Biochemical and Structural Characterization of a Highly Glucose-Tolerant β-Glucosidase from the Termite Reticulitermes perilucifugus
Abstract
1. Introduction
2. Results and Discussion
2.1. Sequence and Structural Analyses of RpBgl8
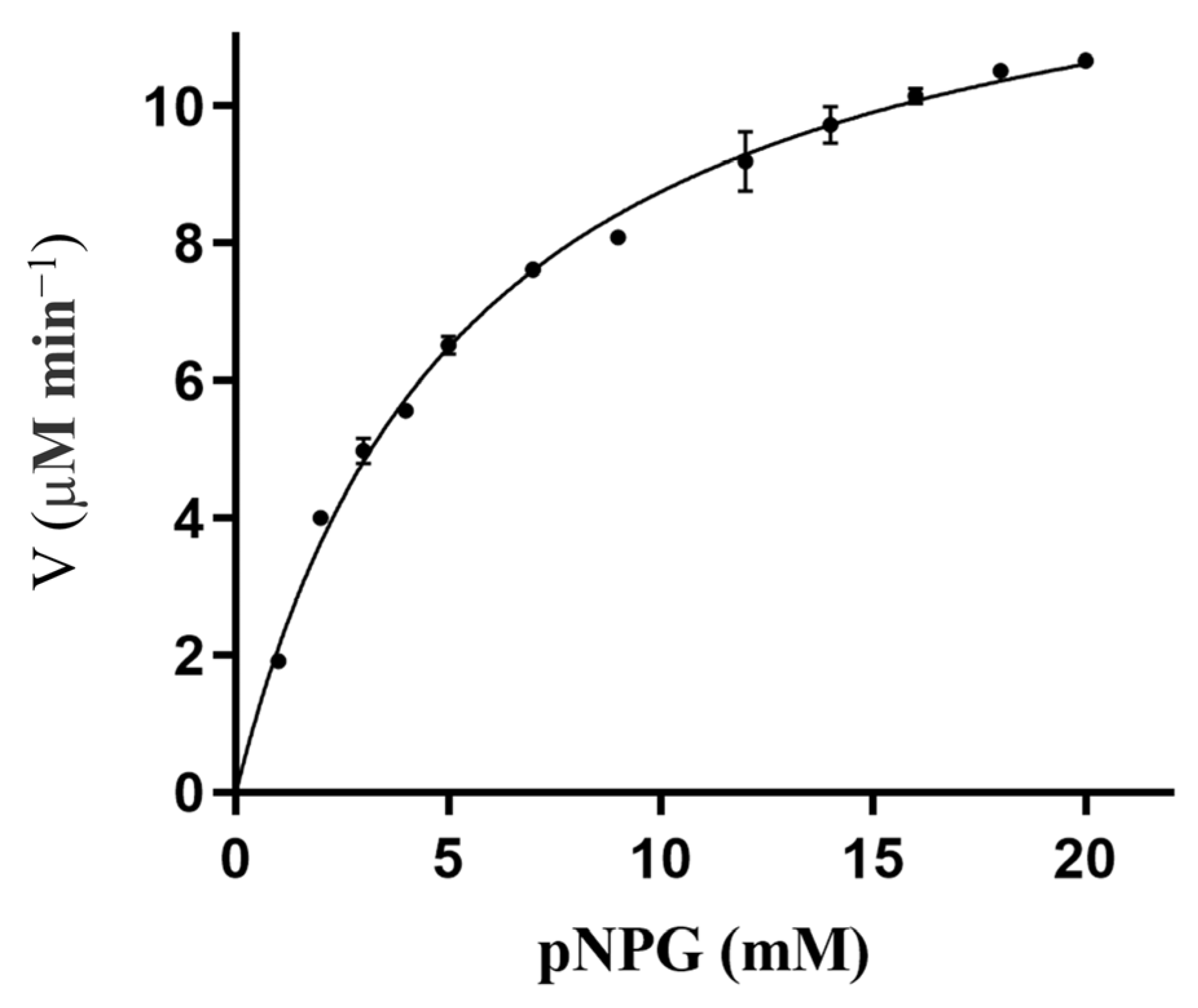
2.2. Cloning, Purification, and Characterization of RpBgl8
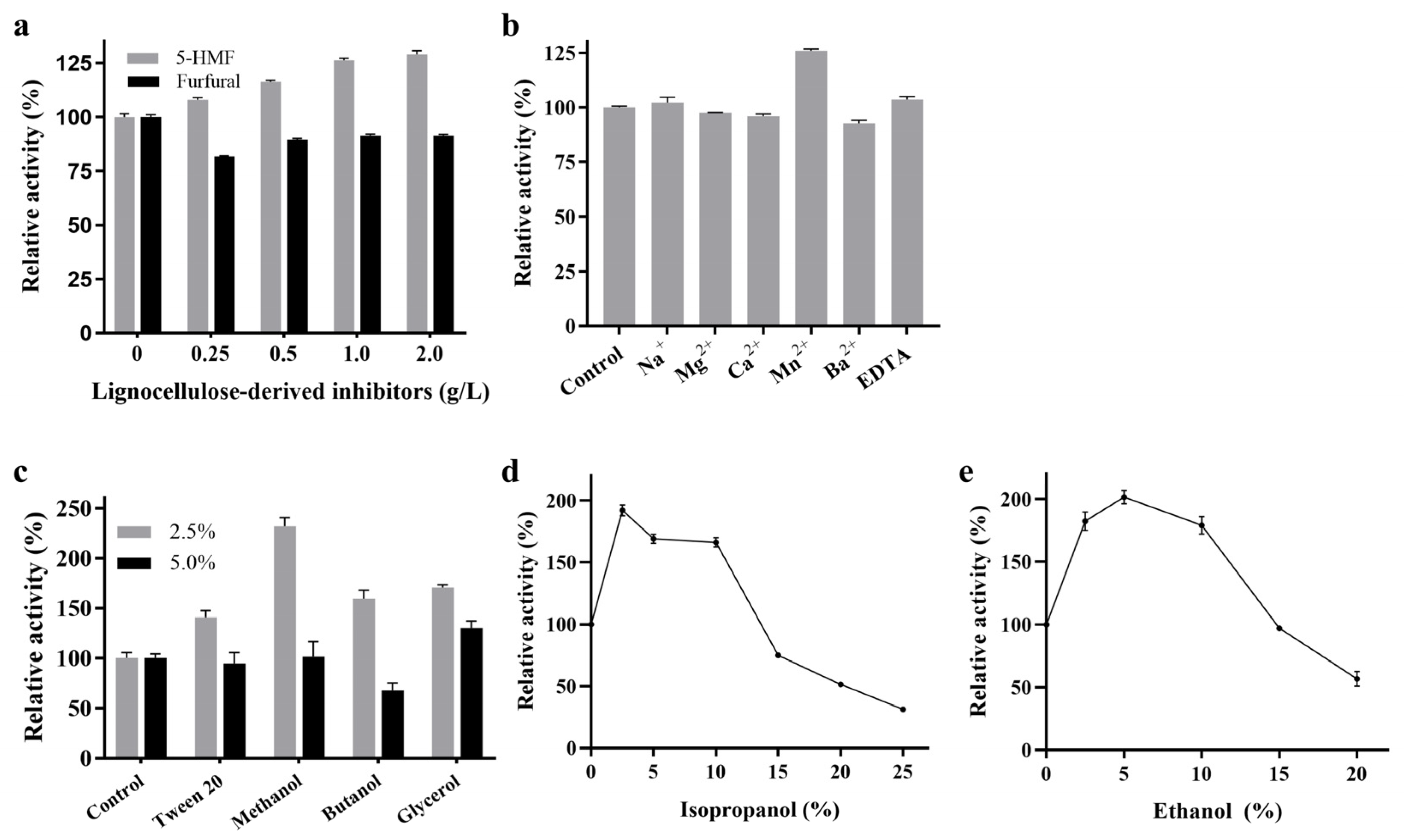
2.3. Effects of Chemicals on the Activity of RpBgl8
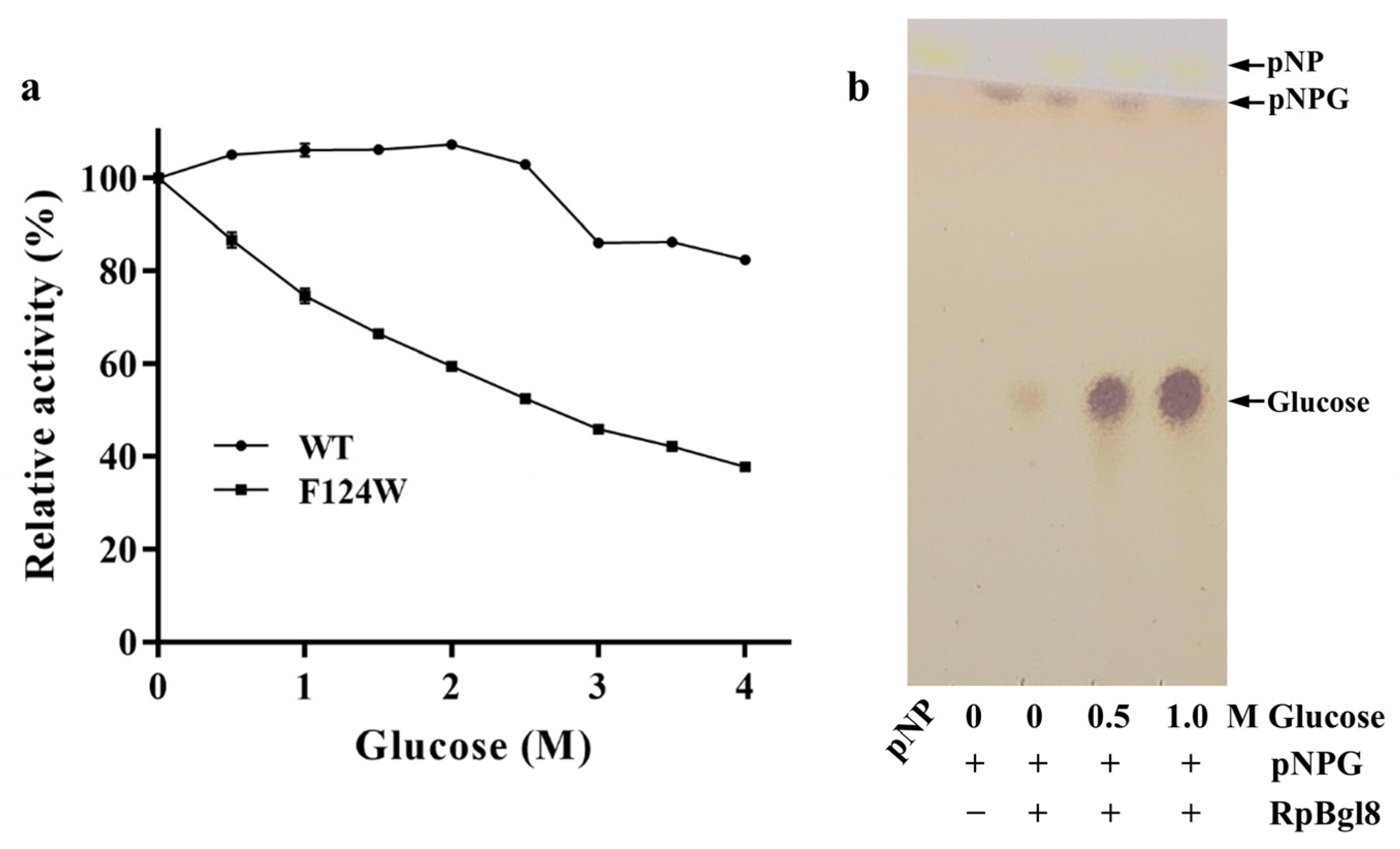
2.4. Glucose Tolerance of RpBgl8
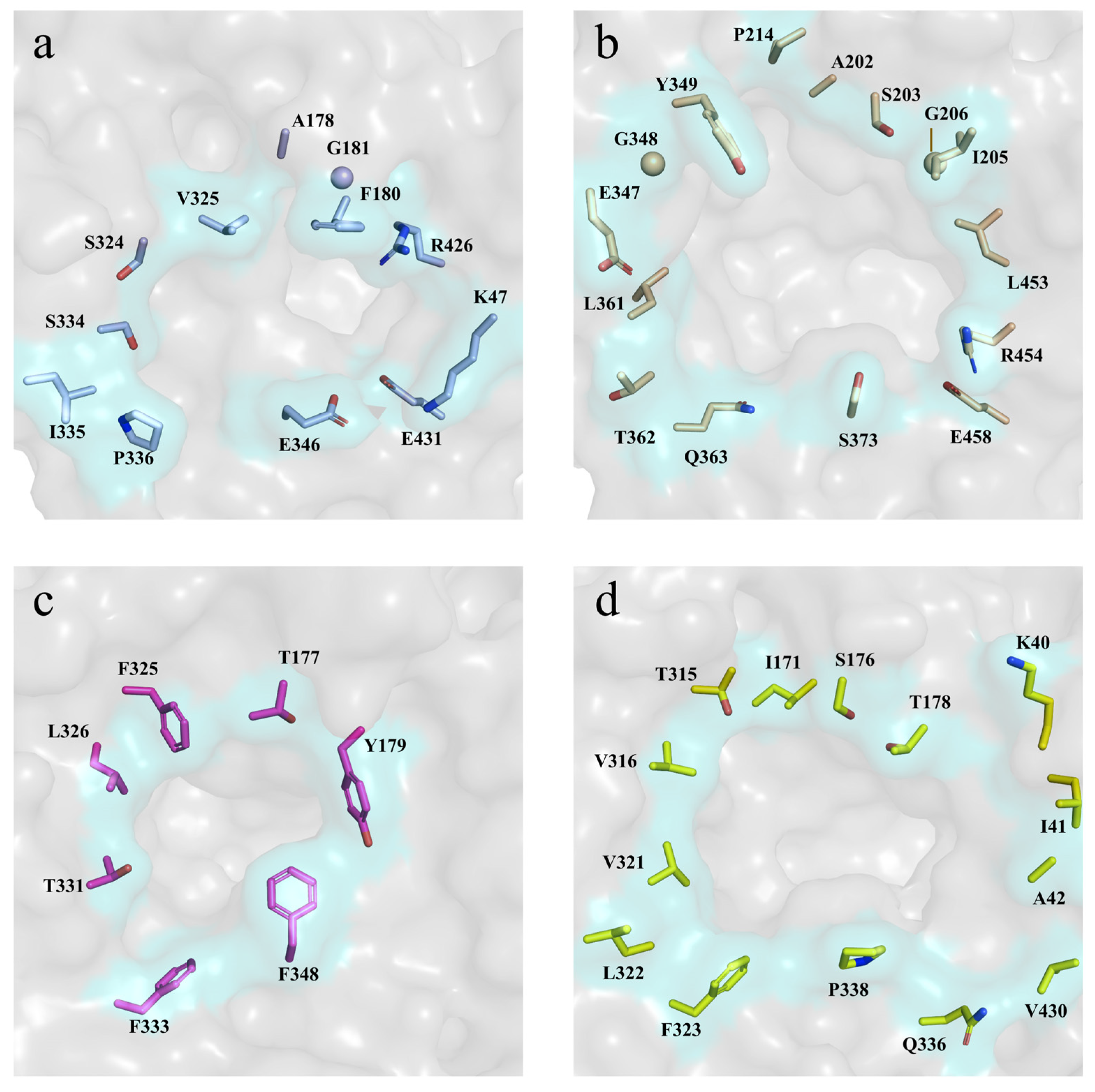
2.5. Molecular Basis for the High Glucose Tolerance of RpBgl8
3. Materials and Methods
3.1. Materials
3.2. Cloning of RpBgl8
3.3. Site-Directed Mutagenesis of RpBgl8
3.4. Expression and Purification of Recombinant RpBgl8
3.5. Enzyme Activity and Stability Assays
3.6. Effects of Chemicals on the Activity of RpBgl8c
3.7. Glucose Tolerance Analysis
3.8. Sequence and Structure Analyses of RpBgl8
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mankar, A.R.; Pandey, A.; Modak, A.; Pant, K.K. Pretreatment of lignocellulosic biomass: A review on recent advances. Bioresour. Technol. 2021, 334, 125235. [Google Scholar] [CrossRef]
- Zajki-Zechmeister, K.; Eibinger, M.; Nidetzky, B. Enzyme synergy in transient clusters of endo- and exocellulase enables a multilayer mode of processive depolymerization of cellulose. ACS Catal. 2022, 12, 10984–10994. [Google Scholar] [CrossRef] [PubMed]
- Kuusk, S.; Väljamäe, P. When substrate inhibits and inhibitor activates: Implications of β-glucosidases. Biotechnol. Biofuels 2017, 10, 7. [Google Scholar] [CrossRef] [PubMed]
- Ketudat Cairns, J.R.; Esen, A. β-Glucosidases. Cell. Mol. Life Sci. 2010, 67, 3389–3405. [Google Scholar] [CrossRef]
- Salgado, J.C.S.; Meleiro, L.P.; Carli, S.; Ward, R.J. Glucose tolerant and glucose stimulated β-glucosidases—A review. Bioresour. Technol. 2018, 267, 704–713. [Google Scholar] [CrossRef] [PubMed]
- de Giuseppe, P.O.; Souza, T.d.A.C.B.; Souza, F.H.M.; Zanphorlin, L.M.; Machado, C.B.; Ward, R.J.; Jorge, J.A.; Furriel, R.d.P.M.; Murakami, M.T. Structural basis for glucose tolerance in GH1 β-glucosidases. Acta Crystallogr. Sect. D 2014, 70, 1631–1639. [Google Scholar] [CrossRef]
- Pang, P.; Cao, L.-c.; Liu, Y.-h.; Xie, W.; Wang, Z. Structures of a glucose-tolerant β-glucosidase provide insights into its mechanism. J. Struct. Biol. 2017, 198, 154–162. [Google Scholar] [CrossRef]
- Li, S.; Cao, L.; Yang, X.; Wu, X.; Xu, S.; Liu, Y. Simultaneously optimizing multiple properties of β-glucosidase Bgl6 using combined (semi-)rational design strategies and investigation of the underlying mechanisms. Bioresour. Technol. 2023, 374, 128792. [Google Scholar] [CrossRef]
- Zhu, Q.; Huang, Y.; Yang, Z.; Wu, X.; Zhu, Q.; Zheng, H.; Zhu, D.; Lv, Z.; Yin, Y. A recombinant thermophilic and glucose-tolerant GH1 β-glucosidase derived from hehua hot spring. Molecules 2024, 29, 1017. [Google Scholar] [CrossRef]
- Scharf, M.E.; Tartar, A. Termite digestomes as sources for novel lignocellulases. Biofuel. Bioprod. Bior. 2008, 2, 540–552. [Google Scholar] [CrossRef]
- Ni, J.; Tokuda, G. Lignocellulose-degrading enzymes from termites and their symbiotic microbiota. Biotechnol. Adv. 2013, 31, 838–850. [Google Scholar] [CrossRef]
- Brune, A. Symbiotic digestion of lignocellulose in termite guts. Nat. Rev. Microbiol. 2014, 12, 168–180. [Google Scholar] [CrossRef]
- Uchima, C.A.; Tokuda, G.; Watanabe, H.; Kitamoto, K.; Arioka, M. Heterologous expression and characterization of a glucose-stimulated β-glucosidase from the termite Neotermes koshunensis in Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2011, 89, 1761–1771. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Chi, S.; Yun, C.; Shen, Y.; Tokuda, G.; Ni, J. Molecular cloning and characterization of an endogenous digestive β-glucosidase from the midgut of the fungus-growing termite Macrotermes barneyi. Insect Mol. Biol. 2012, 21, 604–614. [Google Scholar] [CrossRef]
- Jiang, S.; Li, J.; Cao, C.; Shen, Y.; Ni, J. Molecular modification of β-glucosidase from the midgut of Macrotermes barneyi. Chin. J. Biotechnol. 2018, 34, 1081–1090. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Davies, G.; Henrissat, B. Structures and mechanisms of glycosyl hydrolases. Structure 1995, 3, 853–859. [Google Scholar] [CrossRef]
- Teugjas, H.; Väljamäe, P. Selecting β-glucosidases to support cellulases in cellulose saccharification. Biotechnol. Biofuels 2013, 6, 105. [Google Scholar] [CrossRef]
- Jönsson, L.J.; Martín, C. Pretreatment of lignocellulose: Formation of inhibitory by-products and strategies for minimizing their effects. Bioresour. Technol. 2016, 199, 103–112. [Google Scholar] [CrossRef]
- Guo, H.; Zhao, Y.; Chang, J.-S.; Lee, D.-J. Inhibitor formation and detoxification during lignocellulose biorefinery: A review. Bioresour. Technol. 2022, 361, 127666. [Google Scholar] [CrossRef]
- Raheja, Y.; Singh, V.; Sharma, G.; Tsang, A.; Chadha, B.S. A thermostable and inhibitor resistant β-glucosidase from Rasamsonia emersonii for efficient hydrolysis of lignocellulosics biomass. Bioprocess. Biosyst. Eng. 2024, 47, 567–582. [Google Scholar] [CrossRef]
- Huang, X.; Zhao, Y.; Dai, Y.; Wu, G.; Shao, Z.; Zeng, Q.; Liu, Z. Cloning and biochemical characterization of a glucosidase from a marine bacterium Aeromonas sp. HC11e. World J. Microbiol. Biotechnol. 2012, 28, 3337–3344. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Li, Z.; Huo, Y.-Y.; Bao, L.; Gao, B.; Xiao, P.; Hu, X.; Xu, X.-W.; Li, J. Structural and functional insights into CmGH1, a novel GH39 family β-glucosidase from deep-sea bacterium. Front. Microbiol. 2019, 10, 2922. [Google Scholar] [CrossRef]
- Chen, T.; Yang, W.; Guo, Y.; Yuan, R.; Xu, L.; Yan, Y. Enhancing catalytic performance of β-glucosidase via immobilization on metal ions chelated magnetic nanoparticles. Enzyme Microb. Technol. 2014, 63, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-A.; Zhou, Y.; Qin, Y.; Liu, D.; Zhao, X. Evaluation of the action of Tween 20 non-ionic surfactant during enzymatic hydrolysis of lignocellulose: Pretreatment, hydrolysis conditions and lignin structure. Bioresour. Technol. 2018, 269, 329–338. [Google Scholar] [CrossRef]
- Batra, J.; Mishra, S. Organic solvent tolerance and thermostability of a β-glucosidase co-engineered by random mutagenesis. J. Mol. Catal. B Enzym. 2013, 96, 61–66. [Google Scholar] [CrossRef]
- Wu, J.; Geng, A.; Xie, R.; Wang, H.; Sun, J. Characterization of cold adapted and ethanol tolerant β-glucosidase from Bacillus cellulosilyticus and its application for directed hydrolysis of cellobiose to ethanol. Int. J. Biol. Macromol. 2018, 109, 872–879. [Google Scholar] [CrossRef]
- Gomes-Pepe, E.S.; Machado Sierra, E.G.; Pereira, M.R.; Castellane, T.C.L.; Lemos, E.G.d.M. Bg10: A novel metagenomics alcohol-tolerant and glucose-stimulated GH1 β-Glucosidase suitable for lactose-free milk preparation. PLoS ONE 2016, 11, e0167932. [Google Scholar] [CrossRef]
- Fang, W.; Yang, Y.; Zhang, X.; Yin, Q.; Zhang, X.; Wang, X.; Fang, Z.; Yazhong, X. Improve ethanol tolerance of β-glucosidase Bgl1A by semi-rational engineering for the hydrolysis of soybean isoflavone glycosides. J. Biotechnol. 2016, 227, 64–71. [Google Scholar] [CrossRef]
- Karnaouri, A.; Topakas, E.; Paschos, T.; Taouki, I.; Christakopoulos, P. Cloning, expression and characterization of an ethanol tolerant GH3 β-glucosidase from Myceliophthora thermophila. PeerJ 2013, 1, e46. [Google Scholar] [CrossRef]
- Cao, L.; Li, S.; Huang, X.; Qin, Z.; Kong, W.; Xie, W.; Liu, Y. Enhancing the thermostability of highly active and glucose-tolerant β-glucosidase Ks5A7 by directed evolution for good performance of three properties. J. Agric. Food. Chem. 2018, 66, 13228–13235. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.K.; Datta, S. β-Glucosidase from the hyperthermophilic archaeon Thermococcus sp. is a salt-tolerant enzyme that is stabilized by its reaction product glucose. Appl. Microbiol. Biotechnol. 2016, 100, 8399–8409. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.K.; Das, S.; Konar, S.; Ghorai, P.K.; Das, R.; Datta, S. Elucidating the regulation of glucose tolerance in a β-glucosidase from Halothermothrix orenii by active site pocket engineering and computational analysis. Int. J. Biol. Macromol. 2020, 156, 621–632. [Google Scholar] [CrossRef]
- Meleiro, L.P.; Salgado, J.C.S.; Maldonado, R.F.; Carli, S.; Moraes, L.A.B.; Ward, R.J.; Jorge, J.A.; Furriel, R.P.M. Engineering the GH1 β-glucosidase from Humicola insolens: Insights on the stimulation of activity by glucose and xylose. PLoS ONE 2017, 12, e0188254. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, T.; Miyazaki, K.; Yaoi, K. Characterization of a novel β-glucosidase from a compost microbial metagenome with strong transglycosylation activity. J. Biol. Chem. 2013, 288, 18325–18334. [Google Scholar] [CrossRef]
- Uchiyama, T.; Yaoi, K.; Miyazaki, K. Glucose-tolerant β-glucosidase retrieved from a Kusaya gravy metagenome. Front Microbiol. 2015, 6, 548. [Google Scholar] [CrossRef]
- Lee, H.-L.; Chang, C.-K.; Jeng, W.-Y.; Wang, A.H.-J.; Liang, P.-H. Mutations in the substrate entrance region of β-glucosidase from Trichoderma reesei improve enzyme activity and thermostability. Protein Eng. Des. Sel. 2012, 25, 733–740. [Google Scholar] [CrossRef]
- Weis, W.I.; Drickamer, K. Structural basis of lectin-carbohydrate recognition. Annu. Rev. Biochem. 1996, 65, 441–473. [Google Scholar] [CrossRef]
- Fisher, C.L.; Pei, G.K. Modification of a PCR-based site-directed mutagenesis method. BioTechniques 1997, 23, 570–574. [Google Scholar] [CrossRef]
- Yu, J.; Zhou, Y.; Tanaka, I.; Yao, M. Roll: A new algorithm for the detection of protein pockets and cavities with a rolling probe sphere. Bioinformatics 2009, 26, 46–52. [Google Scholar] [CrossRef]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Goldenzweig, A.; Goldsmith, M.; Hill, S.E.; Gertman, O.; Laurino, P.; Ashani, Y.; Dym, O.; Unger, T.; Albeck, S.; Prilusky, J.; et al. Automated structure- and sequence-based design of proteins for high bacterial expression and stability. Mol. Cell 2016, 63, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Niu, K.; Yan, L.; Ge, J.; Ni, Z.; Liu, Z.; Liu, Q.; Wang, Z.; Du, Z.; Wu, Z.; Lou, Z.; et al. Artificial trimerization of β-glucosidase for enhanced thermostability and activity via computational redesign. Int. J. Biol. Macromol. 2025, 286, 138275. [Google Scholar] [CrossRef]
- Ertelt, M.; Schlegel, P.; Beining, M.; Kaysser, L.; Meiler, J.; Schoeder, C.T. HyperMPNN—A general strategy to design thermostable proteins learned from hyperthermophiles. bioRxiv 2024. [Google Scholar] [CrossRef]
- Jiang, K.; Yan, Z.; Di Bernardo, M.; Sgrizzi, S.R.; Villiger, L.; Kayabolen, A.; Kim, B.J.; Carscadden, J.K.; Hiraizumi, M.; Nishimasu, H.; et al. Rapid in silico directed evolution by a protein language model with EVOLVEpro. Science 2025, 387, eadr6006. [Google Scholar] [CrossRef]


| β-Glucosidase | Glucose Tolerance | Residue 124 1 | Reference |
|---|---|---|---|
| RpBgl8 | 82% activity at 4.0 M glucose | F | This study |
| O08324 | 100% activity at 4.0 M glucose | F | [32] |
| Bgl6 | 50% activity at 3.5 M glucose | W | [7] |
| Ks5A7 | 50% activity at 1.35 M glucose | W | [31] |
| B8CYA8 | 110% activity at 1.5 M glucose | W | [33] |
| G1NKBG | 92% activity at 1.0 M glucose | W | [13] |
| HiBG | 100% activity at 0.45 M glucose | W | [34] |
| Primer | Sequence |
|---|---|
| RpBgl8-NdeI-F | CGCCATATGAGCGCCCTGAAGTTCCC |
| RpBgl8-XbaI-R | TGCTCTAGAATGGCTGGTTTCCACGGCC |
| F124W-F | GTATCACTGGGACCTGCCGCAACCGCTG |
| F124W-R | GCAGGTCCCAGTGATACATGGTGATCATC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mao, G.; Song, M.; Li, H.; Lin, J.; Wang, K.; Liu, Q.; Su, Z.; Zhang, H.; Su, L.; Xie, H.; et al. Biochemical and Structural Characterization of a Highly Glucose-Tolerant β-Glucosidase from the Termite Reticulitermes perilucifugus. Int. J. Mol. Sci. 2025, 26, 3118. https://doi.org/10.3390/ijms26073118
Mao G, Song M, Li H, Lin J, Wang K, Liu Q, Su Z, Zhang H, Su L, Xie H, et al. Biochemical and Structural Characterization of a Highly Glucose-Tolerant β-Glucosidase from the Termite Reticulitermes perilucifugus. International Journal of Molecular Sciences. 2025; 26(7):3118. https://doi.org/10.3390/ijms26073118
Chicago/Turabian StyleMao, Guotao, Ming Song, Hao Li, Junhan Lin, Kai Wang, Qian Liu, Zengping Su, Hongsen Zhang, Lijuan Su, Hui Xie, and et al. 2025. "Biochemical and Structural Characterization of a Highly Glucose-Tolerant β-Glucosidase from the Termite Reticulitermes perilucifugus" International Journal of Molecular Sciences 26, no. 7: 3118. https://doi.org/10.3390/ijms26073118
APA StyleMao, G., Song, M., Li, H., Lin, J., Wang, K., Liu, Q., Su, Z., Zhang, H., Su, L., Xie, H., & Song, A. (2025). Biochemical and Structural Characterization of a Highly Glucose-Tolerant β-Glucosidase from the Termite Reticulitermes perilucifugus. International Journal of Molecular Sciences, 26(7), 3118. https://doi.org/10.3390/ijms26073118







