Catalytic Efficiency Improvement in Cellobiohydrolase I by Cross-Species Domain Exchange Engineering
Abstract
1. Introduction
2. Results and Discussion
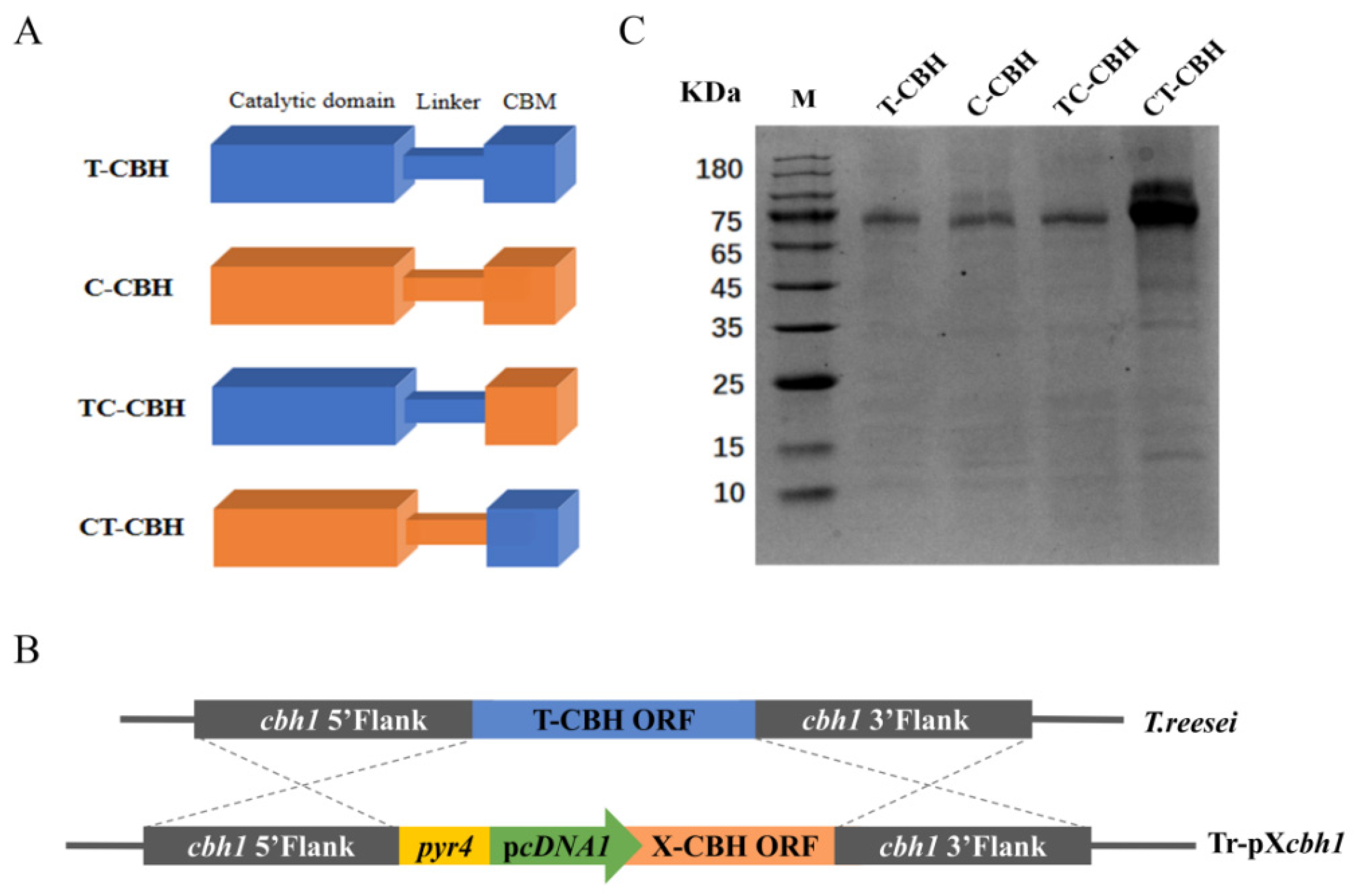
2.1. Recombinant Expression of Two CBHIs and Their Chimeric Mutants
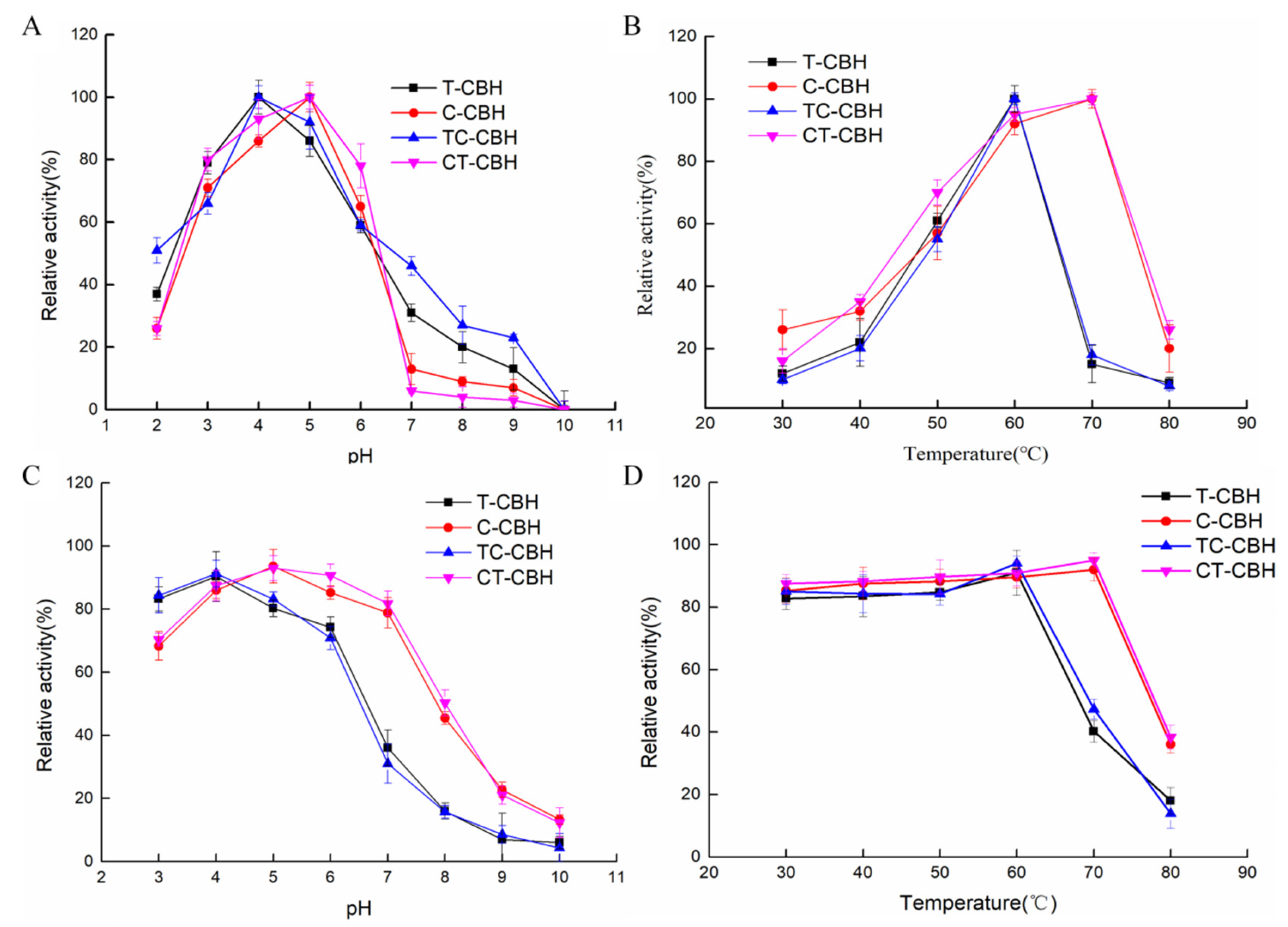
2.2. The Mutant CBHIs Have Similar pH and Temperature Profiles to Their Corresponding Wild-Types
2.3. Kinetic Analysis of the CBHIs on MUC
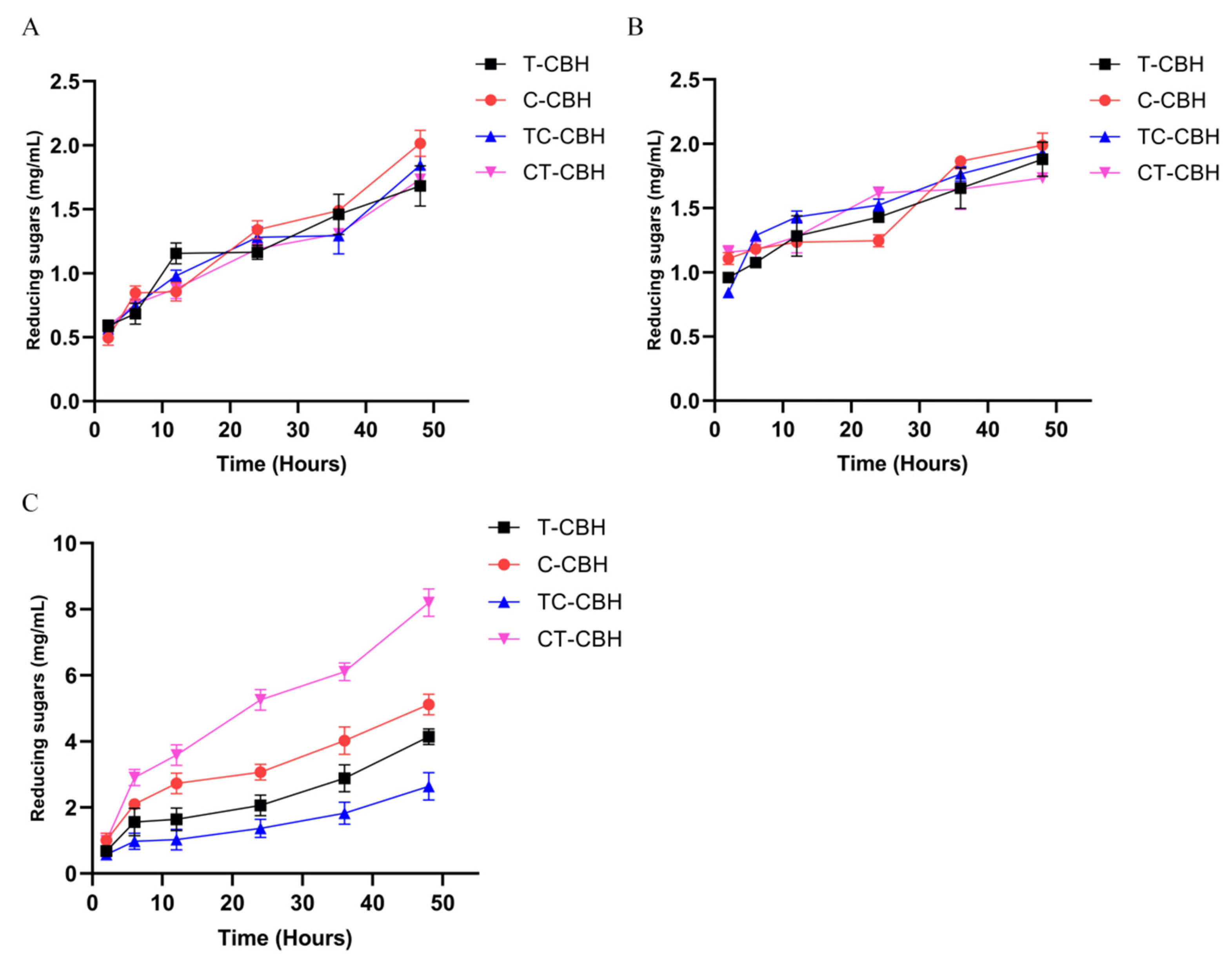
2.4. Domain Swapping Had Different Effects on the Catalytic Performance of the CBHIs on Cellulose
2.5. Hydrolysis of Different Biomass by the Four Recombinant CBHIs
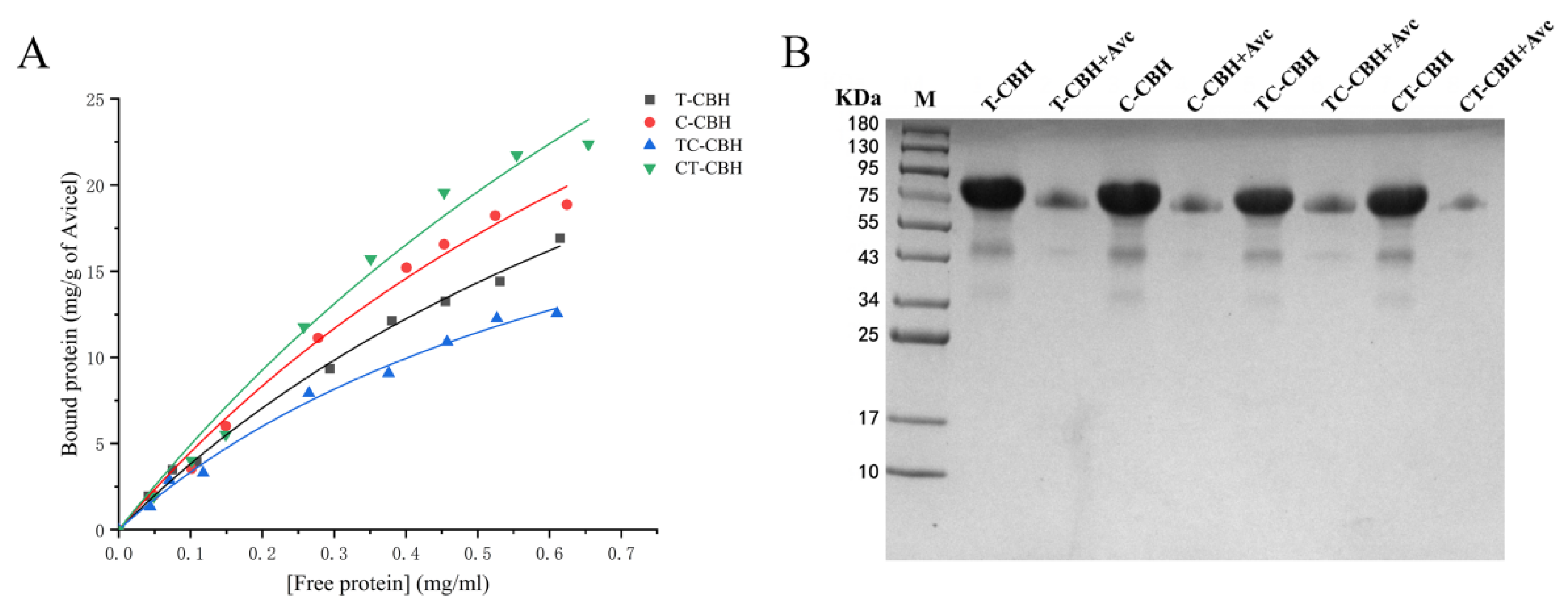
2.6. Binding of Recombinant CBHIs on Crystalline Cellulose
3. Materials and Methods
3.1. Microbial Strains and Growth Conditions
3.2. Transforamtion of T. reesei
3.3. Construction of the Recombinant T. reesei Strains
3.4. Expression and Purification of the CBHIs
3.5. Assay of the Enzyme Activities
3.6. Effects of the pH and Temperature on the Activity of CBHIs
3.7. Hydrolysis of Biomasses
3.8. Binding of Insoluble Cellulose
3.9. Statistical Significance Tests
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Aqeel, A.; Ahmed, Z.; Akram, F.; Abbas, Q.; Ikram-ul-Haq. Cloning, expression and purification of cellobiohydrolase gene from Caldicellulosiruptor bescii for efficient saccharification of plant biomass. Int. J. Biol. Macromol. 2024, 271, 132525. [Google Scholar] [CrossRef]
- Gao, L.; Gao, F.; Wang, L.; Geng, C.; Chi, L.; Zhao, J.; Qu, Y. N-glycoform diversity of cellobiohydrolase I from Penicillium decumbens and synergism of nonhydrolytic glycoform in cellulose degradation. J. Biol. Chem. 2012, 287, 15906–15915. [Google Scholar] [CrossRef] [PubMed]
- Khanday, M.A.; Shergujri, D.A.; Noor, A.; Fayaz, S.; Raza, S.N.; Mir, R.H.; Khan, N.A. Cellulose and Cellulose Nanomaterials: Recent Research and Applications in Medical Field. Comb. Chem. High Throughput Screen. 2025; Epub ahead of print. [Google Scholar] [CrossRef]
- Han, C.; Li, W.; Hua, C.; Sun, F.; Bi, P.; Wang, Q. Enhancement of catalytic activity and thermostability of a thermostable cellobiohydrolase from Chaetomium thermophilum by site-directed mutagenesis. Int. J. Biol. Macromol. 2018, 116, 691–697. [Google Scholar] [CrossRef] [PubMed]
- Wahab, A.F.F.A.; Abdul Karim, N.A.; Ling, J.G.; Hasan, N.S.; Yong, H.Y.; Bharudin, I.; Kamaruddin, S.; Abu Bakar, F.D.; Murad, A.M.A. Functional characterisation of cellobiohydrolase I (Cbh1) from Trichoderma virens UKM1 expressed in Aspergillus niger. Protein Expr. Purif. 2019, 154, 52–61. [Google Scholar] [CrossRef]
- Jiang, X.; Du, J.; He, R.; Zhang, Z.; Qi, F.; Huang, J.; Qin, L. Improved Production of Majority Cellulases in Trichoderma reesei by Integration of cbh1 Gene From Chaetomium thermophilum. Front. Microbiol. 2020, 11, 1633. [Google Scholar] [CrossRef] [PubMed]
- Haviland, Z.K.; Nong, D.; Zexer, N.; Tien, M.; Anderson, C.T.; Hancock, W.O. Lignin impairs Cel7A degradation of in vitro lignified cellulose by impeding enzyme movement and not by acting as a sink. Biotechnol. Biofuels Bioprod. 2024, 17, 7. [Google Scholar] [CrossRef]
- Mou, H.; Tang, L.; Wu, T.; Feng, L.; Liu, Y. Study on the mechanism of lignin non-productive adsorption on cellobiohydrolase. Int. J. Biol. Macromol. 2024, 273, 133003. [Google Scholar] [CrossRef]
- Pramanik, S.; Semenova, M.V.; Rozhkova, A.M.; Zorov, I.N.; Korotkova, O.; Sinitsyn, A.P.; Davari, M.D. An engineered cellobiohydrolase I for sustainable degradation of lignocellulosic biomass. Biotechnol. Bioeng. 2021, 118, 4014–4027. [Google Scholar] [CrossRef]
- Dodda, S.R.; Hossain, M.; Mondal, S.; Das, S.; Khator Jain, S.; Aikat, K.; Mukhopadhyay, S.S. The S-S bridge mutation between the A2 and A4 loops (T416C-I432C) of Cel7A of Aspergillus fumigatus enhances catalytic activity and thermostability. Appl. Environ. Microbiol. 2024, 90, e0232923. [Google Scholar] [CrossRef]
- Dotsenko, A.S.; Dotsenko, G.S.; Rozhkova, A.M.; Zorov, I.N.; Sinitsyn, A.P. Rational design and structure insights for thermostability improvement of Penicillium verruculosum Cel7A cellobiohydrolase. Biochimie 2020, 176, 103–109. [Google Scholar] [CrossRef]
- Wei, H.; Wang, W.; Alahuhta, M.; Vander Wall, T.; Baker, J.O.; Taylor, L.E.; Decker, S.R.; Himmel, M.E.; Zhang, M. Engineering towards a complete heterologous cellulase secretome in Yarrowia lipolytica reveals its potential for consolidated bioprocessing. Biotechnol. Biofuels 2014, 7, 148. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Alahuhta, M.; Wei, H.; Knoshaug, E.P.; Wang, W.; Baker, J.O.; Vander Wall, T.; Himmel, M.E.; Zhang, M. Expression of an endoglucanase-cellobiohydrolase fusion protein in Saccharomyces cerevisiae, Yarrowia lipolytica, and Lipomyces starkeyi. Biotechnol. Biofuels 2018, 11, 322. [Google Scholar] [CrossRef] [PubMed]
- Alahuhta, M.; Xu, Q.; Knoshaug, E.P.; Wang, W.; Wei, H.; Amore, A.; Baker, J.O.; Vander Wall, T.; Himmel, M.E.; Zhang, M. Chimeric cellobiohydrolase I expression, activity, and biochemical properties in three oleaginous yeast. Biotechnol. Biofuels 2021, 14, 6. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhang, X.; Li, X.; Zhao, J.; Liu, G.; Gao, B.; Qu, Y. The cellulose binding region in Trichoderma reesei cellobiohydrolase I has a higher capacity in improving crystalline cellulose degradation than that of Penicillium oxalicum. Bioresour. Technol. 2018, 266, 19–25. [Google Scholar] [CrossRef]
- Lill, A.; Herbst, A.; Langhans, M.; Paech, S.; Hamacher, K.; Biesalski, M.; Meckel, T.; Schmitz, K. Investigating Cellulose Binding of Peptides Derived from Carbohydrate Binding Module 1. Biomacromolecules 2024, 25, 5902–5908. [Google Scholar] [CrossRef]
- Nummi, M.; Niku-Paavola, M.L.; Lappalainen, A.; Enari, T.M.; Raunio, V. Cellobiohydrolase from Trichoderma reesei. Biochem. J. 1983, 215, 677–683. [Google Scholar] [CrossRef]
- Uzbas, F.; Sezerman, U.; Hartl, L.; Kubicek, C.P.; Seiboth, B. A homologous production system for Trichoderma reesei secreted proteins in a cellulase-free background. Appl. Microbiol. Biotechnol. 2012, 93, 1601–1608. [Google Scholar] [CrossRef]
- Voutilainen, S.P.; Puranen, T.; Siika-aho, M.; Lappalainen, A.; Alapuranen, M.; Kallio, J.; Hooman, S.; Viikari, L.; Vehmaanperä, J.; Koivula, A. Cloning, expression, and characterization of novel thermostable family 7 cellobiohydrolases. Biotechnol. Bioeng. 2008, 101, 515–528. [Google Scholar] [CrossRef]
- Chen, Y.; Sun, H.; Chen, H.; Wu, J.; Huang, J.; Jiang, X.; Qin, L. Enhancing cellulase production in Neurospora crassa through combined deletion of the phospholipase D-encoding gene pla-7 and modulation of transcription factor CLR-2 expression. Int. J. Biol. Macromol. 2025, 307, 141944. [Google Scholar] [CrossRef]
- Chu, Y.; Hao, Z.; Wang, K.; Tu, T.; Huang, H.; Wang, Y.; Bai, Y.G.; Wang, Y.; Luo, H.; Yao, B.; et al. The GH10 and GH48 dual-functional catalytic domains from a multimodular glycoside hydrolase synergize in hydrolyzing both cellulose and xylan. Biotechnol. Biofuels 2019, 12, 279. [Google Scholar] [CrossRef]
- Penttilä, M.; Nevalainen, H.; Rättö, M.; Salminen, E.; Knowles, J. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 1987, 61, 155–164. [Google Scholar] [CrossRef]
- Bailey, M.J.; Tähtiharju, J. Efficient cellulase production by Trichoderma reesei in continuous cultivation on lactose medium with a computer-controlled feeding strategy. Appl. Microbiol. Biotechnol. 2003, 62, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Yi, Z.; Su, X.; Revindran, V.; Mackie, R.I.; Cann, I. Molecular and biochemical analyses of CbCel9A/Cel48A, a highly secreted multi-modular cellulase by Caldicellulosiruptor bescii during growth on crystalline cellulose. PLoS ONE 2013, 8, e84172. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Han, J.; Li, Y.; Liu, J.; Gan, L.; Long, M. Promoting cellulase and hemicellulase production from Trichoderma orientalis EU7-22 by overexpression of transcription factors Xyr1 and Ace3. Bioresour. Technol. 2020, 296, 122355. [Google Scholar] [CrossRef]
- Yoshida, S.; Mackie, R.I.; Cann, I.K.O. Biochemical and domain analyses of FSUAxe6B, a modular acetyl xylan esterase, identify a unique carbohydrate binding module in Fibrobacter succinogenes S85. J. Bacteriol. 2010, 192, 483–493. [Google Scholar] [CrossRef]
- Kyriacou, A.; Neufeld, R.J.; MacKenzie, C.R. Effect of physical parameters on the adsorption characteristics of fractionated Trichoderma reesei cellulase components. Enzym. Microb. Technol. 1988, 10, 675–681. [Google Scholar] [CrossRef]
| Enzyme | Specific Activity (U/µM) | KM (mmol/L) | kcat (min−1) | kcat/KM (1/min)/(mmol/L) |
|---|---|---|---|---|
| T-CBH | 282.5 ± 21.12 ns | 1.3 | 77.5 | 61.6 |
| C-CBH | 320.2 ± 35.23 ns | 1.1 | 93.7 | 88.5 |
| TC-CBH | 301.3 ± 20.68 ns | 1.2 | 172.5 | 141.8 |
| CT-CBH | 734.5 ± 18.83 **** | 0.7 | 115.1 | 168.9 |
| Enzyme | PASC | Avicel a (µM Glucose Equivalents/min/µM Enzyme) | Filter Paper b (µM Glucose Equivalents/min/µM Enzyme) | ||||
|---|---|---|---|---|---|---|---|
| kcat (s−1) | Km (mg/mL) | kcat (s−1)/Km (mg/mL) | |||||
| T-CBH | 2.0 × 10−1 | 5.6 × 10−2 | 3.7 | 3.8 ± 0.4 |  | 9.9 ± 1.9 ns | |
| C-CBH | 3.3 × 10−1 | 7.3 × 10−2 | 4.5 | 3.1 ± 0.4 |  | 10.0 ± 1.9 ns | |
| TC-CBH | 3.2 × 10−1 | 1.2 × 10−1 | 2.7 | 2.0 ± 0.1 | 7.5 ± 2.2 ns | ||
| CT-CBH | 2.1 × 10−1 | 3.5 × 10−2 | 6.0 | 2.0 ± 0.2 | 11.7 ± 2.4 ns | ||
| Protein | KP (mg/mL) | qmax (mg Protein/g Avicel) |
|---|---|---|
| T-CBH | 0.9 ± 0.2 ns | 45.9 ± 7.9 |
| C-CBH | 0.8 ± 0.3 ns | 57.6 ± 13.4 |
| TC-CBH | 1.3 ± 0.3 ns | 29.1 ± 4.6 |
| CT-CBH | 0.7 ± 0.3 ns | 77.0 ± 26.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, J.; Jiang, X.; Li, A.; Li, J.; Su, X.; Huang, J.; Qin, L. Catalytic Efficiency Improvement in Cellobiohydrolase I by Cross-Species Domain Exchange Engineering. Int. J. Mol. Sci. 2025, 26, 4024. https://doi.org/10.3390/ijms26094024
Xue J, Jiang X, Li A, Li J, Su X, Huang J, Qin L. Catalytic Efficiency Improvement in Cellobiohydrolase I by Cross-Species Domain Exchange Engineering. International Journal of Molecular Sciences. 2025; 26(9):4024. https://doi.org/10.3390/ijms26094024
Chicago/Turabian StyleXue, Jing, Xianzhang Jiang, Anjing Li, Jiaxin Li, Xiaoyun Su, Jianzhong Huang, and Lina Qin. 2025. "Catalytic Efficiency Improvement in Cellobiohydrolase I by Cross-Species Domain Exchange Engineering" International Journal of Molecular Sciences 26, no. 9: 4024. https://doi.org/10.3390/ijms26094024
APA StyleXue, J., Jiang, X., Li, A., Li, J., Su, X., Huang, J., & Qin, L. (2025). Catalytic Efficiency Improvement in Cellobiohydrolase I by Cross-Species Domain Exchange Engineering. International Journal of Molecular Sciences, 26(9), 4024. https://doi.org/10.3390/ijms26094024






