Loop Dynamics Mediate Thermal Adaptation of Two Xylanases from Marine Bacteria
Abstract
:1. Introduction
2. Results
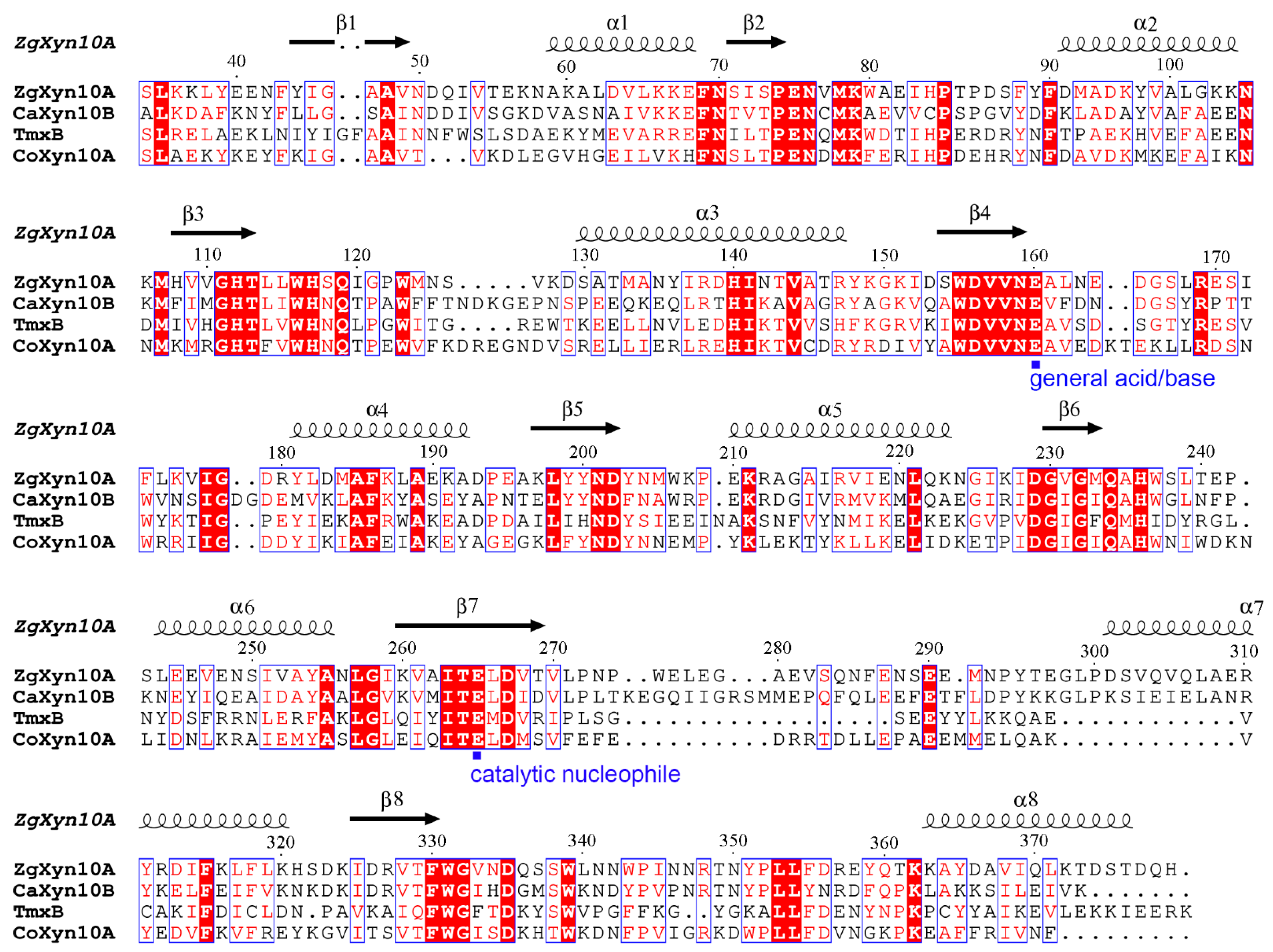
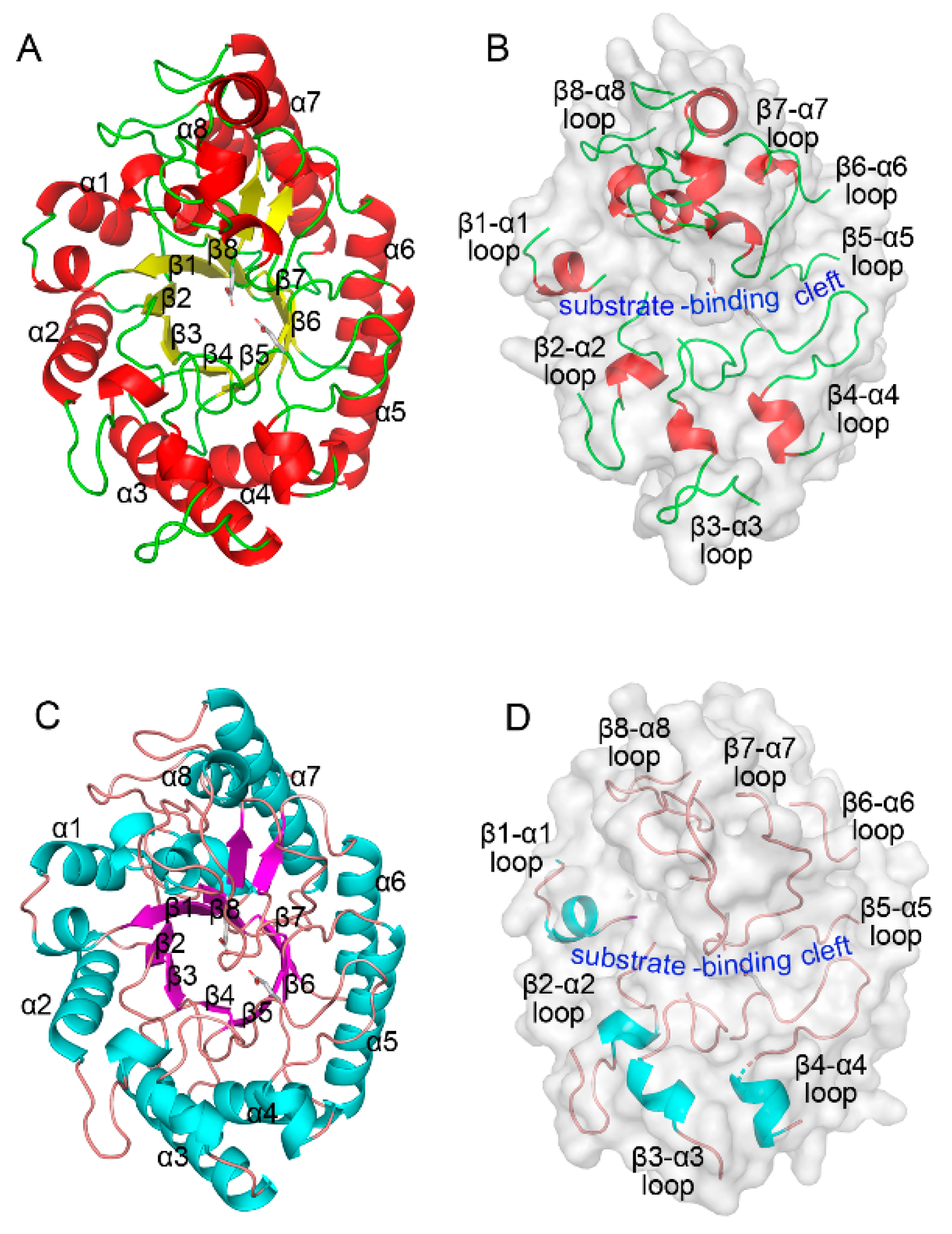
2.1. Sequence and Structural Features of ZgXyn10A and CaXyn10B
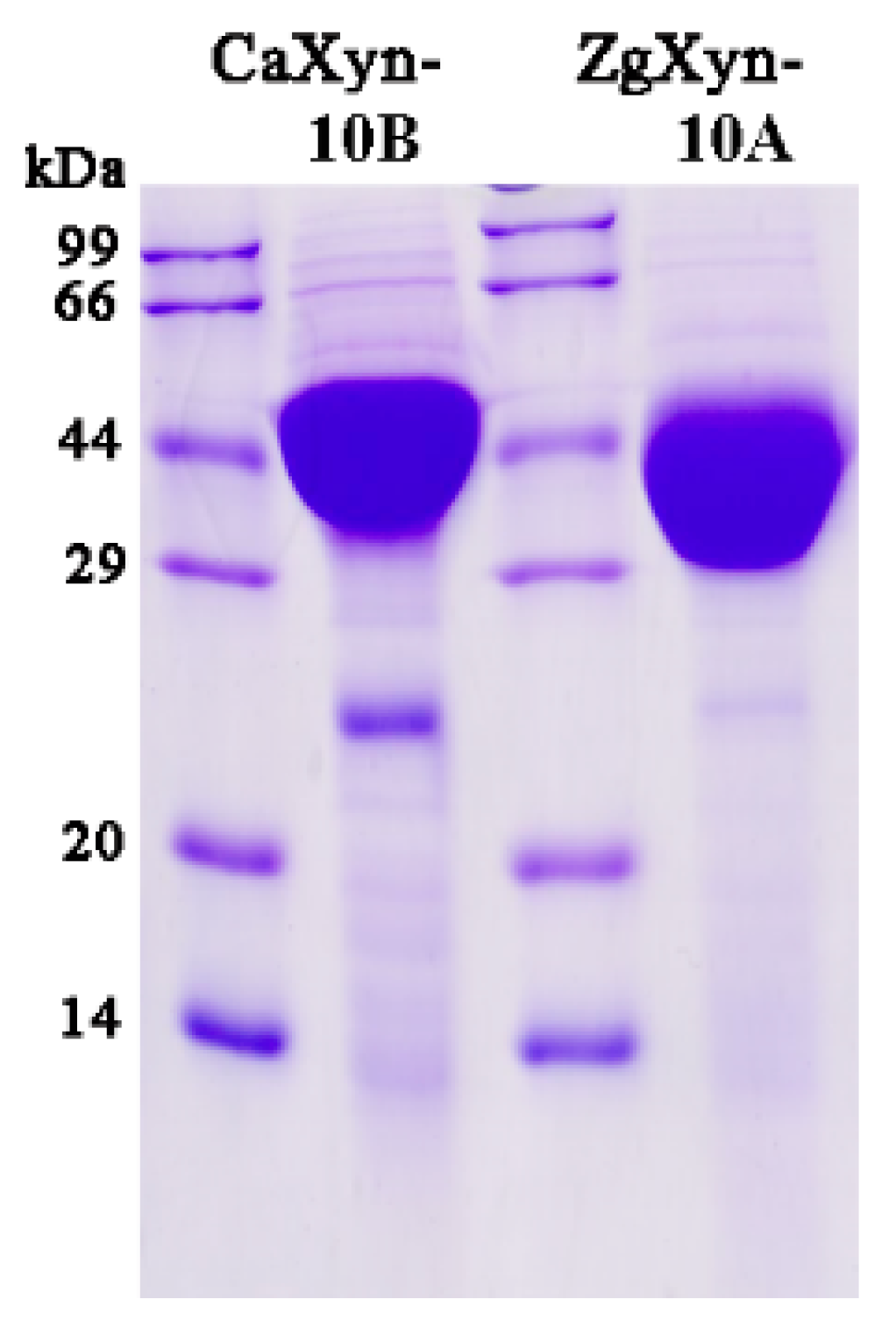
2.2. Enzymatic Properties
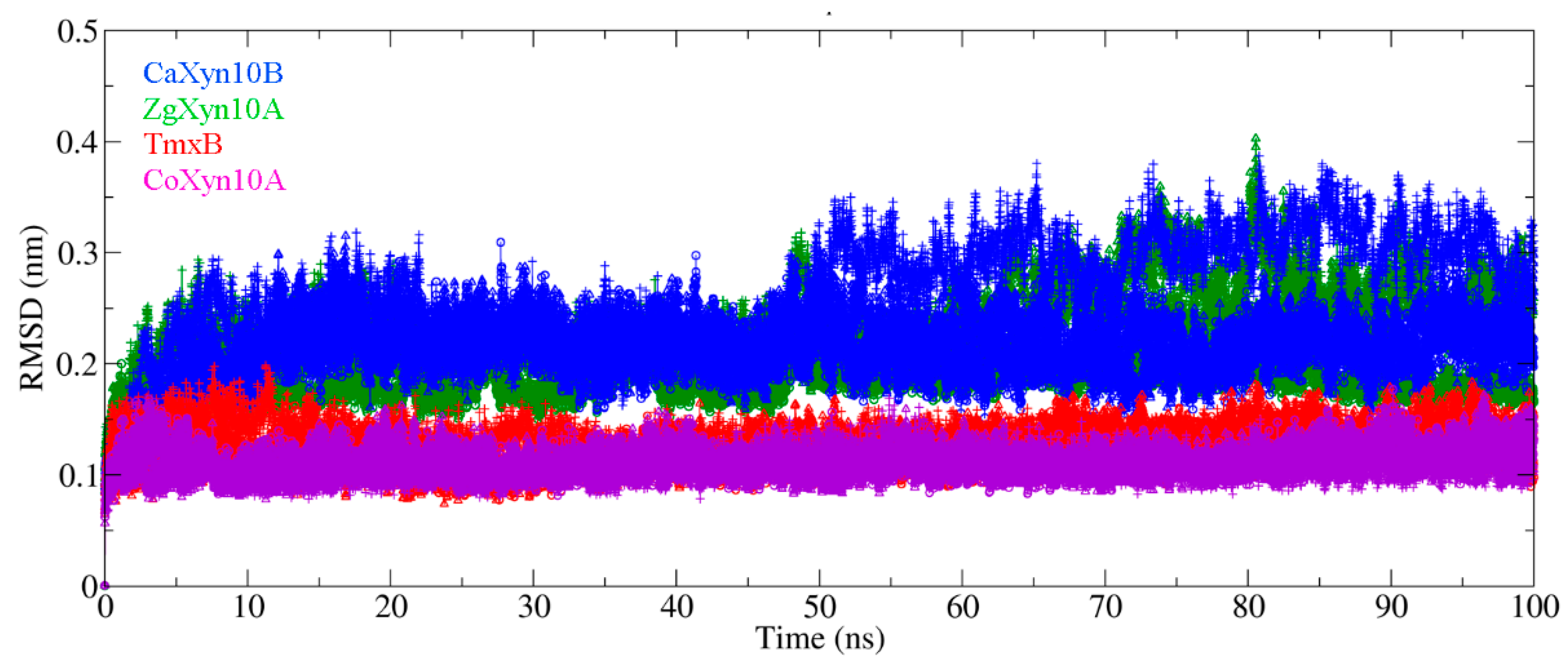
2.3. Loop Dynamics in Substrate-Bonding Cleft Are Determinants of Thermos Properties
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cloning and Recombinant Expression
4.3. Protein Purification
4.4. Enzymatic Assays
4.5. Structural Analysis
4.6. Molecular Dynamics (MD) Simulations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kaur, D.; Joshi, A.; Sharma, V.; Batra, N.; Sharma, A.K. An insight into microbial sources, classification, and industrial applications of xylanases: A rapid review. Biotechnol. Appl. Biochem. 2023, 70, 1489–1503. [Google Scholar] [PubMed]
- Biely, P.; Singh, S.; Puchart, V. Towards enzymatic breakdown of complex plant xylan structures: State of the art. Biotechnol. Adv. 2016, 34, 1260–1274. [Google Scholar] [PubMed]
- Verma, D.; Satyanarayana, T. Molecular approaches for ameliorating microbial xylanases. Bioresour. Technol. 2012, 117, 360–367. [Google Scholar]
- Siddiqui, K.S. Some like it hot, some like it cold: Temperature dependent biotechnological applications and improvements in extremophilic enzymes. Biotechnol. Adv. 2015, 33, 1912–1922. [Google Scholar]
- Parvizpour, S.; Hussin, N.; Shamsir, M.S.; Razmara, J. Psychrophilic enzymes: Structural adaptation, pharmaceutical and industrial applications. Appl. Microbiol. Biotechnol. 2021, 105, 899–907. [Google Scholar] [PubMed]
- Kumar, V.; Dangi, A.K.; Shukla, P. Engineering thermostable microbial xylanases toward its industrial applications. Mol. Biotechnol. 2018, 60, 226–235. [Google Scholar]
- Trincone, A. Marine biocatalysts: Enzymatic features and applications. Mar. Drugs 2011, 9, 478–499. [Google Scholar] [CrossRef]
- Ghattavi, S.; Homaei, A. Marine enzymes: Classification and application in various industries. Int. J. Biol. Macromol. 2023, 230, 123136. [Google Scholar]
- Barbeyron, T.; L’Haridon, S.; Corre, E.; Kloareg, B.; Potin, P. Zobellia galactanivorans gen. nov., sp. nov., a marine species of Flavobacteriaceae isolated from a red alga. Int. J. Syst. Evol. Microbiol. 2001, 51, 985–997. [Google Scholar]
- Naretto, A.; Fanuel, M.; Ropartz, D.; Rogniaux, H.; Larocque, R.; Czjzek, M.; Tellier, C.; Michel, G. The agar-specific hydrolase ZgAgaC from the marine bacterium Zobellia galactanivorans defines a new GH16 protein subfamily. J. Biol. Chem. 2019, 294, 6923–6939. [Google Scholar]
- Jouanneau, D.; Klau, L.J.; Larocque, R.; Jaffrennou, A.; Duval, G.; Le Duff, N.; Roret, T.; Jeudy, A.; Aachmann, F.L.; Czjzek, M.; et al. Structure-function analysis of a new PL17 oligoalginate lyase from the marine bacterium Zobellia galactanivorans DsijT. Glycobiology 2021, 31, 1364–1377. [Google Scholar] [CrossRef] [PubMed]
- Abt, B.; Lu, M.; Misra, M.; Han, C.; Nolan, M.; Lucas, S.; Hammon, N.; Deshpande, S.; Cheng, J.F.; Tapia, R.; et al. Complete genome sequence of Cellulophaga algicola type strain (IC166). Stand. Genom. Sci. 2011, 4, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Zhang, Y.; Yang, J. Biochemical characterization of a new β-agarase from Cellulophaga algicola. Int. J. Mol. Sci. 2019, 20, 2143. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.; Wefers, D. Chromatographic analysis of alginate degradation by five recombinant alginate lyases from Cellulophaga algicola. Food Chem. 2019, 299, 125142. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, W.; Han, F. Expression and characterization of a cold-adapted, thermotolerant GH5 endoglucanase from Cellulophaga algicola. Biotechnol. Lett. 2016, 38, 285–290. [Google Scholar] [CrossRef]
- Dorival, J.; Ruppert, S.; Gunnoo, M.; Orłowski, A.; Chapelais-Baron, M.; Dabin, J.; Labourel, A.; Thompson, D.; Michel, G.; Czjzek, M.; et al. The laterally acquired GH5 ZgEngAGH5_4 from the marine bacterium Zobellia galactanivorans is dedicated to hemicellulose hydrolysis. Biochem. J. 2018, 475, 3609–3628. [Google Scholar] [CrossRef]
- Winterhalter, C.; Liebl, W. Two extremely thermostable xylanases of the hyperthermophilic bacterium Thermotoga maritima MSB8. Appl. Environ. Microbiol. 1995, 61, 1810–1815. [Google Scholar] [CrossRef]
- Liu, X.; Liu, T.; Zhang, Y.; Xin, F.; Mi, S.; Wen, B.; Gu, T.; Shi, X.; Wang, F.; Sun, L. Structural insights into the thermophilic adaptation mechanism of endo-1,4-β-xylanase from Caldicellulosiruptor owensensis. J. Agric. Food Chem. 2018, 66, 187–193. [Google Scholar]
- Bilal, M.; Iqbal, H.M.N. Tailoring enzyme microenvironment: State-of-the-art strategy to fulfill the quest for efficient bio-catalysis. Biotechnol. Adv. 2023, 62, 108074. [Google Scholar] [CrossRef]
- Basit, A.; Liu, J.; Rahim, K.; Jiang, W.; Lou, H. Thermophilic xylanases: From bench to bottle. Crit. Rev. Biotechnol. 2018, 38, 989–1002. [Google Scholar] [CrossRef]
- Zhao, F.; Yu, C.M.; Sun, H.N.; Zhao, L.S.; Ding, H.T.; Cao, H.Y.; Chen, Y.; Qin, Q.L.; Zhang, Y.Z.; Li, P.Y.; et al. A novel class of xylanases specifically degrade marine red algal β1,3/1,4-mixed-linkage xylan. J. Biol. Chem. 2023, 299, 105116. [Google Scholar]
- Dutschei, T.; Beidler, I.; Bartosik, D.; Seeßelberg, J.M.; Teune, M.; Bäumgen, M.; Ferreira, S.Q.; Heldmann, J.; Nagel, F.; Krull, J.; et al. Marine Bacteroidetes enzymatically digest xylans from terrestrial plants. Environ. Microbiol. 2023, 25, 1713–1727. [Google Scholar] [CrossRef]
- Hung, K.S.; Liu, S.M.; Fang, T.Y.; Tzou, W.S.; Lin, F.P.; Sun, K.H.; Tang, S.J. Characterization of a salt-tolerant xylanase from Thermoanaerobacterium saccharolyticum NTOU1. Biotechnol. Lett. 2011, 33, 1441–1447. [Google Scholar]
- Huang, X.; Lin, J.; Ye, X.; Wang, G. Molecular characterization of a thermophilic and salt- and alkaline-tolerant xylanase from Planococcus sp. SL4, a strain isolated from the sediment of a soda lake. J. Microbiol. Biotechnol. 2015, 25, 662–671. [Google Scholar] [PubMed]
- Bai, W.; Xue, Y.; Zhou, C.; Ma, Y. Cloning, expression and characterization of a novel salt-tolerant xylanase from Bacillus sp. SN5. Biotechnol. Lett. 2012, 34, 2093–2099. [Google Scholar] [PubMed]
- Santiago, M.; Ramírez-Sarmiento, C.A.; Zamora, R.A.; Parra, L.P. Discovery, molecular mechanisms, and industrial applications of cold-active enzymes. Front. Microbiol. 2016, 7, 1408. [Google Scholar]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar]
- Chong, W.; Zhang, Z.; Li, Z.; Meng, S.; Nian, B.; Hu, Y. Hook loop dynamics engineering transcended the barrier of activity-stability trade-off and boosted the thermostability of enzymes. Int. J. Biol. Macromol. 2024, 278 Pt 4, 134953. [Google Scholar]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar]
- Studier, F.W. Stable expression clones and auto-induction for protein production in E. coli. Methods Mol. Biol. 2014, 1091, 17–32. [Google Scholar]
- Kruger, N.J. The Bradford method for protein quantitation. Methods Mol. Biol. 1994, 32, 9–15. [Google Scholar] [PubMed]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D., Jr. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar]


| ZgXyn10A | TmxB | CoXyn10A | |
|---|---|---|---|
| CaXyn10B | 42%, 1.56 Å | 27%, 1.25 Å | 35%, 1.67 Å |
| ZgXyn10A | / | 30%, 1.30 Å | 33%, 1.78 Å |
| TmxB | / | / | 34%, 1.52 Å |
| Enzyme | Vmax (U/mg) | Km (mg/mL) | kcat (s−1) | kcat/Km (mL/s/mg) |
|---|---|---|---|---|
| CaXyn10B | 119.86 | 16.00 | 163.69 | 10.23 |
| ZgXyn10A | 1532.11 | 8.84 | 1017.65 | 115.12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhuang, J.; Zhang, Y.; Wang, Y.; Han, Z.; Yang, J. Loop Dynamics Mediate Thermal Adaptation of Two Xylanases from Marine Bacteria. Int. J. Mol. Sci. 2025, 26, 3215. https://doi.org/10.3390/ijms26073215
Zhuang J, Zhang Y, Wang Y, Han Z, Yang J. Loop Dynamics Mediate Thermal Adaptation of Two Xylanases from Marine Bacteria. International Journal of Molecular Sciences. 2025; 26(7):3215. https://doi.org/10.3390/ijms26073215
Chicago/Turabian StyleZhuang, Jinhua, Yuxi Zhang, Yawei Wang, Zhenggang Han, and Jiangke Yang. 2025. "Loop Dynamics Mediate Thermal Adaptation of Two Xylanases from Marine Bacteria" International Journal of Molecular Sciences 26, no. 7: 3215. https://doi.org/10.3390/ijms26073215
APA StyleZhuang, J., Zhang, Y., Wang, Y., Han, Z., & Yang, J. (2025). Loop Dynamics Mediate Thermal Adaptation of Two Xylanases from Marine Bacteria. International Journal of Molecular Sciences, 26(7), 3215. https://doi.org/10.3390/ijms26073215





