Xing Y, Li G, Wang X, Zhou X, Lu Z, Ma L, Yang D. Decoding PHR-Orchestrated Stress Adaptation: A Genome-Wide Integrative Analysis of Transcriptional Regulation Under Abiotic Stress in Eucalyptus grandis. International Journal of Molecular Sciences. 2025; 26(7):2958. https://doi.org/10.3390/ijms26072958
Chicago/Turabian StyleXu, Huiming, Yifan Xing, Guangyou Li, Xin Wang, Xu Zhou, Zhaohua Lu, Liuyin Ma, and Deming Yang. 2025. "Decoding PHR-Orchestrated Stress Adaptation: A Genome-Wide Integrative Analysis of Transcriptional Regulation Under Abiotic Stress in Eucalyptus grandis" International Journal of Molecular Sciences 26, no. 7: 2958. https://doi.org/10.3390/ijms26072958
APA StyleXu, H., Xing, Y., Li, G., Wang, X., Zhou, X., Lu, Z., Ma, L., & Yang, D. (2025). Decoding PHR-Orchestrated Stress Adaptation: A Genome-Wide Integrative Analysis of Transcriptional Regulation Under Abiotic Stress in Eucalyptus grandis. International Journal of Molecular Sciences, 26(7), 2958. https://doi.org/10.3390/ijms26072958







