A Rapid Approach for Identifying Cell Lines Lacking Functional Cytidine Deaminase
Abstract
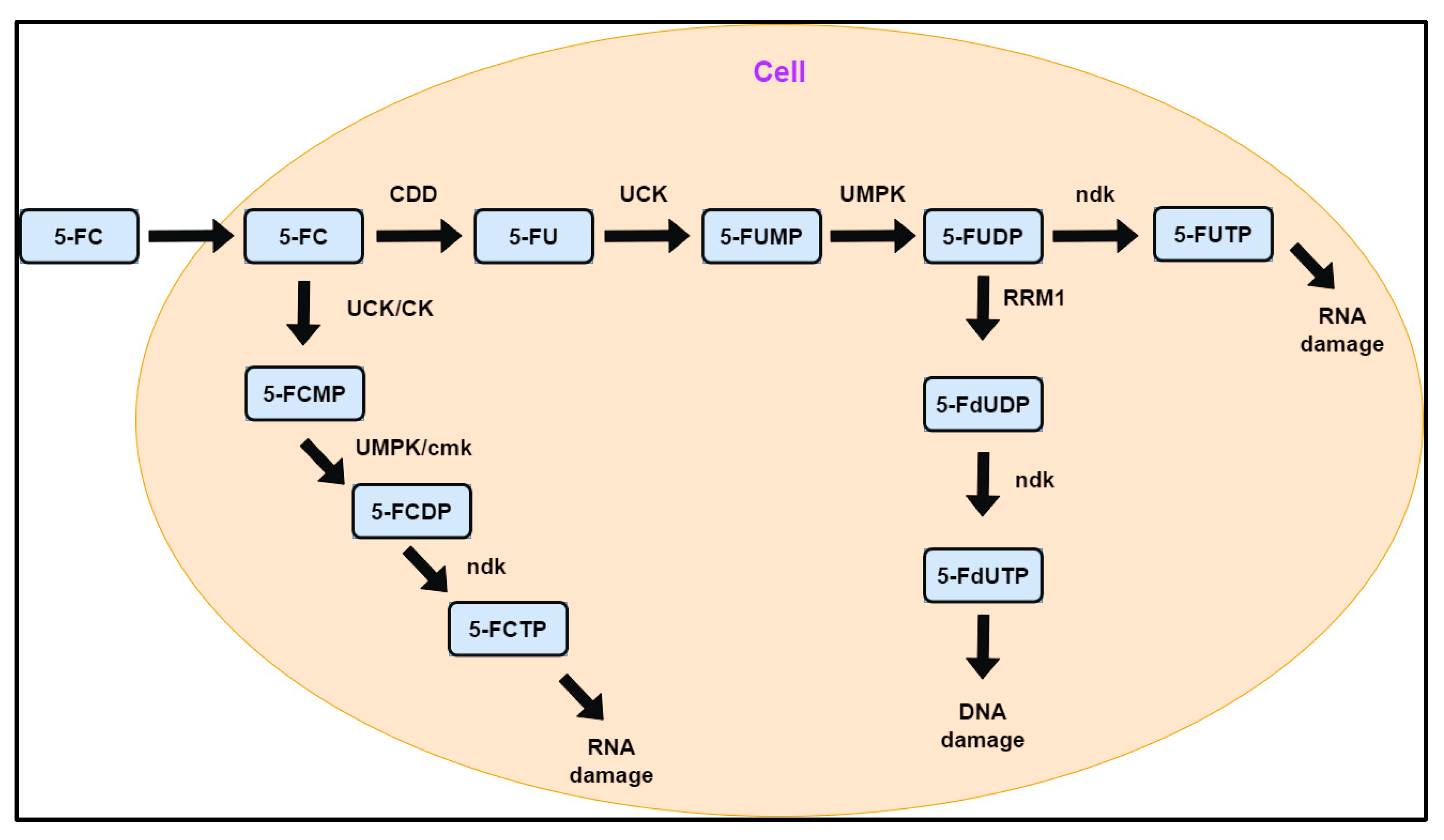
1. Introduction
2. Results and Discussion
2.1. Method Overview
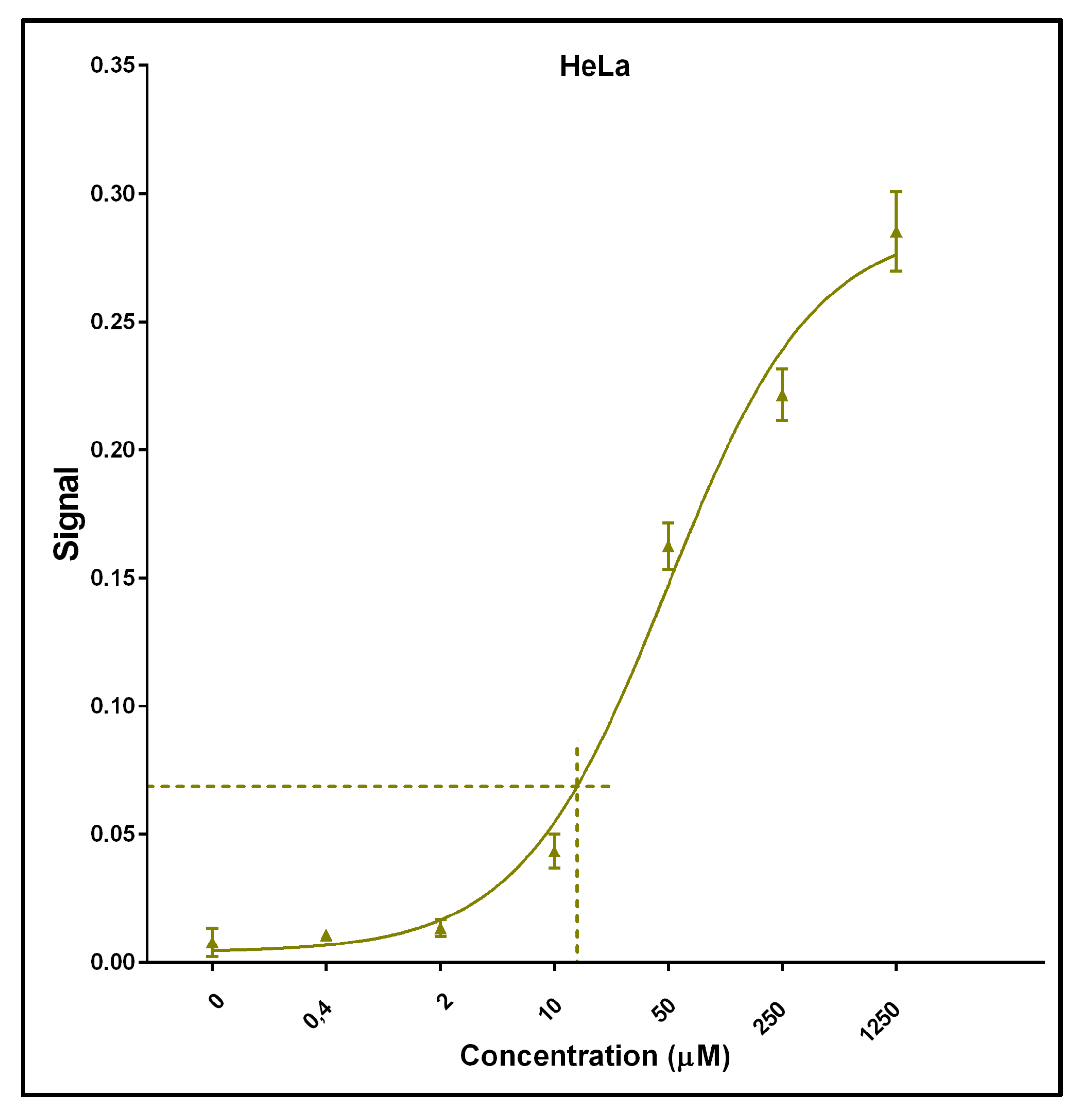
2.2. Method Validation and Optimization
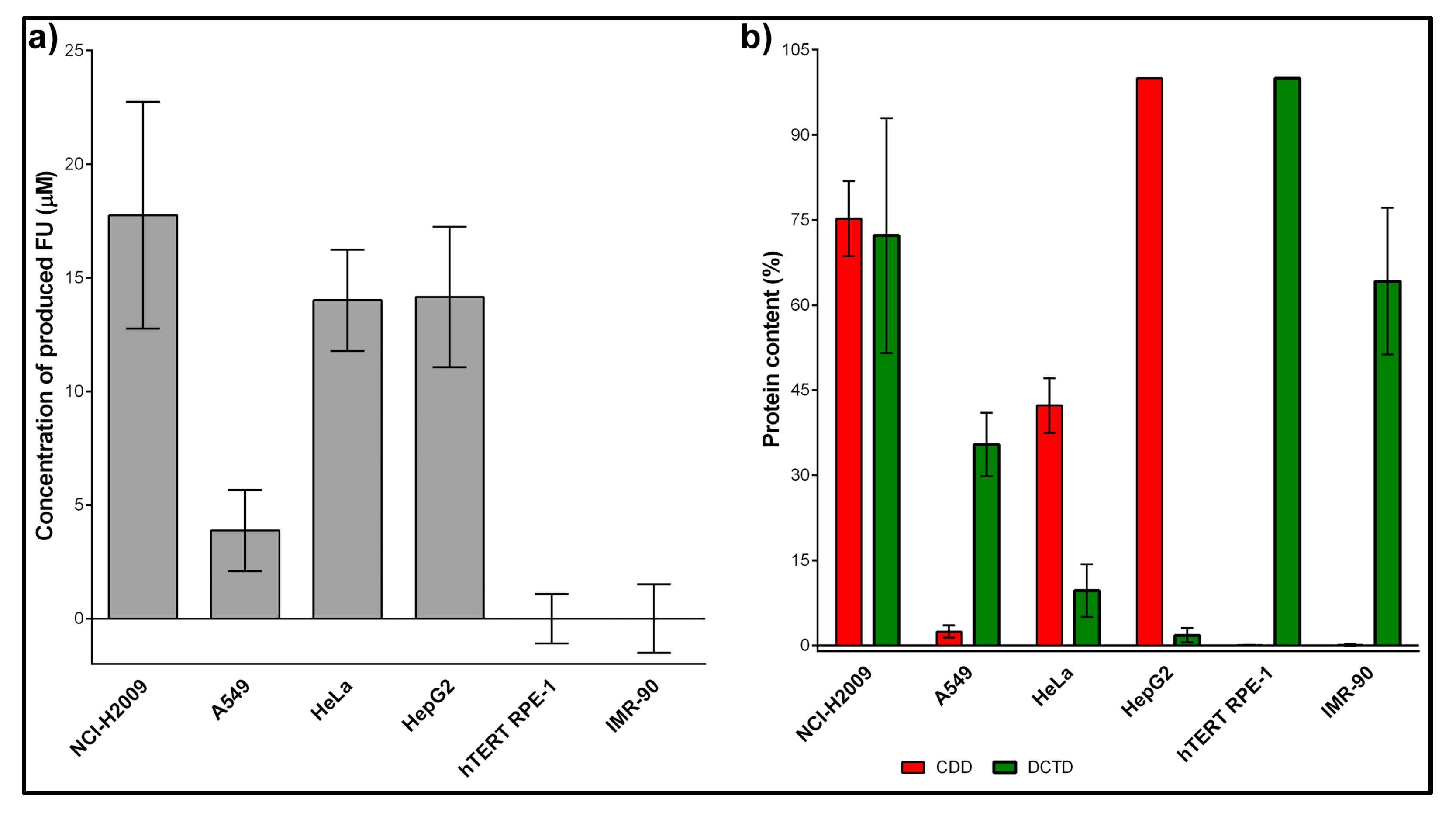
2.3. The Amount of CDD Strongly Correlated with the FU/FC Cytotoxicity Ratio
3. Materials and Methods
3.1. Cell Cultures
3.2. Tested Fixation Protocols
3.3. FC and FU Treatment and FU Detection
3.4. Cytotoxicity Assay
3.5. Data Acquisition, Computational Approach, and Data Evaluation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CDD | Cytidine deaminase |
| DCTD | Deoxycytidine monophosphate deaminase |
| FdUMP | 5-fluoro-2′-deoxyuridine monophosphate |
| EdC | 5-ethynyl-2′-deoxycytidine |
| EdU | 5-ethynyl-2′-deoxyuridine |
| FC | 5-fluorocytidine |
| FU | 5-fluorouridine |
| 5-FUMP | 5-fluorouridine monophosphate |
| 5-FUDP | 5-fluorouridine diphosphate |
| 5-FUTP | 5-fluorouridine triphosphate |
| 5-FdUDP | 5-fluorodeoxyuridine diphosphate |
| 5-FdUTP | 5-fluorodeoxyuridine triphosphate |
| 5-FCMP | 5-fluorocytidine monophosphate |
| 5-FCDP | 5-fluorocytidine diphosphate |
| 5-FCTP | 5-fluorocytidine triphosphate |
| UCK | Uridine kinase |
| UMPK | UMP-CMP kinase |
| ndk | Nucleoside-diphosphate kinase |
| RRM1 | Ribonucleoside-diphosphate reductase subunit M1 |
| CK | Cytidine kinase |
| cmk | CMP/dCMP kinase |
| 5-Fur | 5-fluorouracil |
| DMEM | Dulbecco’s modified Eagle’s medium |
| EMEM | Eagle’s minimum essential medium |
References
- Frances, A.; Cordelier, P. The Emerging Role of Cytidine Deaminase in Human Diseases: A New Opportunity for Therapy? Mol. Ther. 2020, 28, 357–366. [Google Scholar] [CrossRef]
- de Vos, D.; van Overveld, W. Decitabine: A historical review of the development of an epigenetic drug. Ann. Hematol. 2005, 84 (Suppl. S1), 3–8. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Salavaggione, O.E.; Ji, Y.; Pelleymounter, L.L.; Eckloff, B.W.; Wieben, E.D.; Ames, M.M.; Weinshilboum, R.M. Gemcitabine pharmacogenomics: Cytidine deaminase and deoxycytidylate deaminase gene resequencing and functional genomics. Clin. Cancer Res. 2006, 12, 1794–1803. [Google Scholar] [CrossRef]
- Hamada, A.; Kawaguchi, T.; Nakano, M. Clinical pharmacokinetics of cytarabine formulations. Clin. Pharmacokinet. 2002, 41, 705–718. [Google Scholar] [CrossRef]
- Heinemann, V.; Xu, Y.Z.; Chubb, S.; Sen, A.; Hertel, L.W.; Grindey, G.B.; Plunkett, W. Cellular elimination of 2′,2′-difluorodeoxycytidine 5′-triphosphate: A mechanism of self-potentiation. Cancer Res. 1992, 52, 533–539. [Google Scholar]
- Lamba, J.K. Pharmacogenomics of cytarabine in childhood leukemia. Pharmacogenomics 2011, 12, 1629–1632. [Google Scholar] [CrossRef]
- Xu, Y.Z.; Plunkett, W. Modulation of deoxycytidylate deaminase in intact human leukemia cells. Action of 2′,2′-difluorodeoxycytidine. Biochem. Pharmacol. 1992, 44, 1819–1827. [Google Scholar] [CrossRef]
- de Sousa Cavalcante, L.; Monteiro, G. Gemcitabine: Metabolism and molecular mechanisms of action, sensitivity and chemoresistance in pancreatic cancer. Eur. J. Pharmacol. 2014, 741, 8–16. [Google Scholar] [CrossRef]
- Mini, E.; Nobili, S.; Caciagli, B.; Landini, I.; Mazzei, T. Cellular pharmacology of gemcitabine. Ann. Oncol. 2006, 17 (Suppl. S5), v7–v12. [Google Scholar] [CrossRef]
- Hamed, S.S.; Straubinger, R.M.; Jusko, W.J. Pharmacodynamic modeling of cell cycle and apoptotic effects of gemcitabine on pancreatic adenocarcinoma cells. Cancer Chemother. Pharmacol. 2013, 72, 553–563. [Google Scholar] [CrossRef]
- Grant, S. Ara-C: Cellular and molecular pharmacology. Adv. Cancer Res. 1998, 72, 197–233. [Google Scholar] [CrossRef] [PubMed]
- Issa, J.P.; Gharibyan, V.; Cortes, J.; Jelinek, J.; Morris, G.; Verstovsek, S.; Talpaz, M.; Garcia-Manero, G.; Kantarjian, H.M. Phase II study of low-dose decitabine in patients with chronic myelogenous leukemia resistant to imatinib mesylate. J. Clin. Oncol. 2005, 23, 3948–3956. [Google Scholar] [CrossRef]
- Jones, P.A.; Taylor, S.M. Cellular differentiation, cytidine analogs and DNA methylation. Cell 1980, 20, 85–93. [Google Scholar] [CrossRef]
- Budman, D.R.; Meropol, N.J.; Reigner, B.; Creaven, P.J.; Lichtman, S.M.; Berghorn, E.; Behr, J.; Gordon, R.J.; Osterwalder, B.; Griffin, T. Preliminary studies of a novel oral fluoropyrimidine carbamate: Capecitabine. J. Clin. Oncol. 1998, 16, 1795–1802. [Google Scholar] [CrossRef]
- Miwa, M.; Ura, M.; Nishida, M.; Sawada, N.; Ishikawa, T.; Mori, K.; Shimma, N.; Umeda, I.; Ishitsuka, H. Design of a novel oral fluoropyrimidine carbamate, capecitabine, which generates 5-fluorouracil selectively in tumours by enzymes concentrated in human liver and cancer tissue. Eur. J. Cancer 1998, 34, 1274–1281. [Google Scholar] [CrossRef]
- Ishikawa, T.; Sekiguchi, F.; Fukase, Y.; Sawada, N.; Ishitsuka, H. Positive correlation between the efficacy of capecitabine and doxifluridine and the ratio of thymidine phosphorylase to dihydropyrimidine dehydrogenase activities in tumors in human cancer xenografts. Cancer Res. 1998, 58, 685–690. [Google Scholar]
- Verweij, J. Rational design of new tumoractivated cytotoxic agents. Oncology 1999, 57 (Suppl. S1), 9–15. [Google Scholar] [CrossRef]
- Terranova-Barberio, M.; Roca, M.S.; Zotti, A.I.; Leone, A.; Bruzzese, F.; Vitagliano, C.; Scogliamiglio, G.; Russo, D.; D’Angelo, G.; Franco, R.; et al. Valproic acid potentiates the anticancer activity of capecitabine in vitro and in vivo in breast cancer models via induction of thymidine phosphorylase expression. Oncotarget 2016, 7, 7715–7731. [Google Scholar] [CrossRef]
- Grem, J.L.; Keith, B. Mechanisms of Action of Cancer Chemotherapeutic Agents: Antimetabolites. In The Cancer Handbook; Alison, M.R., Ed.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2005. [Google Scholar]
- Ligasová, A.; Piskláková, B.; Friedecký, D.; Koberna, K. A new technique for the analysis of metabolic pathways of cytidine analogues and cytidine deaminase activities in cells. Sci. Rep. 2023, 13, 20530. [Google Scholar] [CrossRef]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef]
- Vodenkova, S.; Buchler, T.; Cervena, K.; Veskrnova, V.; Vodicka, P.; Vymetalkova, V. 5-fluorouracil and other fluoropyrimidines in colorectal cancer: Past, present and future. Pharmacol. Ther. 2020, 206, 107447. [Google Scholar] [CrossRef] [PubMed]
- Ligasová, A.; Koberna, K. Quantification of fixed adherent cells using a strong enhancer of the fluorescence of DNA dyes. Sci. Rep. 2019, 9, 8701. [Google Scholar] [CrossRef]
- Cvačková, Z.; Albring, K.F.; Koberna, K.; Ligasová, A.; Huber, O.; Raška, I.; Staněk, D. Pontin is localized in nucleolar fibrillar centers. Chromosoma 2008, 117, 487–497. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ligasová, A.; Frydrych, I.; Piskláková, B.; Friedecký, D.; Koberna, K. The kinetics of uracil-N-glycosylase distribution inside replication foci. Sci. Rep. 2025, 15, 3026. [Google Scholar] [CrossRef]
- Ligasová, A.; Vydržalová, M.; Burianová, R.; Bručková, L.; Večeřová, R.; Janošťáková, A.; Koberna, K. A New Sensitive Method for the Detection of Mycoplasmas Using Fluorescence Microscopy. Cells 2019, 8, 1510. [Google Scholar] [CrossRef]
- Ligasová, A.; Rosenberg, I.; Bocková, M.; Homola, J.; Koberna, K. Anchored linear oligonucleotides: The effective tool for the real-time measurement of uracil DNA glycosylase activity. Open Biol. 2021, 11, 210136. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Carpenter, A.E.; Jones, T.R.; Lamprecht, M.R.; Clarke, C.; Kang, I.H.; Friman, O.; Guertin, D.A.; Chang, J.H.; Lindquist, R.A.; Moffat, J.; et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7, R100. [Google Scholar] [CrossRef]
- Kamentsky, L.; Jones, T.R.; Fraser, A.; Bray, M.A.; Logan, D.J.; Madden, K.L.; Ljosa, V.; Rueden, C.; Eliceiri, K.W.; Carpenter, A.E. Improved structure, function and compatibility for CellProfiler: Modular high-throughput image analysis software. Bioinformatics 2011, 27, 1179–1180. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
| Ctrl vs. 50 µM FU | Ctrl vs. 50 µM FC | |
|---|---|---|
| NCI-H2009 | 0.00214 | 0.00269 |
| A549 | 0.00054 | 0.00028 |
| HeLa | 0.00003 | 0.00012 |
| HepG2 | 0.00084 | 0.00002 |
| hTERT RPE-1 | 0.00152 | 0.16752 |
| IMR-90 | 0.00297 | 0.54762 |
| Pearson Coefficient | p-Value | |
|---|---|---|
| DCTD | −0.678 | 0.14 |
| hENT1 | −0.143 | 0.79 |
| hENT2 | −0.729 | 0.10 |
| CNT1 | −0.105 | 0.84 |
| CNT2 | −0.671 | 0.15 |
| CNT3 | −0.503 | 0.31 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ligasová, A.; Kociánová, M.; Koberna, K. A Rapid Approach for Identifying Cell Lines Lacking Functional Cytidine Deaminase. Int. J. Mol. Sci. 2025, 26, 3344. https://doi.org/10.3390/ijms26073344
Ligasová A, Kociánová M, Koberna K. A Rapid Approach for Identifying Cell Lines Lacking Functional Cytidine Deaminase. International Journal of Molecular Sciences. 2025; 26(7):3344. https://doi.org/10.3390/ijms26073344
Chicago/Turabian StyleLigasová, Anna, Markéta Kociánová, and Karel Koberna. 2025. "A Rapid Approach for Identifying Cell Lines Lacking Functional Cytidine Deaminase" International Journal of Molecular Sciences 26, no. 7: 3344. https://doi.org/10.3390/ijms26073344
APA StyleLigasová, A., Kociánová, M., & Koberna, K. (2025). A Rapid Approach for Identifying Cell Lines Lacking Functional Cytidine Deaminase. International Journal of Molecular Sciences, 26(7), 3344. https://doi.org/10.3390/ijms26073344





