Anti-Obesity Properties of a Novel Probiotic Strain of Latilactobacillus sakei CNTA 173 in Caenorhabditis elegans
Abstract
1. Introduction
2. Results and Discussion
2.1. L. sakei CNTA 173 Identification, Growth, and Culture
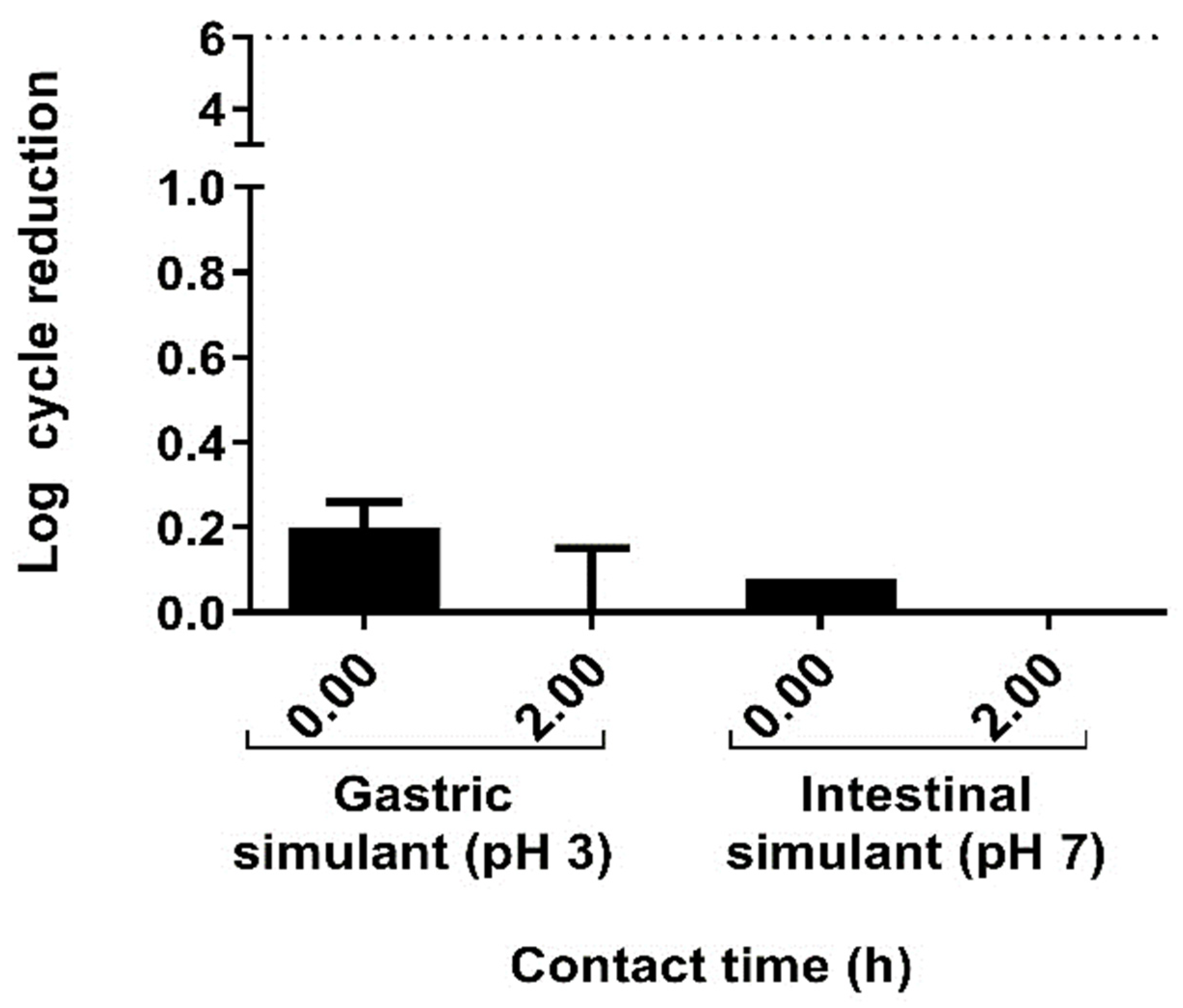
2.2. The In Vitro Characterization of L. sakei CNTA 173 Demonstrates Its Suitability as a Probiotic Strain
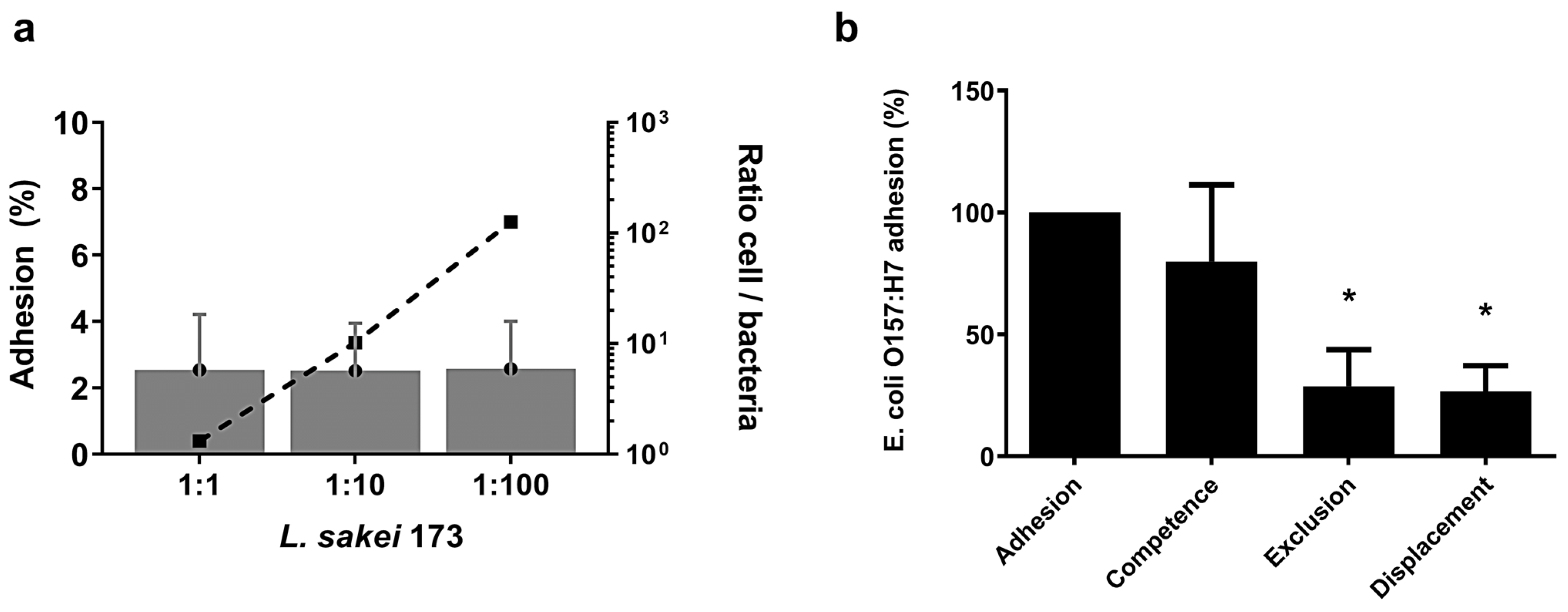
2.3. L. sakei CNTA 173 Reduces the Adhesion of the Pathogenic E. coli O157:H7 In Vitro
2.4. L. sakei CNTA 173 Exhibits Immunomodulatory Capacity In Vitro
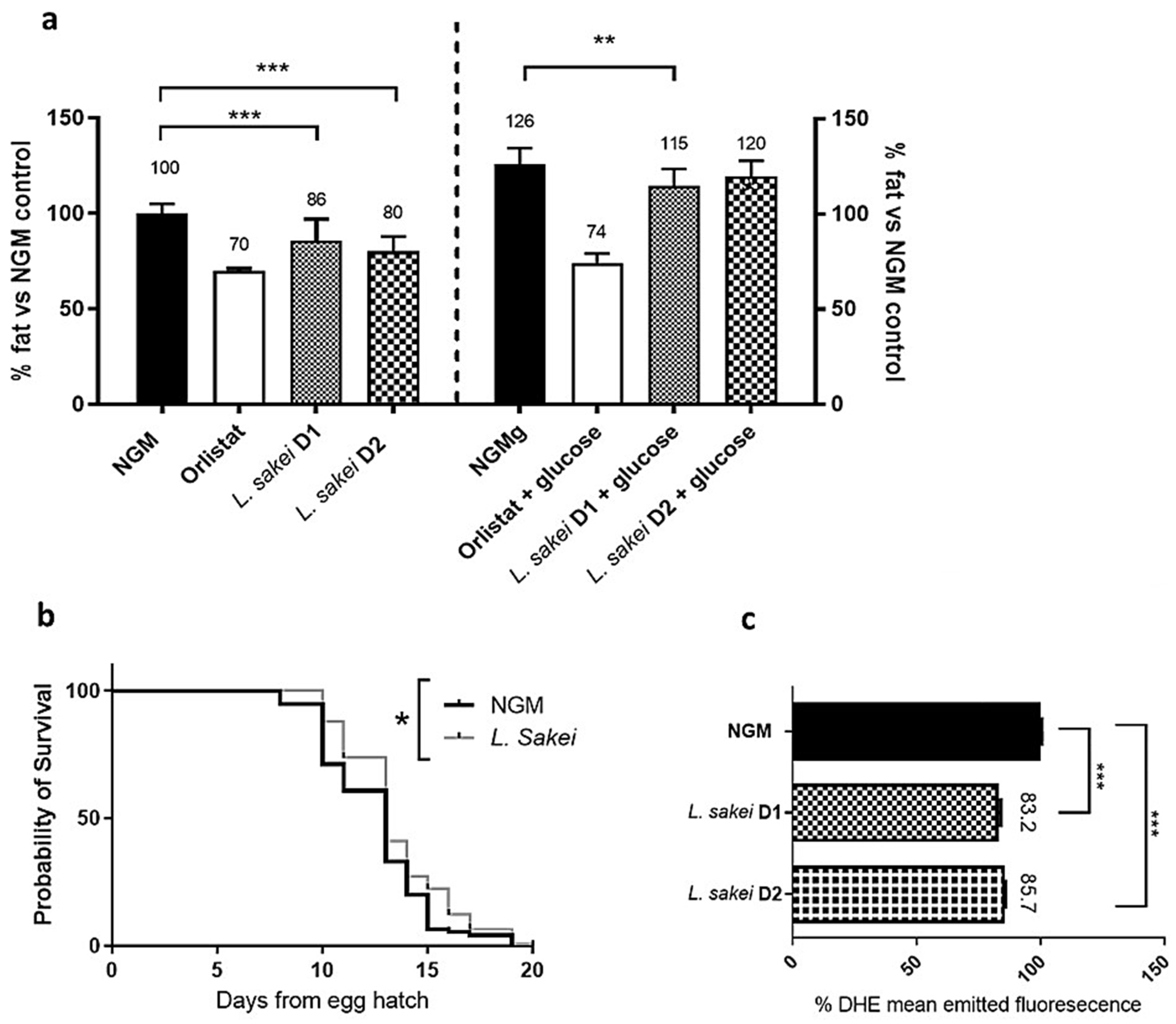
2.5. L. sakei CNTA 173 Reduces Fat Accumulation, Enhances Oxidative Stress Response, and Increases Lifespan in C. elegans
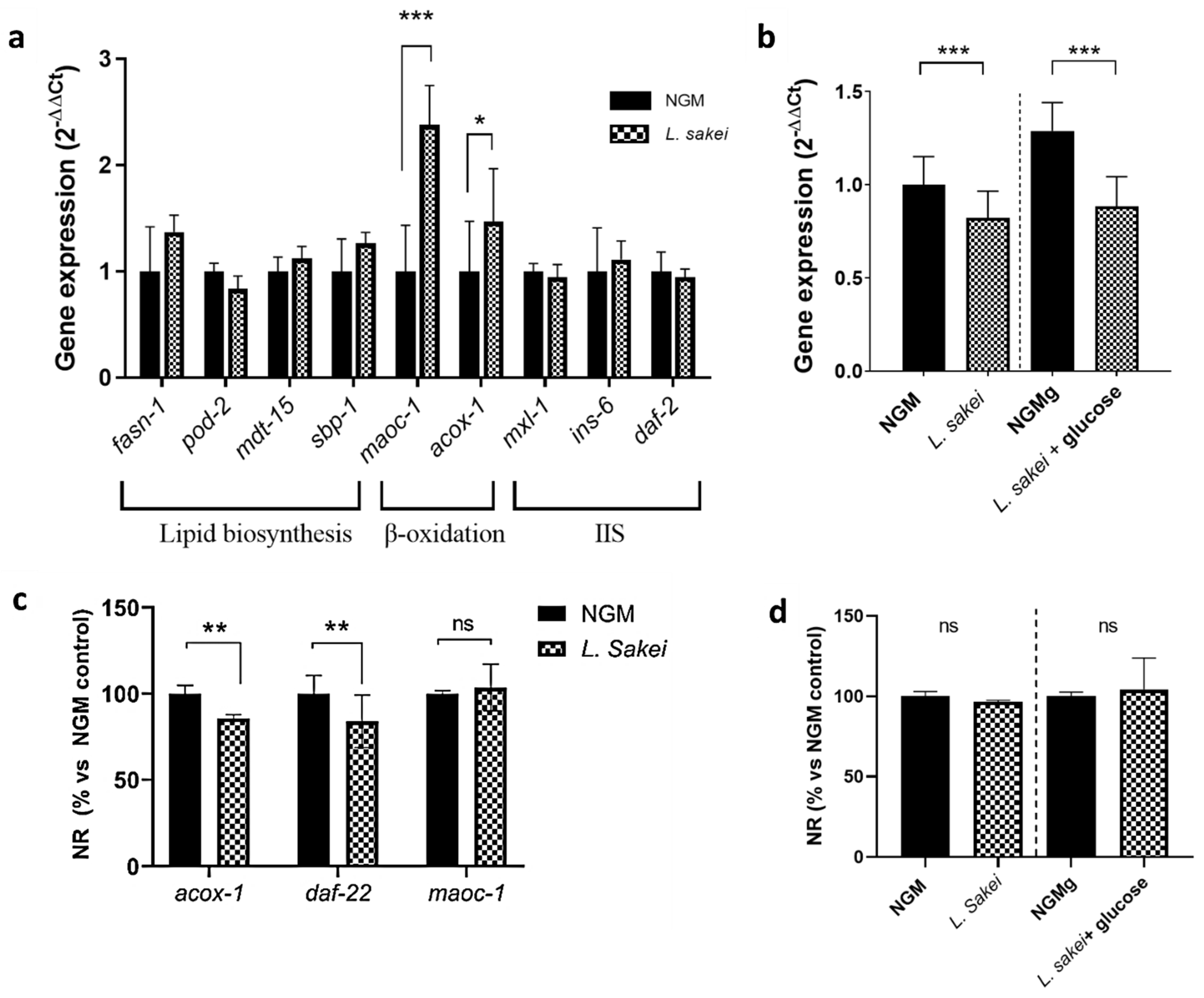
2.6. L. sakei CNTA 173 Modulates Lipid and Carbohydrate Metabolism in C. elegans
3. Materials and Methods
3.1. Bacterial Strain and Growth Conditions
3.1.1. Growth Conditions
3.1.2. Growth Kinetic Determination
3.2. Assessment of Antibiotic Resistance
3.3. Resistance to Simulated Gastrointestinal Conditions
3.4. Ability of the Potential Probiotics to Produce Short-Chain Fatty Acids (SCFAs)
3.5. Assessment of β-Galactosidase Activity
3.6. Ability to Form a Biofilm
3.7. Qualitative Determination of Bile Salts Hydrolase Activity (BSH Activity)
3.8. Adhesion
3.8.1. Bacteria and Growth Conditions
3.8.2. Caco-2 Cell Culture
3.8.3. Adhesion Assays on Caco-2 Cells
3.9. Immunomodulation and Cell Viability
3.9.1. Bacterial Culture
3.9.2. Cell Culture and Maintenance
3.9.3. Cell Viability Assays
3.9.4. Cytokine Quantification
3.10. Statistical Analysis of In Vitro Probiotic Characterization
3.11. L. sakei CNTA 173 In Vivo Activities in C. elegans
3.11.1. Nematode Culture and Experimental Design
3.11.2. Nile Red and DHE Staining
3.11.3. Image Acquisition and Quantification
3.11.4. C. elegans Lifespan Analysis
3.11.5. Egg Laying
3.11.6. C. elegans RNA Extraction and Quantitative PCR Analysis
3.11.7. Statistical Analysis of C. elegans Determinations
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ambroselli, D.; Masciulli, F.; Romano, E.; Catanzaro, G.; Besharat, Z.M.; Massari, M.C.; Ferretti, E.; Migliaccio, S.; Izzo, L.; Ritieni, A.; et al. New Advances in Metabolic Syndrome, from Prevention to Treatment: The Role of Diet and Food. Nutrients 2023, 15, 640. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yun, J.M.; Kim, M.K.; Kwon, O.; Cho, B. Lactobacillus gasseri BNR17 Supplementation Reduces the Visceral Fat Accumulation and Waist Circumference in Obese Adults: A Randomized, Double-Blind, Placebo-Controlled Trial. J. Med. Food 2018, 21, 454–461. [Google Scholar] [CrossRef]
- Homayouni, A.; Bagheri, N.; Mohammad-Alizadeh-Charandabi, S.; Kashani, N.; Mobaraki-Asl, N.; Mirghafurvand, M.; Asgharian, H.; Ansari, F.; Pourjafar, H. Prevention of Gestational Diabetes Mellitus (GDM) and Probiotics: Mechanism of Action: A Review. Curr. Diabetes Rev. 2019, 16, 538–545. [Google Scholar] [CrossRef]
- Sánchez, B.; Delgado, S.; Blanco-Míguez, A.; Lourenço, A.; Gueimonde, M.; Margolles, A. Probiotics, gut microbiota, and their influence on host health and disease. Mol. Nutr. Food Res. 2017, 61, 1600240. [Google Scholar] [CrossRef]
- Abenavoli, L.; Scarpellini, E.; Colica, C.; Boccuto, L.; Salehi, B.; Sharifi-Rad, J.; Aiello, V.; Romano, B.; De Lorenzo, A.; Izzo, A.A.; et al. Gut Microbiota and Obesity: A Role for Probiotics. Nutrients 2019, 11, 2690. [Google Scholar] [CrossRef]
- Cai, Y.; Liu, P.; Zhou, X.; Yuan, J.; Chen, Q. Probiotics therapy show significant improvement in obesity and neurobehavioral disorders symptoms. Front. Cell. Infect. Microbiol. 2023, 13, e1178399. [Google Scholar] [CrossRef]
- Lim, S.-M.; Jeong, J.-J.; Woo, K.H.; Han, M.J.; Kim, D.-H. Lactobacillus sakei OK67 ameliorates high-fat diet–induced blood glucose intolerance and obesity in mice by inhibiting gut microbiota lipopolysaccharide production and inducing colon tight junction protein expression. Nutr. Res. 2016, 36, 337–348. [Google Scholar] [CrossRef]
- Jung, J.-Y.; Shin, J.-S.; Lee, S.-G.; Rhee, Y.K.; Cho, C.-W.; Hong, H.-D.; Lee, K.-T. Lactobacillus sakei K040706 evokes immunostimulatory effects on macrophages through TLR 2-mediated activation. Int. Immunopharmacol. 2015, 28, 88–96. [Google Scholar] [CrossRef]
- Won, S.-M.; Chen, S.; Lee, S.Y.; Lee, K.E.; Park, K.W.; Yoon, J.-H. Lactobacillus sakei ADM14 Induces Anti-Obesity Effects and Changes in Gut Microbiome in High-Fat Diet-Induced Obese Mice. Nutrients 2020, 12, 3703. [Google Scholar] [CrossRef]
- Kim, S.-K.; Guevarra, R.B.; Kim, Y.-T.; Kwon, J.; Kim, H.; Cho, J.H.; Kim, H.B.; Lee, J.-H. Role of Probiotics in Human Gut Microbiome-Associated Diseases. J. Microbiol. Biotechnol. 2019, 29, 1335–1340. [Google Scholar] [CrossRef]
- Li, H.-Y.; Zhou, D.-D.; Gan, R.-Y.; Huang, S.-Y.; Zhao, C.-N.; Shang, A.; Xu, X.-Y.; Li, H.-B. Effects and mechanisms of probiotics, prebiotics, synbiotics, and postbiotics on metabolic diseases targeting gut microbiota: A narrative review. Nutrients 2021, 13, 3211. [Google Scholar] [CrossRef]
- Goyache, I.; Yavorov-Dayliev, D.; Milagro, F.I.; Aranaz, P. Caenorhabditis elegans as a Screening Model for Probiotics with Properties against Metabolic Syndrome. Int. J. Mol. Sci. 2024, 25, 1321. [Google Scholar] [CrossRef]
- Tjørve, K.M.C.; Tjørve, E. The use of Gompertz models in growth analyses, and new Gompertz-model approach: An addition to the Unified-Richards family. PLoS ONE 2017, 12, e0178691. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP); Rychen, G.; Aquilina, G.; Azimonti, G.; Bampidis, V.; de Lourdes Bastos, M.; Bories, G.; Chesson, A.; Cocconcelli, P.S.; Flachowsky, G.; et al. Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018, 16, e05206. [Google Scholar] [CrossRef]
- da Silva, M.N.; Tagliapietra, B.L.; Flores, V.D.A.; dos Santos Richards, N.S.P. In vitro test to evaluate survival in the gastrointestinal tract of commercial probiotics. Curr. Res. Food Sci. 2021, 4, 320–325. [Google Scholar] [CrossRef] [PubMed]
- May, K.S.; Hartigh, L.J.D. Gut Microbial-Derived Short Chain Fatty Acids: Impact on Adipose Tissue Physiology. Nutrients 2023, 15, 272. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Gu, M.; Werlinger, P.; Cho, J.-H.; Cheng, J.; Suh, J.-W. Lactobacillus sakei MJM60958 as a Potential Probiotic Alleviated Non-Alcoholic Fatty Liver Disease in Mice Fed a High-Fat Diet by Modulating Lipid Metabolism, Inflammation, and Gut Microbiota. Int. J. Mol. Sci. 2022, 23, 13436. [Google Scholar] [CrossRef]
- Hernández, M.A.G.; Canfora, E.E.; Jocken, J.W.E.; Blaak, E.E. The Short-Chain Fatty Acid Acetate in Body Weight Control and Insulin Sensitivity. Nutrients 2019, 11, 1943. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Salas-Jara, M.J.; Ilabaca, A.; Vega, M.; García, A. Biofilm Forming Lactobacillus: New Challenges for the Development of Probiotics. Microorganisms 2016, 4, 35. [Google Scholar] [CrossRef] [PubMed]
- Vasudha, M.; Prashantkumar, C.S.; Bellurkar, M.; Kaveeshwar, V.; Gayathri, D. Probiotic potential of β-galactosidase-producing lactic acid bacteria from fermented milk and their molecular characterization. Biomed. Rep. 2023, 18, 23. [Google Scholar] [CrossRef]
- Tsai, C.-C.; Lin, P.-P.; Hsieh, Y.-M.; Zhang, Z.-Y.; Wu, H.-C.; Huang, C.-C. Cholesterol-lowering potentials of lactic acid bacteria based on bile-salt hydrolase activity and effect of potent strains on cholesterol metabolism in vitro and in vivo. Sci. World J. 2014, 2014, 690752. [Google Scholar] [CrossRef]
- Lee, Y.K.; Lim, C.Y.; Teng, W.L.; Ouwehand, A.C.; Tuomola, E.M.; Salminen, S. Quantitative approach in the study of adhesion of lactic acid bacteria to intestinal cells and their competition with enterobacteria. Appl. Environ. Microbiol. 2000, 66, 3692–3697. [Google Scholar] [CrossRef] [PubMed]
- Tumola, E.M.; Salminen, S.J. Adhesion of some probiotic and dairy Lactobacillus strains to Caco-2 cell cultures. Int. J. Food Microbiol. 1998, 41, 45–51. [Google Scholar] [CrossRef]
- Matijašić, B.B.; Narat, M.; Peternel, M.Z.; Rogelj, I. Ability of Lactobacillus gasseri K7 to inhibit Escherichia coli adhesion in vitro on Caco-2 cells and ex vivo on pigs’ jejunal tissue. Int. J. Food Microbiol. 2006, 107, 92–96. [Google Scholar] [CrossRef]
- Todoriki, K.; Mukai, T.; Sato, S.; Toba, T. Inhibition of adhesion of food-borne pathogens to Caco-2 cells by Lactobacillus strains. J. Appl. Microbiol. 2001, 91, 154–159. [Google Scholar] [CrossRef]
- Bernet, M.F.; Brassart, D.; Neeser, J.R.; Servin, A.L. Lactobacillus acidophilus LA 1 binds to cultured human intestinal cell lines and inhibits cell attachment and cell invasion by enterovirulent bacteria. Gut 1994, 35, 483–489. [Google Scholar] [CrossRef]
- Coconnier, M.-H.; Bernet, M.-F.; Kernéis, S.; Chauvière, G.; Fourniat, J.; Servin, A.L. Inhibition of adhesion of enteroinvasive pathogens to human intestinal Caco-2 cells by Lactobacillus acidophilus strain LB decreases bacterial invasion. FEMS Microbiol. Lett. 1993, 110, 299–305. [Google Scholar] [CrossRef][Green Version]
- Lievin, V.; Peiffer, I.; Hudault, S.; Rochat, F.; Brassart, D.; Neeser, J.-R.; Servin, A.L. Bifidobacterium strains from resident infant human gastrointestinal microflora exert antimicrobial activity. Gut 2000, 47, 646–652. [Google Scholar] [CrossRef]
- Gagnon, M.; Kheadr, E.E.; Dabour, N.; Richard, D.; Fliss, I. Effect of Bifidobacterium thermacidophilum probiotic feeding on enterohemorrhagic Escherichia coli O157:H7 infection in BALB/c mice. Int. J. Food Microbiol. 2006, 111, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.-Y.; Li, C.; Qin, Y.-Q.; Yin, R.-L.; Du, S.-W.; Ye, F.; Liu, H.-F.; Wang, M.-P.; Sun, Y.; Li, X.; et al. Lactobacilli reduce chemokine il-8 production in response to TNF-α and Salmonella challenge of Caco-2 cells. BioMed Res. Int. 2013, 2013, 925219. [Google Scholar] [CrossRef]
- Panpetch, W.; Spinler, J.K.; Versalovic, J.; Tumwasorn, S. Characterization of Lactobacillus salivarius strains B37 and B60 capable of inhibiting IL-8 production in Helicobacter pylori-stimulated gastric epithelial cells. BMC Microbiol. 2016, 16, 242. [Google Scholar] [CrossRef]
- Cooper, J.F.; Van Raamsdonk, J.M. Modeling Parkinson’s Disease in C. elegans. J. Park. Dis. 2018, 8, 17–32. [Google Scholar] [CrossRef]
- Zhang, G.; Wang, Z.; Du, Z.; Zhang, H. mTOR Regulates Phase Separation of PGL Granules to Modulate Their Autophagic Degradation. Cell 2018, 174, 1492–1506.e22. [Google Scholar] [CrossRef] [PubMed]
- Kranz, A.; Anastassiadis, K. The role of SETD1A and SETD1B in development and disease. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194578. [Google Scholar] [CrossRef]
- González-Aguilera, C.; Palladino, F.; Askjaer, P. C. elegans epigenetic regulation in development and aging. Briefings Funct. Genom. 2013, 13, 223–234. [Google Scholar] [CrossRef]
- An, L.; Fu, X.; Chen, J.; Ma, J. Application of Caenorhabditis elegans in Lipid Metabolism Research. Int. J. Mol. Sci. 2023, 24, 1173. [Google Scholar] [CrossRef] [PubMed]
- Komura, T.; Takemoto, A.; Kosaka, H.; Suzuki, T.; Nishikawa, Y. Prolonged Lifespan, Improved Perception, and Enhanced Host Defense of Caenorhabditis elegans by Lactococcus cremoris subsp. cremoris. Microbiol. Spectr. 2022, 10, e0045421. [Google Scholar] [CrossRef]
- Kwon, G.; Lee, J.; Lim, Y.-H. Dairy Propionibacterium extends the mean lifespan of Caenorhabditis elegans via activation of the innate immune system. Sci. Rep. 2016, 6, 31713. [Google Scholar] [CrossRef]
- Kumar, S.; Praneet, N.S.; Suchiang, K. Lactobacillus brevis MTCC 1750 enhances oxidative stress resistance and lifespan extension with improved physiological and functional capacity in Caenorhabditis elegans via the DAF-16 pathway. Free. Radic. Res. 2022, 56, 555–571. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Shin, M.; Ryu, S.; Yun, B.; Oh, S.; Park, D.-J.; Kim, Y. Evaluation of Probiotic Characteristics of Newly Isolated Lactic Acid Bacteria from Dry-Aged Hanwoo Beef. Food Sci. Anim. Resour. 2021, 41, 468–480. [Google Scholar] [CrossRef]
- Yoo, J.; Lee, J.; Zhang, M.; Mun, D.; Kang, M.; Yun, B.; Kim, Y.-A.; Kim, S.; Oh, S. Enhanced γ-aminobutyric acid and sialic acid in fermented deer antler velvet and immune promoting effects. J. Anim. Sci. Technol. 2022, 64, 166–182. [Google Scholar] [CrossRef] [PubMed]
- Ai, L.; Luo, D.; Wang, H.; Liu, X.; Yang, M.; Tian, F.; Qin, S.; Liu, J.; Li, Y. Ameliorative effects of Bifidobacterium longum peptide-1 on benzo(α)pyrene induced oxidative damages via daf-16 in Caenorhabditis elegans. Cell Stress Chaperon 2023, 28, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Juan, C.A.; de la Lastra, J.M.P.; Plou, F.J.; Pérez-Lebeña, E. The Chemistry of Reactive Oxygen Species (ROS) Revisited: Outlining Their Role in Biological Macromolecules (DNA, Lipids and Proteins) and Induced Pathologies. Int. J. Mol. Sci. 2021, 22, 4642. [Google Scholar] [CrossRef]
- Berk, Ş.; Cetin, A.; Özdemir, Ö.Ü.; Pektaş, A.N.; Yurtcu, N.; Dastan, S.D. The Combination of Metformin and High Glucose Increased Longevity of Caenorhabditis elegans a DAF-16/FOXO-Independent Manner: Cancer/Diabetic Model via C. elegans. Front. Endocrinol. 2024, 15, 1435098. [Google Scholar] [CrossRef]
- Yavorov-Dayliev, D.; Milagro, F.I.; Ayo, J.; Oneca, M.; Aranaz, P. Pediococcus acidilactici CECT9879 (pA1c) Counteracts the Effect of a High-Glucose Exposure in C. elegans by Affecting the Insulin Signaling Pathway (IIS). Int. J. Mol. Sci. 2022, 23, 2689. [Google Scholar] [CrossRef]
- Martorell, P.; Llopis, S.; González, N.; Chenoll, E.; López-Carreras, N.; Aleixandre, A.; Chen, Y.; Karoly, E.D.; Ramón, D.; Genovés, S. Probiotic Strain Bifidobacterium animalis subsp. Lactis CECT 8145 Reduces Fat Content and Modulates Lipid Metabolism and Antioxidant Response in Caenorhabditis elegans. J. Agric. Food Chem. 2016, 64, 3462–3472. [Google Scholar] [CrossRef]
- Balaguer, F.; Enrique, M.; Llopis, S.; Barrena, M.; Navarro, V.; Álvarez, B.; Chenoll, E.; Ramón, D.; Tortajada, M.; Martorell, P. Lipoteichoic acid from Bifidobacterium animalis subsp. lactis BPL1: A novel postbiotic that reduces fat deposition via IGF-1 pathway. Microb. Biotechnol. 2022, 15, 805–816. [Google Scholar] [CrossRef]
- Gu, M.; Werlinger, P.; Cho, J.-H.; Jang, N.; Choi, S.S.; Suh, J.-W.; Cheng, J. Lactobacillus pentosus MJM60383 Inhibits Lipid Accumulation in Caenorhabditis elegans Induced by Enterobacter cloacae and Glucose. Int. J. Mol. Sci. 2022, 24, 280. [Google Scholar] [CrossRef]
- ISO 10932:2010; Milk and Milk Products—Determination of the Minimal Inhibitory Concentration (MIC) of Antibiotics Applicable to Bifidobacteria and Non-Enterococcal Lactic Acid Bacteria (LAB). ISO: Geneva, Switzerland, 2010. Available online: https://www.iso.org/standard/46434.html (accessed on 21 January 2025).
- Brodkorb, A.; Egger, L.; Alminger, M.; Alvito, P.; Assunção, R.; Ballance, S.; Bohn, T.; Bourlieu-Lacanal, C.; Boutrou, R.; Carrière, F.; et al. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef] [PubMed]
- Valdés-Varela, L.; Ruas-Madiedo, P.; Gueimonde, M. In vitro fermentation of different fructo-oligosaccharides by Bifidobacterium strains for the selection of synbiotic combinations. Int. J. Food Microbiol. 2017, 242, 19–23. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, K.M.O.; Vieira, A.D.S.; Buriti, F.C.A.; Nascimento, J.C.F.D.; de Melo, M.E.S.; Bruno, L.M.; Borges, M.d.F.; Rocha, C.R.C.; Lopes, A.C.d.S.; Franco, B.D.G.d.M.; et al. Artisanal Coalho cheeses as source of beneficial Lactobacillus plantarum and Lactobacillus rhamnosus strains. Dairy Sci. Technol. 2015, 95, 209–230. [Google Scholar] [CrossRef]
- Kubota, H.; Senda, S.; Nomura, N.; Tokuda, H.; Uchiyama, H. Biofilm formation by lactic acid bacteria and resistance to environmental stress. J. Biosci. Bioeng. 2008, 106, 381–386. [Google Scholar] [CrossRef]
- Dashkevicz, M.P.; Feighner, S.D. Development of a differential medium for bile salt hydrolase-active Lactobacillus spp. Appl. Environ. Microbiol. 1989, 55, 11–16. [Google Scholar] [CrossRef]
- Gagnon, M.; Kheadr, E.E.; Le Blay, G.; Fliss, I. In vitro inhibition of Escherichia coli O157:H7 by bifidobacterial strains of human origin. Int. J. Food Microbiol. 2004, 92, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Repetto, G.; del Peso, A.; Zurita, J.L. Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nat. Protoc. 2008, 3, 1125–1131. [Google Scholar] [CrossRef]
- Navarro-Herrera, D.; Aranaz, P.; Eder-Azanza, L.; Zabala, M.; Hurtado, C.; Romo-Hualde, A.; Martínez, J.A.; González-Navarro, C.J.; Vizmanos, J.L. Dihomo-gamma-linolenic acid induces fat loss in C. elegans in an omega-3-independent manner by promoting peroxisomal fatty acid β-oxidation. Food Funct. 2018, 9, 1621–1637. [Google Scholar] [CrossRef]
- Aranaz, P.; Navarro-Herrera, D.; Zabala, M.; Romo-Hualde, A.; López-Yoldi, M.; Vizmanos, J.L.; Milagro, F.I.; González-Navarro, C.J. Phenolic Compounds Reduce the Fat Content in Caenorhabditis elegans by Affecting Lipogenesis, Lipolysis, and Different Stress Responses. Pharmaceuticals 2020, 13, 355. [Google Scholar] [CrossRef]
- Aranaz, P.; Peña, A.; Vettorazzi, A.; Fabra, M.J.; Martínez-Abad, A.; López-Rubio, A.; Pera, J.; Parladé, J.; Castellari, M.; Milagro, F.I.; et al. Grifola frondosa (Maitake) Extract Reduces Fat Accumulation and Improves Health Span in C. elegans through the DAF-16/FOXO and SKN-1/NRF2 Signalling Pathways. Nutrients 2021, 13, 3968. [Google Scholar] [CrossRef]


| Strain | MIC Values Against Antibiotics (mg/L) | |||||||
|---|---|---|---|---|---|---|---|---|
| Gm | Km | Sm | Tc | Cl | Cm | Am | Em | |
| L. sakei CNTA 173 | 4 | 16 | 64–16 | 2 | 0.062–0.032 | 2 | 4 | 0.125 |
| MIC cut-off values * | ≤16 | ≤64 | ≤64 | ≤8 | ≤1 | ≤4 | ≤4 | ≤1 |
| Medium | Concentration (g/L) |
|---|---|
| Defined medium without adding carbon source | 1.46 ± 0.01 |
| Defined medium with glucose (2%) | 0.40 ± 0.04 |
| Defined medium with Synergy 1 (2%) | 1.06 ± 0.10 |
| Defined medium with P95 (2%) | 1.33 ± 0.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goyache, I.; Valdés-Varela, L.; Virto, R.; López-Yoldi, M.; López-Giral, N.; Sánchez-Vicente, A.; Milagro, F.I.; Aranaz, P. Anti-Obesity Properties of a Novel Probiotic Strain of Latilactobacillus sakei CNTA 173 in Caenorhabditis elegans. Int. J. Mol. Sci. 2025, 26, 3286. https://doi.org/10.3390/ijms26073286
Goyache I, Valdés-Varela L, Virto R, López-Yoldi M, López-Giral N, Sánchez-Vicente A, Milagro FI, Aranaz P. Anti-Obesity Properties of a Novel Probiotic Strain of Latilactobacillus sakei CNTA 173 in Caenorhabditis elegans. International Journal of Molecular Sciences. 2025; 26(7):3286. https://doi.org/10.3390/ijms26073286
Chicago/Turabian StyleGoyache, Ignacio, Lorena Valdés-Varela, Raquel Virto, Miguel López-Yoldi, Noelia López-Giral, Ana Sánchez-Vicente, Fermín I. Milagro, and Paula Aranaz. 2025. "Anti-Obesity Properties of a Novel Probiotic Strain of Latilactobacillus sakei CNTA 173 in Caenorhabditis elegans" International Journal of Molecular Sciences 26, no. 7: 3286. https://doi.org/10.3390/ijms26073286
APA StyleGoyache, I., Valdés-Varela, L., Virto, R., López-Yoldi, M., López-Giral, N., Sánchez-Vicente, A., Milagro, F. I., & Aranaz, P. (2025). Anti-Obesity Properties of a Novel Probiotic Strain of Latilactobacillus sakei CNTA 173 in Caenorhabditis elegans. International Journal of Molecular Sciences, 26(7), 3286. https://doi.org/10.3390/ijms26073286







