Biomarkers of Skeletal Muscle Atrophy Based on Atrogenes Evaluation: A Systematic Review and Meta-Analysis Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Criteria for Eligibility
2.1.1. Types of Studies and Interventions
2.1.2. Characteristics of the Animal Model
2.1.3. Characteristics of the Variables Measured
2.2. Selection Strategy
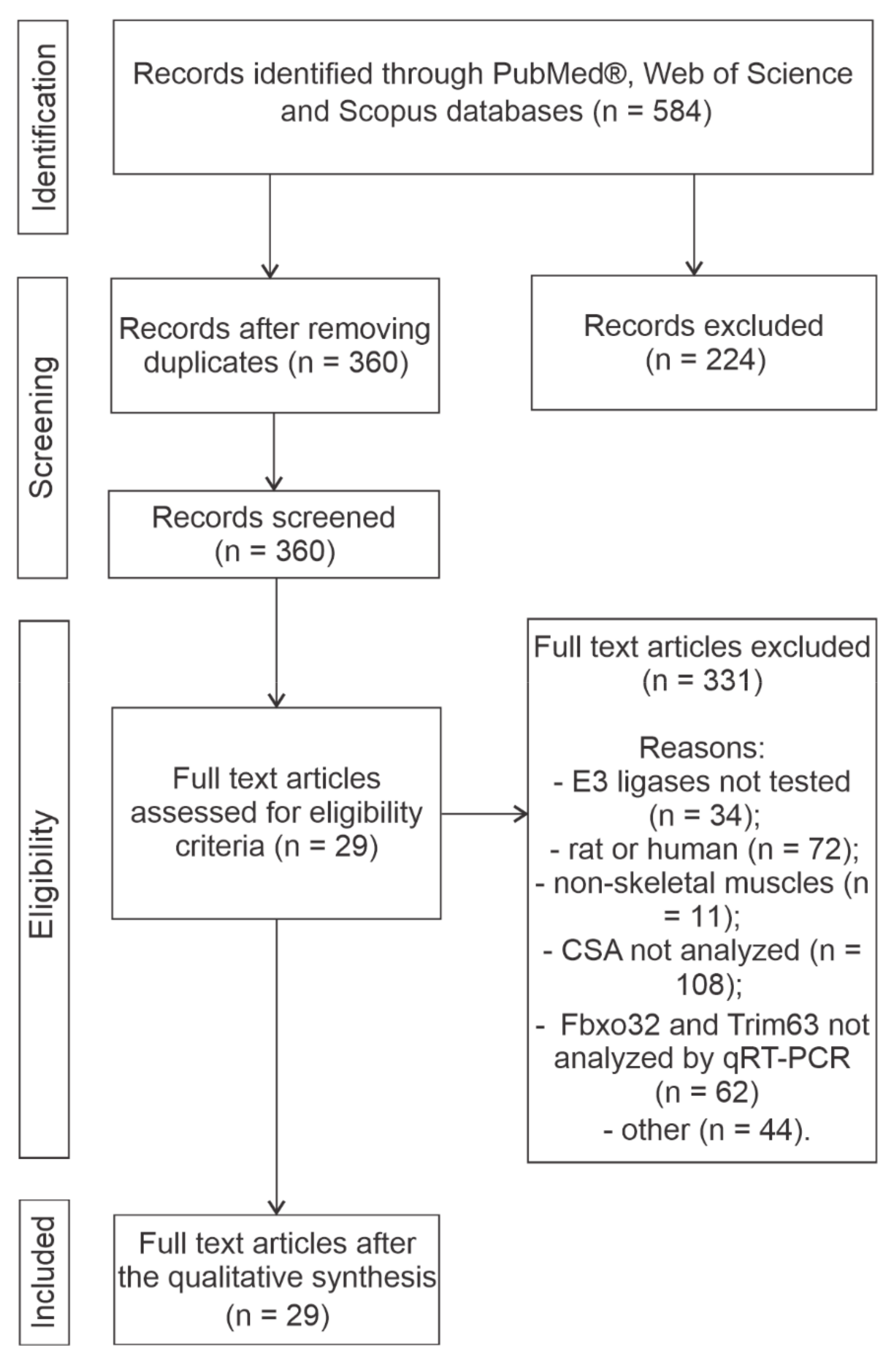
2.3. Study Selection
2.4. Data Extraction
2.5. Bias Risk Assessment and Degree of Between-Rater Agreements
2.6. Consistency of the Selected Studies
2.7. Effect Size
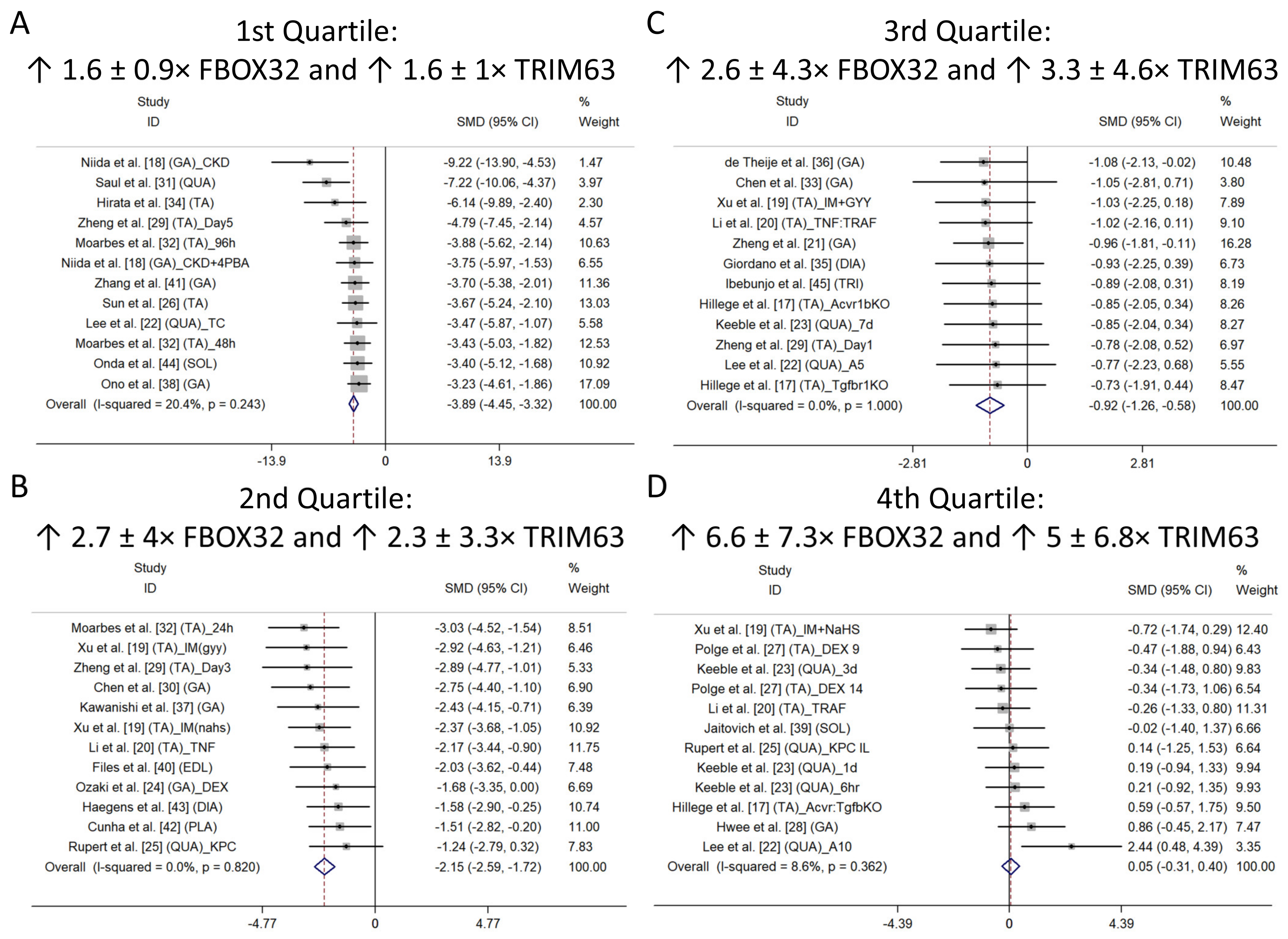
Subgroup Analysis
3. Results
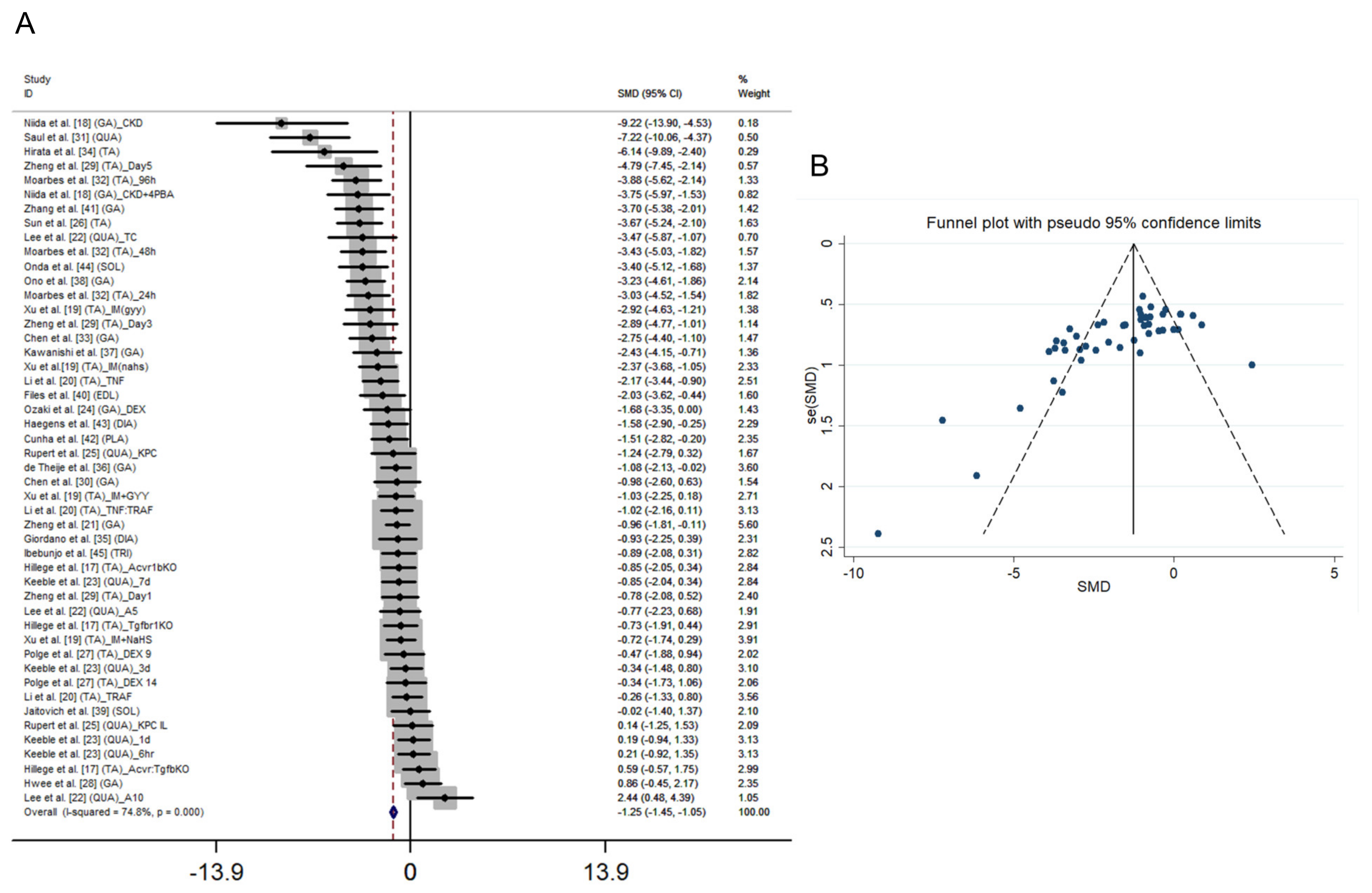
3.1. Main Characteristics and Quality of the Selected Studies
3.2. Main Effect, Heterogeneity and Risk of Bias in the Studies
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
Appendix A
Appendix B
Appendix B.1
| Cohorts | Studies | n_ctrl | CSA_ctrl (μm²) | SD_ctrl (μm²) | n_injury | CSA_injury (μm²) | SD_injury (μm²) |
|---|---|---|---|---|---|---|---|
| 1 | Hillege et al. [13] (TA)_Acvr1bKO | 6.0 | 423.82 | 94.991 | 6.0 | 330.47 | 122.132 |
| 2 | Hillege et al. [13] (TA)_Tgfbr1KO | 6.0 | 423.82 | 94.991 | 6.0 | 294.18 | 230.693 |
| 3 | Hillege et al. [13] (TA)_Acvr:TgfbKO | 6.0 | 423.82 | 94.991 | 6.0 | 532.96 | 244.263 |
| 4 | Niida et al. [14] (GA)_CKD | 5.0 | 2014.16 | 125.399 | 5.0 | 1134.38 | 50.043 |
| 5 | Niida et al. [14] (GA)_CKD + 4PBA | 5.0 | 2014.16 | 125.399 | 5.0 | 1543.86 | 125.399 |
| 6 | Xu et al. [15] (TA)_IM(gyy) | 6.0 | 1469.09 | 260.454 | 6.0 | 823.03 | 173.007 |
| 7 | Xu et al. [15] (TA)_IM + GYY | 6.0 | 1469.09 | 260.454 | 6.0 | 1221.11 | 217.196 |
| 8 | Xu et al. [15] (TA)_IM(nahs) | 8.0 | 1442.61 | 300.747 | 8.0 | 814.2 | 224.747 |
| 9 | Xu et al. [15] (TA)_IM + NaHS | 8.0 | 1442.61 | 300.747 | 8.0 | 1185.79 | 400.647 |
| 10 | Li et al. [16] (TA)_TNF | 8.0 | 2488.72 | 938.43 | 8.0 | 980.25 | 293.23 |
| 11 | Li et al. [16] (TA)_TRAF | 8.0 | 2488.72 | 938.43 | 6.0 | 2271.16 | 636.74 |
| 12 | Li et al. [16] (TA)_TNF:TRAF | 8.0 | 2488.72 | 938.43 | 6.0 | 1734.38 | 259.87 |
| 13 | Zheng et al. [17] (TA)_Day1 | 5.0 | 1024.56 | 171.797 | 5.0 | 916.56 | 92.931 |
| 14 | Zheng et al. [17] (TA)_Day3 | 5.0 | 1111.77 | 187.271 | 5.0 | 613.35 | 156.279 |
| 15 | Zheng et al. [17] (TA)_Day5 | 5.0 | 1108.31 | 140.805 | 5.0 | 596.03 | 54.918 |
| 16 | Lee et al. [18] (QUA)_TC | 4.0 | 1664.65 | 87.480 | 4.0 | 1365.76 | 84.940 |
| 17 | Lee et al. [18] (QUA)_A5 | 4.0 | 1664.65 | 87.480 | 4.0 | 1579.39 | 129.320 |
| 18 | Lee et al. [18] (QUA)_A10 | 4.0 | 1664.65 | 87.480 | 4.0 | 1877.97 | 87.480 |
| 19 | Keeble et al. [19] (QUA)_6 h | 6.0 | 2355.43 | 548.51 | 6.0 | 2463.02 | 451.48 |
| 20 | Keeble et al. [19] (QUA)_1 d | 6.0 | 2011.53 | 301.63 | 6.0 | 2129.68 | 806.92 |
| 21 | Keeble et al. [19] (QUA)_3 d | 6.0 | 2398.66 | 743.52 | 6.0 | 2150.82 | 720.46 |
| 22 | Keeble et al. [19] (QUA)_7 d | 6.0 | 2194.04 | 171.95 | 6.0 | 2000 | 274.74 |
| 23 | Ozaki et al. [20] (GA)_DEX | 4.0 | 1098.46 | 41.020 | 4.0 | 850.59 | 205.120 |
| 24 | Rupert et al. [21] (QUA)_KPC | 4.0 | 952.13 | 258.68 | 4.0 | 645.58 | 236.92 |
| 25 | Rupert et al. [21] (QUA)_KPC IL | 4.0 | 952.13 | 258.68 | 4.0 | 997.72 | 380.62 |
| 26 | Sun et al. [22] (TA) | 9.0 | 657.49 | 55.72 | 9.0 | 479.12 | 40.25 |
| 27 | Polge et al. [23] (TA)_DEX 9 | 4.5 | 5.66 | 3.967 | 4.5 | 4.14 | 2.227 |
| 28 | Polge et al. [23] (TA)_DEX 14 | 4.5 | 5.66 | 3.967 | 4.5 | 4.39 | 3.585 |
| 29 | Hwee et al. [24] (GA) | 5.0 | 1203.71 | 737.679 | 5.0 | 1680.9 | 266.852 |
| 30 | Zheng et al. [25] (GA) | 12.0 | 2247.05 | 254.230 | 12.0 | 2042.35 | 162.986 |
| 31 | Chen et al. [26] (GA) | 3.0 | 3228.5 | 2359.24 | 3.0 | 1339.22 | 952.84 |
| 32 | Saul et al. [27] (QUA) | 8.0 | 3970.4 | 236.174 | 8.0 | 901.69 | 553.127 |
| 33 | Moarbes et al. [28] (TA)_24 h | 8.0 | 613.73 | 138.508 | 8.0 | 274.95 | 76.594 |
| 34 | Moarbes et al. [28] (TA)_48 h | 8.0 | 552.13 | 124.875 | 8.0 | 206.3 | 69.240 |
| 35 | Moarbes et al. [28] (TA)_96 h | 8.0 | 611.13 | 132.229 | 8.0 | 191.46 | 76.594 |
| 36 | Chen et al. [29] (GA) | 6.0 | 589.68 | 143.64 | 6.0 | 268.69 | 81.27 |
| 37 | Hirata et al. [30] (TA) | 4.0 | 2032.4 | 150.800 | 4.0 | 1243.3 | 101.200 |
| 38 | Giordano et al. [31] (DIA) | 5.0 | 1313.5 | 564.607 | 5.0 | 933.5 | 131.034 |
| 39 | de Theije et al. [32] (GA) | 8.0 | 2177.19 | 374.795 | 8.0 | 1852.45 | 204.778 |
| 40 | Kawanishi et al. [33] (GA) | 5.5 | 1797.87 | 365.899 | 5.5 | 1079.19 | 202.345 |
| 41 | Ono et al. [34] (GA) | 10.0 | 2059.04 | 355.883 | 10.0 | 1226.93 | 75.831 |
| 42 | Jaitovich et al. [35] (SOL) | 4.5 | 14,618.62 | 174,981.861 | 4.5 | 8596.9 | 491,930.137 |
| 43 | Files et al. [36] (EDL) | 5.0 | 1621.02 | 77.793 | 5.0 | 1236 | 257.081 |
| 44 | Zhang et al. [37] (GA) | 8.0 | 64,680.6 | 10,758.6 | 8.0 | 32,946.6 | 5624.4 |
| 45 | Cunha et al. [38] (PLA) | 6.0 | 2039.34 | 505.942 | 6.0 | 1431.14 | 265.010 |
| 46 | Haegens et al. [39] (DIA) | 6.0 | 58,684.9 | 6622.35 | 6.0 | 48,480.6 | 6330.8 |
| 47 | Onda et al. [40] (SOL) | 7.0 | 1840.84 | 317.252 | 7.0 | 1029.06 | 115.355 |
| 48 | Ibebunjo et al. [41] (TRI) | 6.0 | 92,392.5 | 16,487.515 | 6.0 | 80,200.5 | 10,265.812 |
Appendix B.2
| Cohorts | Studies | n_ctrl | MAFbx_ctrl (Relative Levels) | SD_ctrl (Relative Levels) | n_injury | MAFbx_injury (Relative Levels) | SD_injury (Relative Levels) |
|---|---|---|---|---|---|---|---|
| 1 | Hillege et al. [13] (TA)_Acvr1bKO | 6.0 | 0.088 | 0.152 | 6.0 | 0.088 | 0.135 |
| 2 | Hillege et al. [13] (TA)_Tgfbr1KO | 6.0 | 0.088 | 0.152 | 6.0 | 0.088 | 0.066 |
| 3 | Hillege et al. [13] (TA)_Acvr:TgfbKO | 6.0 | 0.088 | 0.152 | 6.0 | 0.29 | 0.255 |
| 4 | Niida et al. [14] (GA)_CKD | 5.0 | 0.98 | 1.14 | 5.0 | 2.58 | 2.594 |
| 5 | Niida et al. [14] (GA)_CKD + 4PBA | 5.0 | 0.98 | 1.14 | 5.0 | 1.35 | 1.006 |
| 6 | Xu et al. [15] (TA)_IM(gyy) | 6.0 | 0.803 | 1.2 | 6.0 | 5.05 | 4.777 |
| 7 | Xu et al. [15] (TA)_IM + GYY | 6.0 | 0.803 | 1.2 | 6.0 | 2.29 | 3.677 |
| 8 | Xu et al. [15] (TA)_IM(nahs) | 8.0 | 0.9 | 2.404 | 8.0 | 5.202 | 5.091 |
| 9 | Xu et al. [15] (TA)_IM + NaHS | 8.0 | 0.9 | 2.404 | 8.0 | 1.95 | 4.384 |
| 10 | Li et al. [16] (TA)_TNF | 8.0 | 1.05 | 0.86 | 8.0 | 2.17 | 1.06 |
| 11 | Li et al. [16] (TA)_TRAF | 8.0 | 1.05 | 0.86 | 6.0 | 0.89 | 0.29 |
| 12 | Li et al. [16] (TA)_TNF:TRAF | 8.0 | 1.05 | 0.86 | 6.0 | 0.72 | 0.68 |
| 13 | Zheng et al. [17] (TA)_Day1 | 5.0 | 0.95 | 0.52 | 5.0 | 3.84 | 0.96 |
| 14 | Zheng et al. [17] (TA)_Day3 | 5.0 | 0.98 | 0.58 | 5.0 | 1.52 | 1.34 |
| 15 | Zheng et al. [17] (TA)_Day5 | 5.0 | 1.02 | 0.64 | 5.0 | 1.52 | 1.68 |
| 16 | Lee et al. [18] (QUA)_TC | 4.0 | 0.96 | 1.006 | 4.0 | 4.85 | 5.121 |
| 17 | Lee et al. [18] (QUA)_A5 | 4.0 | 0.96 | 1.006 | 4.0 | 1.73 | 1.766 |
| 18 | Lee et al. [18] (QUA)_A10 | 4.0 | 0.96 | 1.006 | 4.0 | 1.29 | 3.198 |
| 19 | Keeble et al. [19] (QUA)_6 h | 6.0 | 1 | 0.79 | 6.0 | 0.93 | 0.61 |
| 20 | Keeble et al. [19] (QUA)_1 d | 6.0 | 1 | 0.78 | 6.0 | 1.62 | 0.57 |
| 21 | Keeble et al. [19] (QUA)_3 d | 6.0 | 1.07 | 1.33 | 6.0 | 2.2 | 2.52 |
| 22 | Keeble et al. [19] (QUA)_7 d | 6.0 | 1.02 | 0.7 | 6.0 | 0.92 | 0.25 |
| 23 | Ozaki et al. [20] (GA)_DEX | 4.0 | 1.01 | 0.514 | 4.0 | 2.96 | 3.87 |
| 24 | Rupert et al. [21] (QUA)_KPC | 4.0 | 1.13 | 1.7 | 4.0 | 17.65 | 3.46 |
| 25 | Rupert et al. [21] (QUA)_KPC IL | 4.0 | 1.13 | 1.7 | 4.0 | 3.65 | 4.4 |
| 26 | Sun et al. [22] (TA) | 9.0 | 1 | 0.081 | 9.0 | 3.1 | 0.768 |
| 27 | Polge et al. [23] (TA)_DEX 9 | 4.5 | 1.7 | 2.715 | 4.5 | 4.63 | 6.64 |
| 28 | Polge et al. [23] (TA)_DEX 14 | 4.5 | 1.7 | 2.715 | 4.5 | 4.35 | 5.091 |
| 29 | Hwee et al. [24] (GA) | 5.0 | 1.34 | 2.12 | 5.0 | 2.34 | 2.3 |
| 30 | Zheng et al. [25] (GA) | 12.0 | 0.99 | 0.686 | 12.0 | 1.46 | 0.682 |
| 31 | Chen et al. [26] (GA) | 3.0 | 1.02 | 0.92 | 3.0 | 3.95 | 4.05 |
| 32 | Saul et al. [27] (QUA) | 8.0 | 2.26 | 0.933 | 8.0 | 15.41 | 11.399 |
| 33 | Moarbes et al. [28] (TA)_24 h | 8.0 | 0.86 | 0 | 8.0 | 13.63 | 5.996 |
| 34 | Moarbes et al. [28] (TA)_48 h | 8.0 | 0.86 | 0 | 8.0 | 6.28 | 3.564 |
| 35 | Moarbes et al. [28] (TA)_96 h | 8.0 | 0.86 | 0 | 8.0 | 21.5 | 5.798 |
| 36 | Chen et al. [29] (GA) | 6.0 | 1.52 | 0.37 | 6.0 | 20.64 | 2.51 |
| 37 | Hirata et al. [30] (TA) | 4.0 | 19.05 | 4.24 | 4.0 | 24.01 | 1.72 |
| 38 | Giordano et al. [31] (DIA) | 5.0 | 1 | 0.16 | 5.0 | 1.17 | 0.16 |
| 39 | de Theije et al. [32] (GA) | 8.0 | 6.42 | 2.432 | 8.0 | 21.3 | 4.13 |
| 40 | Kawanishi et al. [33] (GA) | 5.5 | 2.05 | 0.61 | 5.5 | 12.96 | 8.841 |
| 41 | Ono et al. [34] (GA) | 10.0 | 4.23 | 2.308 | 10.0 | 22.3 | 1.897 |
| 42 | Jaitovich et al. [35] (SOL) | 4.5 | - | - | 4.5 | - | - |
| 43 | Files et al. [36] (EDL) | 5.0 | 0.79 | 0.447 | 5.0 | 12.9 | 9.168 |
| 44 | Zhang et al. [37] (GA) | 8.0 | 2.45 | 0.33 | 8.0 | 22.75 | 4.1 |
| 45 | Cunha et al. [38] (PLA) | 6.0 | 8.76 | 0 | 6.0 | 14.02 | 1.788 |
| 46 | Haegens et al. [39] (DIA) | 6.0 | 4.63 | 0.53 | 6.0 | 12.24 | 4.17 |
| 47 | Onda et al. [40] (SOL) | 7.0 | 4.3 | 1.746 | 7.0 | 13.03 | 1.931 |
| 48 | Ibebunjo et al. [41] (TRI) | 6.0 | 3.77 | 0.49 | 6.0 | 3.64 | 0.49 |
Appendix B.3
| Cohorts | Studies | n_ctrl | MuRF-1_ctrl (Relative Levels) | SD_ctrl (Relative Levels) | n_injury | MuRF-1_injury (Relative Levels) | SD_injury (Relative Levels) |
|---|---|---|---|---|---|---|---|
| 1 | Hillege et al. [13] (TA)_Acvr1bKO | 6.0 | 0.19 | 0.118 | 6.0 | 0.19 | 0.149 |
| 2 | Hillege et al. [13] (TA)_Tgfbr1KO | 6.0 | 0.19 | 0.118 | 6.0 | 0.19 | 0.083 |
| 3 | Hillege et al. [13] (TA)_Acvr:TgfbKO | 6.0 | 0.19 | 0.118 | 6.0 | 0.28 | 0.203 |
| 4 | Niida et al. [14] (GA)_CKD | 5.0 | 1 | 1.297 | 5.0 | 3.27 | 3.935 |
| 5 | Niida et al. [14] (GA)_CKD + 4PBA | 5.0 | 1 | 1.297 | 5.0 | 1.67 | 1.409 |
| 6 | Xu et al. [15] (TA)_IM(gyy) | 6.0 | 1.099 | 1.347 | 6.0 | 5.102 | 5.511 |
| 7 | Xu et al. [15] (TA)_IM + GYY | 6.0 | 1.099 | 1.347 | 6.0 | 2.25 | 3.16 |
| 8 | Xu et al. [15] (TA)_IM(nahs) | 8.0 | 1.099 | 2.271 | 8.0 | 4.6 | 7.354 |
| 9 | Xu et al. [15] (TA)_IM + NaHS | 8.0 | 1.099 | 2.271 | 8.0 | 2.4 | 5.063 |
| 10 | Li et al. [16] (TA)_TNF | 8.0 | 0.98 | 0.22 | 8.0 | 4.13 | 1.86 |
| 11 | Li et al. [16] (TA)_TRAF | 8.0 | 0.98 | 0.22 | 8.0 | 2.63 | 1.27 |
| 12 | Li et al. [16] (TA)_TNF:TRAF | 8.0 | 0.98 | 0.22 | 8.0 | 1.56 | 2.16 |
| 13 | Zheng et al. [17] (TA)_Day1 | 5.0 | 0.96 | 0.64 | 5.0 | 3.38 | 2.32 |
| 14 | Zheng et al. [17] (TA)_Day3 | 5.0 | 0.96 | 0.44 | 5.0 | 0.87 | 0.7 |
| 15 | Zheng et al. [17] (TA)_Day5 | 5.0 | 0.96 | 0.28 | 5.0 | 0.87 | 0.19 |
| 16 | Lee et al. [18] (QUA)_TC | 4.0 | 1 | 1.342 | 4.0 | 7.53 | 12.947 |
| 17 | Lee et al. [18] (QUA)_A5 | 4.0 | 1 | 1.342 | 4.0 | 2.49 | 4.025 |
| 18 | Lee et al. [18] (QUA)_A10 | 4.0 | 1 | 1.342 | 4.0 | 1.69 | 2.214 |
| 19 | Keeble et al. [19] (QUA)_6 h | 6.0 | 0.96 | 0.32 | 6.0 | 0.82 | 0.54 |
| 20 | Keeble et al. [19] (QUA)_1 d | 6.0 | 1.05 | 0.98 | 6.0 | 1.66 | 0.52 |
| 21 | Keeble et al. [19] (QUA)_3 d | 6.0 | 1 | 0.74 | 6.0 | 3.33 | 2.75 |
| 22 | Keeble et al. [19] (QUA)_7 d | 6.0 | 1.07 | 1.61 | 6.0 | 1.26 | 2.39 |
| 23 | Ozaki et al. [20] (GA)_DEX | 4.0 | 1.06 | 0.563 | 4.0 | 2.92 | 3.87 |
| 24 | Rupert et al. [21] (QUA)_KPC | 4.0 | 1.13 | 1.6 | 4.0 | 19.95 | 7.77 |
| 25 | Rupert et al. [21] (QUA)_KPC IL | 4.0 | 1.13 | 1.6 | 4.0 | 1.44 | 1.14 |
| 26 | Sun et al. [22] 14 (TA) | 9.0 | 1 | 0.09 | 9.0 | 3.72 | 0.67 |
| 27 | Polge et al. [23] (TA)_DEX 9 | 4.5 | 1.73 | 3.649 | 4.5 | 4.23 | 4.582 |
| 28 | Polge et al. [23] (TA)_DEX 14 | 4.5 | 1.73 | 3.649 | 4.5 | 3.14 | 3.967 |
| 29 | Hwee et al. [24] (GA) | 5.0 | 1.37 | 2.08 | 5.0 | 0.05 | 0.1 |
| 30 | Zheng et al. [25] (GA) | 12.0 | 1.003 | 0.26 | 12.0 | 1.627 | 0.197 |
| 31 | Chen et al. [26] (GA) | 3.0 | 0.96 | 0.846 | 3.0 | 3.45 | 3.4 |
| 32 | Saul et al. [27] (QUA) | 8.0 | 3.11 | 1.131 | 8.0 | 16.14 | 13.096 |
| 33 | Moarbes et al [28] (TA)_24 h | 8.0 | 0.86 | 0 | 8.0 | 20.64 | 7.863 |
| 34 | Moarbes et al [28] (TA)_48 h | 8.0 | 0.86 | 0 | 8.0 | 7.14 | 2.63 |
| 35 | Moarbes et al [28] (TA)_96 h | 8.0 | 0.86 | 0 | 8.0 | 12.5 | 4.299 |
| 36 | Chen et al. [29] (GA) | 6.0 | 1.82 | 0.4 | 6.0 | 17.56 | 3.8 |
| 37 | Hirata et al. [30] (TA) | 4.0 | 18.79 | 0.8 | 4.0 | 24.41 | 1.98 |
| 38 | Giordano et al. [31] (DIA) | 5.0 | 1 | 0.54 | 5.0 | 1.05 | 29.695 |
| 39 | de Theije et al. [32] (GA) | 8.0 | 8.6 | 1.499 | 8.0 | 19.38 | 310.005 |
| 40 | Kawanishi et al. [33] (GA) | 5.5 | 2.05 | 0.61 | 5.5 | 9.26 | 4.644 |
| 41 | Ono et al. [34] (GA) | 10.0 | 7.21 | 3.131 | 10.0 | 24.01 | 3.763 |
| 42 | Jaitovich et al. [35] (SOL) | 4.5 | 3.44 | 0 | 4.5 | 15.74 | 7.573 |
| 43 | Files et al. [36] (EDL) | 5.0 | 1.06 | 5.188 | 5.0 | 13.23 | 10.196 |
| 44 | Zhang et al. [37] (GA) | 8.0 | 3.24 | 0.33 | 8.0 | 22.75 | 4.1 |
| 45 | Cunha et al. [38] (PLA) | 6.0 | 8.76 | 0 | 6.0 | 11.64 | 3.552 |
| 46 | Haegens et al. [39] (DIA) | 6.0 | 2.32 | 1.12 | 6.0 | 12.44 | 8.2 |
| 47 | Onda et al. [40] (SOL) | 7.0 | 5.49 | 2.275 | 7.0 | 13.36 | 5.424 |
| 48 | Ibebunjo et al. [41] (TRI) | 6.0 | 5.56 | 0.637 | 6.0 | 12.54 | 1.127 |
References
- Kotler, D.P. Cachexia. Ann. Intern. Med. 2000, 133, 622–634. [Google Scholar] [CrossRef] [PubMed]
- Ciani, L.; Salinas, P.C. WNTs in the Vertebrate Nervous System: From Patterning to Neuronal Connectivity. Nat. Rev. Neurosci. 2005, 6, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Gouvêa, A.L.; Gracindo Silva, M.; Cabral, B.; Martinez, C.G.; Lauthartte, L.C.; Rodrigues Bastos, W.; Kurtenbach, E. Progressive Resistance Exercise Prevents Muscle Strength Loss Due to Muscle Atrophy Induced by Methylmercury Systemic Intoxication. JCSM Clin. Rep. 2021, 6, 80–92. [Google Scholar] [CrossRef]
- Pacifico, J.; Reijnierse, E.M.; Lim, W.K.; Maier, A.B. The Association between Sarcopenia as a Comorbid Disease and Incidence of Institutionalisation and Mortality in Geriatric Rehabilitation Inpatients: REStORing Health of Acutely Unwell adulTs (RESORT). Gerontology 2022, 68, 498–508. [Google Scholar] [CrossRef]
- Yin, L.; Li, N.; Jia, W.; Wang, N.; Liang, M.; Yang, X.; Du, G. Skeletal Muscle Atrophy: From Mechanisms to Treatments. Pharmacol. Res. 2021, 172, 105807. [Google Scholar] [CrossRef]
- Schiaffino, S.; Dyar, K.A.; Ciciliot, S.; Blaauw, B.; Sandri, M. Mechanisms Regulating Skeletal Muscle Growth and Atrophy. FEBS J. 2013, 280, 4294–4314. [Google Scholar] [CrossRef]
- Bodine, S.C.; Latres, E.; Baumhueter, S.; Lai, V.K.; Nunez, L.; Clarke, B.A.; Poueymirou, W.T.; Panaro, F.J.; Na, E.; Dharmarajan, K.; et al. Identification of Ubiquitin Ligases Required for Skeletal Muscle Atrophy. Science 2001, 294, 1704–1708. [Google Scholar] [CrossRef]
- Glickman, M.H.; Ciechanover, A. The Ubiquitin-Proteasome Proteolytic Pathway: Destruction for the Sake of Construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef]
- Amerik, A.Y.; Hochstrasser, M. Mechanism and Function of Deubiquitinating Enzymes. Biochim. Biophys. Acta 2004, 1695, 189–207. [Google Scholar] [CrossRef]
- Peris-Moreno, D.; Cussonneau, L.; Combaret, L.; Polge, C.; Taillandier, D. Ubiquitin Ligases at the Heart of Skeletal Muscle Atrophy Control. Molecules 2021, 26, 407. [Google Scholar] [CrossRef]
- Fielitz, J.; Kim, M.-S.; Shelton, J.M.; Latif, S.; Spencer, J.A.; Glass, D.J.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. Myosin Accumulation and Striated Muscle Myopathy Result from the Loss of Muscle RING Finger 1 and 3. J. Clin. Investig. 2007, 117, 2486–2495. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; Moher, D.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. PRISMA 2020 explanation and elaboration: Updated guidance and exemplars for reporting systematic reviews. BMJ 2021, 372, n160. [Google Scholar] [CrossRef] [PubMed]
- Hillege, M.M.G.; Shi, A.; Galli, R.A.; Wu, G.; Bertolino, P.; Hoogaars, W.M.H.; Jaspers, R.T. Lack of Tgfbr1 and Acvr1b Synergistically Stimulates Myofibre Hypertrophy and Accelerates Muscle Regeneration. elife 2022, 11, e77610. [Google Scholar] [CrossRef]
- Niida, Y.; Masuda, M.; Adachi, Y.; Yoshizawa, A.; Ohminami, H.; Mori, Y.; Ohnishi, K.; Yamanaka-Okumura, H.; Uchida, T.; Nikawa, T.; et al. Reduction of Stearoyl-CoA Desaturase (SCD) Contributes Muscle Atrophy through the Excess Endoplasmic Reticulum Stress in Chronic Kidney Disease. J. Clin. Biochem. Nutr. 2020, 67, 179–187. [Google Scholar] [CrossRef]
- Xu, M.; Liu, X.; Bao, P.; Wang, Y.J.; Lu, J.; Liu, Y.J. H2S Protects against Immobilization-Induced Muscle Atrophy via Reducing Oxidative Stress and Inflammation. Front. Physiol. 2022, 13, 844539. [Google Scholar] [CrossRef]
- Li, J.; Yi, X.; Yao, Z.; Chakkalakal, J.V.; Xing, L.; Boyce, B.F. TNF Receptor-Associated Factor 6 Mediates TNFα-Induced Skeletal Muscle Atrophy in Mice during Aging: TRAF6 Induces Muscle Loss during Aging. J. Bone Miner. Res. 2020, 35, 1535–1548. [Google Scholar] [CrossRef]
- Zheng, Y.; Dai, H.; Chen, R.; Zhong, Y.; Zhou, C.; Wang, Y.; Zhan, C.; Luo, J. Endoplasmic Reticulum Stress Promotes Sepsis-Induced Muscle Atrophy via Activation of STAT3 and Smad3. J. Cell. Physiol. 2023, 238, 582–596. [Google Scholar] [CrossRef]
- Lee, H.; Heo, J.-W.; Kim, A.-R.; Kweon, M.; Nam, S.; Lim, J.-S.; Sung, M.-K.; Kim, S.-E.; Ryu, J.-H. Z-Ajoene from Crushed Garlic Alleviates Cancer-Induced Skeletal Muscle Atrophy. Nutrients 2019, 11, 2724. [Google Scholar] [CrossRef]
- Keeble, A.R.; Brightwell, C.R.; Latham, C.M.; Thomas, N.T.; Mobley, C.B.; Murach, K.A.; Johnson, D.L.; Noehren, B.; Fry, C.S. Depressed Protein Synthesis and Anabolic Signaling Potentiate ACL Tear-Resultant Quadriceps Atrophy. Am. J. Sports Med. 2023, 51, 81–96. [Google Scholar] [CrossRef]
- Ozaki, Y.; Ohashi, K.; Otaka, N.; Ogawa, H.; Kawanishi, H.; Takikawa, T.; Fang, L.; Tatsumi, M.; Takefuji, M.; Enomoto, T.; et al. Neuron-Derived Neurotrophic Factor Protects against Dexamethasone-Induced Skeletal Muscle Atrophy. Biochem. Biophys. Res. Commun. 2022, 593, 5–12. [Google Scholar] [CrossRef]
- Rupert, J.E.; Narasimhan, A.; Jengelley, D.H.A.; Jiang, Y.; Liu, J.; Au, E.; Silverman, L.M.; Sandusky, G.; Bonetto, A.; Cao, S.; et al. Tumor-Derived IL-6 and Trans-Signaling among Tumor, Fat, and Muscle Mediate Pancreatic Cancer Cachexia. J. Exp. Med. 2021, 218, e20190450. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Gong, Y.; Qiu, J.; Chen, Y.; Ding, F.; Zhao, Q. TRAF6 Inhibition Rescues Dexamethasone-Induced Muscle Atrophy. Int. J. Mol. Sci. 2014, 15, 11126–11141. [Google Scholar] [CrossRef] [PubMed]
- Polge, C.; Aniort, J.; Armani, A.; Claustre, A.; Coudy-Gandilhon, C.; Tournebize, C.; Deval, C.; Combaret, L.; Béchet, D.; Sandri, M.; et al. UBE2E1 Is Preferentially Expressed in the Cytoplasm of Slow-Twitch Fibers and Protects Skeletal Muscles from Exacerbated Atrophy upon Dexamethasone Treatment. Cells 2018, 7, 214. [Google Scholar] [CrossRef]
- Hwee, D.T.; Baehr, L.M.; Philp, A.; Baar, K.; Bodine, S.C. Maintenance of Muscle Mass and Load-Induced Growth in Muscle RING Finger 1 Null Mice with Age. Aging Cell 2014, 13, 92–101. [Google Scholar] [CrossRef]
- Zheng, J.; Gao, J.; Zhang, Q.; Wu, X.; Shen, W.; Sun, M. Sirtuin 3 Deficiency Accelerates Angiotensin II-Induced Skeletal Muscle Atrophy. Connect. Tissue Res. 2020, 61, 586–593. [Google Scholar] [CrossRef]
- Chen, L.; Xu, W.; Yang, Q.; Zhang, H.; Wan, L.; Xin, B.; Zhang, J.; Guo, C. Imperatorin Alleviates Cancer Cachexia and Prevents Muscle Wasting via Directly Inhibiting STAT3. Pharmacol. Res. 2020, 158, 104871. [Google Scholar] [CrossRef]
- Saul, D.; Kosinsky, R.L. Dextran Sodium Sulfate-Induced Colitis as a Model for Sarcopenia in Mice. Inflamm. Bowel Dis. 2020, 26, 56–65. [Google Scholar] [CrossRef]
- Moarbes, V.; Mayaki, D.; Huck, L.; Leblanc, P.; Vassilakopoulos, T.; Petrof, B.J.; Hussain, S.N.A. Differential Regulation of Myofibrillar Proteins in Skeletal Muscles of Septic Mice. Physiol. Rep. 2019, 7, e14248. [Google Scholar] [CrossRef]
- Chen, L.; Chen, L.; Wan, L.; Huo, Y.; Huang, J.; Li, J.; Lu, J.; Xin, B.; Yang, Q.; Guo, C. Matrine Improves Skeletal Muscle Atrophy by Inhibiting E3 Ubiquitin Ligases and Activating the Akt/mTOR/FoxO3α Signaling Pathway in C2C12 Myotubes and Mice. Oncol. Rep. 2019, 42, 479–494. [Google Scholar] [CrossRef]
- Hirata, Y.; Nomura, K.; Senga, Y.; Okada, Y.; Kobayashi, K.; Okamoto, S.; Minokoshi, Y.; Imamura, M.; Takeda, S.; Hosooka, T.; et al. Hyperglycemia Induces Skeletal Muscle Atrophy via a WWP1/KLF15 Axis. JCI Insight 2019, 4, e124952. [Google Scholar] [CrossRef]
- Giordano, C.; Lemaire, C.; Li, T.; Kimoff, R.J.; Petrof, B.J. Autophagy-Associated Atrophy and Metabolic Remodeling of the Mouse Diaphragm after Short-Term Intermittent Hypoxia. PLoS ONE 2015, 10, e0131068. [Google Scholar] [CrossRef] [PubMed]
- de Theije, C.C.; Schols, A.M.W.J.; Lamers, W.H.; Ceelen, J.J.M.; van Gorp, R.H.; Hermans, J.J.R.; Köhler, S.E.; Langen, R.C.J. Glucocorticoid Receptor Signaling Impairs Protein Turnover Regulation in Hypoxia-Induced Muscle Atrophy in Male Mice. Endocrinology 2018, 159, 519–534. [Google Scholar] [CrossRef] [PubMed]
- Kawanishi, N.; Nozaki, R.; Naito, H.; Machida, S. TLR4-Defective (C3H/HeJ) Mice Are Not Protected from Cast Immobilization-Induced Muscle Atrophy. Physiol. Rep. 2017, 5, e13255. [Google Scholar] [CrossRef] [PubMed]
- Ono, T.; Takada, S.; Kinugawa, S.; Tsutsui, H. Curcumin Ameliorates Skeletal Muscle Atrophy in Type 1 Diabetic Mice by Inhibiting Protein Ubiquitination: Curcumin and Diabetic Skeletal Muscle Atrophy. Exp. Physiol. 2015, 100, 1052–1063. [Google Scholar] [CrossRef]
- Jaitovich, A.; Angulo, M.; Lecuona, E.; Dada, L.A.; Welch, L.C.; Cheng, Y.; Gusarova, G.; Ceco, E.; Liu, C.; Shigemura, M.; et al. High CO2 Levels Cause Skeletal Muscle Atrophy via AMP-Activated Kinase (AMPK), FoxO3a Protein, and Muscle-Specific Ring Finger Protein 1 (MuRF1). J. Biol. Chem. 2015, 290, 9183–9194. [Google Scholar] [CrossRef]
- Files, D.C.; Xiao, K.; Zhang, T.; Liu, C.; Qian, J.; Zhao, W.; Morris, P.E.; Delbono, O.; Feng, X. The Posterior Cricoarytenoid Muscle Is Spared from MuRF1-Mediated Muscle Atrophy in Mice with Acute Lung Injury. PLoS ONE 2014, 9, e87587. [Google Scholar] [CrossRef][Green Version]
- Zhang, L.; Tang, H.; Kou, Y.; Li, R.; Zheng, Y.; Wang, Q.; Zhou, X.; Jin, L. MG132-Mediated Inhibition of the Ubiquitin-Proteasome Pathway Ameliorates Cancer Cachexia. J. Cancer Res. Clin. Oncol. 2013, 139, 1105–1115. [Google Scholar] [CrossRef]
- Cunha, T.F.; Bacurau, A.V.N.; Moreira, J.B.N.; Paixão, N.A.; Campos, J.C.; Ferreira, J.C.B.; Leal, M.L.; Negrão, C.E.; Moriscot, A.S.; Wisløff, U.; et al. Exercise Training Prevents Oxidative Stress and Ubiquitin-Proteasome System Overactivity and Reverse Skeletal Muscle Atrophy in Heart Failure. PLoS ONE 2012, 7, e41701. [Google Scholar] [CrossRef]
- Haegens, A.; Schols, A.M.; Gorissen, S.H.; van Essen, A.L.; Snepvangers, F.; Gray, D.A.; Shoelson, S.E.; Langen, R.C. NF-κB Activation and Polyubiquitin Conjugation Are Required for Pulmonary Inflammation-Induced Diaphragm Atrophy. Am. J. Physiol. Lung Cell. Mol. Physiol. 2012, 302, L103–L110. [Google Scholar] [CrossRef]
- Onda, A.; Jiao, Q.; Nagano, Y.; Akimoto, T.; Miyamoto, T.; Minamisawa, S.; Fukubayashi, T. Acupuncture Ameliorated Skeletal Muscle Atrophy Induced by Hindlimb Suspension in Mice. Biochem. Biophys. Res. Commun. 2011, 410, 434–439. [Google Scholar] [CrossRef]
- Ibebunjo, C.; Eash, J.K.; Li, C.; Ma, Q.; Glass, D.J. Voluntary Running, Skeletal Muscle Gene Expression, and Signaling Inversely Regulated by Orchidectomy and Testosterone Replacement. Am. J. Physiol. Endocrinol. Metab. 2011, 300, E327–E340. [Google Scholar] [CrossRef] [PubMed]
- Egger, M.; Davey Smith, G.; Schneider, M.; Minder, C. Bias in Meta-Analysis Detected by a Simple, Graphical Test. BMJ 1997, 315, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Study Quality Assessment Tools Disponível Em. Available online: https://www.nhlbi.nih.gov/health-pro/guidelines/in-develop/cardiovascular-risk-reduction/tools/cohort (accessed on 13 March 2024).
- Cochran, W.G. The combination of estimates from different experiments. Biometrics 1954, 10, 101. [Google Scholar] [CrossRef]
- Higgins, J.P.T.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring Inconsistency in Meta-Analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef]
- Taskin, S.; Stumpf, V.I.; Bachmann, J.; Weber, C.; Martignoni, M.E.; Friedrich, O. Motor Protein Function in Skeletal Abdominal Muscle of Cachectic Cancer Patients. J. Cell. Mol. Med. 2014, 18, 69–79. [Google Scholar] [CrossRef]
- Bonetto, A.; Andersson, D.C.; Waning, D.L. Assessment of Muscle Mass and Strength in Mice. BoneKEy Rep. 2015, 4, 732. [Google Scholar] [CrossRef]
- Akagi, R.; Takai, Y.; Ohta, M.; Kanehisa, H.; Kawakami, Y.; Fukunaga, T. Muscle Volume Compared to Cross-Sectional Area Is More Appropriate for Evaluating Muscle Strength in Young and Elderly Individuals. Age Ageing 2009, 38, 564–569. [Google Scholar] [CrossRef]
- Ceglia, L.; Niramitmahapanya, S.; Price, L.L.; Harris, S.S.; Fielding, R.A.; Dawson-Hughes, B. An Evaluation of the Reliability of Muscle Fiber Cross-Sectional Area and Fiber Number Measurements in Rat Skeletal Muscle. Biol. Proced. Online 2013, 15, 6. [Google Scholar] [CrossRef]
- Reggiani, C.; Schiaffino, S. Muscle hypertrophy and muscle strength: Dependent or independent variables? A provocative review. Eur. J. Transl. Myol. 2020, 30, 9311. [Google Scholar] [CrossRef]
- Jones, E.J.; Bishop, P.A.; Woods, A.K.; Green, J.M. Cross-Sectional Area and Muscular Strength: A Brief Review: A Brief Review. Sports Med. 2008, 38, 987–994. [Google Scholar] [CrossRef]
- Lord, S.O.; Dawson, P.W.J.; Chunthorng-Orn, J.; Ng, J.; Baehr, L.M.; Hughes, D.C.; Sridhar, P.; Knowles, T.; Bodine, S.C.; Lai, Y.-C. Uncovering the Mechanisms of MuRF1-Induced Ubiquitylation and Revealing Similarities with MuRF2 and MuRF3. Biochem. Biophys. Rep. 2024, 37, 101636. [Google Scholar] [CrossRef]
- Kitajima, Y.; Yoshioka, K.; Suzuki, N. The Ubiquitin–Proteasome System in Regulation of the Skeletal Muscle Homeostasis and Atrophy: From Basic Science to Disorders. J. Physiol. Sci. 2020, 70, 40. [Google Scholar] [CrossRef] [PubMed]
- Sterne, J.A.C.; Sutton, A.J.; Ioannidis, J.P.A.; Terrin, N.; Jones, D.R.; Lau, J.; Carpenter, J.; Rücker, G.; Harbord, R.M.; Schmid, C.H.; et al. Recommendations for Examining and Interpreting Funnel Plot Asymmetry in Meta-Analyses of Randomised Controlled Trials. BMJ 2011, 343, d4002. [Google Scholar] [CrossRef] [PubMed]
- Foletta, V.C.; White, L.J.; Larsen, A.E.; Léger, B.; Russell, A.P. The Role and Regulation of MAFbx/Atrogin-1 and MuRF1 in Skeletal Muscle Atrophy. Pflug. Arch. 2011, 461, 325–335. [Google Scholar] [CrossRef]
- Sandri, M.; Sandri, C.; Gilbert, A.; Skurk, C.; Calabria, E.; Picard, A.; Walsh, K.; Schiaffino, S.; Lecker, S.H.; Goldberg, A.L. Foxo Transcription Factors Induce the Atrophy-Related Ubiquitin Ligase Atrogin-1 and Cause Skeletal Muscle Atrophy. Cell 2004, 117, 399–412. [Google Scholar] [CrossRef]
- Stitt, T.N.; Drujan, D.; Clarke, B.A.; Panaro, F.; Timofeyva, Y.; Kline, W.O.; Gonzalez, M.; Yancopoulos, G.D.; Glass, D.J. The IGF-1/PI3K/Akt Pathway Prevents Expression of Muscle Atrophy-Induced Ubiquitin Ligases by Inhibiting FOXO Transcription Factors. Mol. Cell 2004, 14, 395–403. [Google Scholar] [CrossRef]
- Bonuccelli, G.; Sotgia, F.; Capozza, F.; Gazzerro, E.; Minetti, C.; Lisanti, M.P. Localized Treatment with a Novel FDA-Approved Proteasome Inhibitor Blocks the Degradation of Dystrophin and Dystrophin-Associated Proteins in Mdx Mice. Cell Cycle 2007, 6, 1242–1248. [Google Scholar] [CrossRef]
- Assereto, S.; Stringara, S.; Sotgia, F.; Bonuccelli, G.; Broccolini, A.; Pedemonte, M.; Traverso, M.; Biancheri, R.; Zara, F.; Bruno, C.; et al. Pharmacological Rescue of the Dystrophin-Glycoprotein Complex in Duchenne and Becker Skeletal Muscle Explants by Proteasome Inhibitor Treatment. Am. J. Physiol. Cell Physiol. 2006, 290, C577–C582. [Google Scholar] [CrossRef][Green Version]
- Bonuccelli, G.; Sotgia, F.; Schubert, W.; Park, D.S.; Frank, P.G.; Woodman, S.E.; Insabato, L.; Cammer, M.; Minetti, C.; Lisanti, M.P. Proteasome Inhibitor (MG-132) Treatment of Mdx Mice Rescues the Expression and Membrane Localization of Dystrophin and Dystrophin-Associated Proteins. Am. J. Pathol. 2003, 163, 1663–1675. [Google Scholar] [CrossRef]
- Selsby, J.; Morris, C.; Morris, L.; Sweeney, L. A proteasome inhibitor fails to attenuate dystrophic pathology in mdx mice. PLoS Curr. 2012, 4, e4f84a944d8930. [Google Scholar] [CrossRef]
- Adams, V.; Gußen, V.; Zozulya, S.; Cruz, A.; Moriscot, A.; Linke, A.; Labeit, S. Small-Molecule Chemical Knockdown of MuRF1 in Melanoma Bearing Mice Attenuates Tumor Cachexia Associated Myopathy. Cells 2020, 9, 2272. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y. E3 Ubiquitin Ligases as Cancer Targets and Biomarkers. Neoplasia 2006, 8, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Landré, V.; Rotblat, B.; Melino, S.; Bernassola, F.; Melino, G. Screening for E3-Ubiquitin Ligase Inhibitors: Challenges and Opportunities. Oncotarget 2014, 5, 7988–8013. [Google Scholar] [CrossRef]
- Bowen, T.S.; Adams, V.; Werner, S.; Fischer, T.; Vinke, P.; Brogger, M.N.; Mangner, N.; Linke, A.; Sehr, P.; Lewis, J.; et al. Small-molecule Inhibition of MuRF1 Attenuates Skeletal Muscle Atrophy and Dysfunction in Cardiac Cachexia: Inhibition of MuRF1 Prevents Skeletal Muscle Wasting. J. Cachexia Sarcopenia Muscle 2017, 8, 939–953. [Google Scholar] [CrossRef]

| Title | |||
|---|---|---|---|
| Title | 1 | Identify the report as a systematic review. | Lines 2 to 3 |
| Abstract | |||
| Abstract | 2 | See the PRISMA 2020 for Abstracts checklist. | Lines 10 to 25 |
| Introduction | |||
| Rationale | 3 | Describe the rationale for the review in the context of existing knowledge. | Lines 49 to 73 |
| Objectives | 4 | Provide an explicit statement of the objective(s) or question(s) the review addresses. | Lines 74 to 79 |
| Methods | |||
| Eligibility criteria | 5 | Specify the inclusion and exclusion criteria for the review and how studies were grouped for the syntheses. | Lines 85 to 98 |
| Information sources | 6 | Specify all databases, registers, websites, organisations, reference lists and other sources searched or consulted to identify studies. Specify the date when each source was last searched or consulted. | Lines 87 to 89 |
| Search strategy | 7 | Present the full search strategies for all databases, registers and websites, including any filters and limits used. | Lines 100 to 102 |
| Selection process | 8 | Specify the methods used to decide whether a study met the inclusion criteria of the review, including how many reviewers screened each record and each report retrieved, whether they worked independently, and if applicable, details of automation tools used in the process. | Lines 103 to 115 |
| Data collection process | 9 | Specify the methods used to collect data from reports, including how many reviewers collected data from each report, whether they worked independently, any processes for obtaining or confirming data from study investigators, and if applicable, details of automation tools used in the process. | Lines 121 to 125 |
| Data items | 10a | List and define all outcomes for which data were sought. Specify whether all results that were compatible with each outcome domain in each study were sought (e.g., for all measures, time points, analyses), and if not, the methods used to decide which results to collect. | Lines 110 to 114 |
| 10b | List and define all other variables for which data were sought (e.g., participant and intervention characteristics, funding sources). Describe any assumptions made about any missing or unclear information. | Lines 90 to 98 | |
| Study risk of bias assessment | 11 | Specify the methods used to assess risk of bias in the included studies, including details of the tool(s) used, how many reviewers assessed each study, and whether they worked independently, and if applicable, details of automation tools used in the process. | Lines 126 to 129 |
| Effect measures | 12 | Specify for each outcome the effect measure(s) (e.g., risk ratio, mean difference) used in the synthesis or presentation of results. | Lines 138 to 142 |
| Synthesis methods | 13a | Describe the processes used to decide which studies were eligible for each synthesis (e.g., tabulating the study intervention characteristics and comparing against the planned groups for each synthesis (item #5)). | Lines 85 to 120 |
| 13b | Describe any methods required to prepare the data for presentation or synthesis, such as the handling of missing summary statistics, or data conversions. | Lines 122 to 125 | |
| 13c | Describe any methods used to tabulate or visually display results of individual studies and syntheses. | Lines 122 to 125 | |
| 13d | Describe any methods used to synthesize results and provide a rationale for the choice(s). If meta-analysis was performed, describe the model(s), method(s) to identify the presence and extent of statistical heterogeneity, and software package(s) used. | Lines 138 to 142 | |
| 13e | Describe any methods used to explore possible causes of heterogeneity among study results (e.g., subgroup analysis, meta-regression). | Lines 131 to 136 | |
| 13f | Describe any sensitivity analyses conducted to assess robustness of the synthesized results. | Lines 143 to 146 | |
| Reporting bias assessment | 14 | Describe any methods used to assess risk of bias due to missing results in a synthesis (arising from reporting biases). | Lines 126 to 129 |
| Certainty assessment | 15 | Describe any methods used to assess certainty (or confidence) in the body of evidence for an outcome. | Lines 137 to 142 |
| Results | |||
| Study selection | 16a | Describe the results of the search and selection process, from the number of records identified in the search to the number of studies included in the review, ideally using a flow diagram. | Lines 108 to 120 and Section 2.3 |
| 16b | Cite studies that might appear to meet the inclusion criteria, but which were excluded, and explain why they were excluded. | Lines 110 to 114 | |
| Study characteristics | 17 | Cite each included study and present its characteristics. | Lines 152 to 165 and Section 3.1 |
| Risk of bias in studies | 18 | Present assessments of risk of bias for each included study. | Lines 208 to 218 and Section 3.2 |
| Results of individual studies | 19 | For all outcomes, present, for each study (a) summary statistics for each group (where appropriate) and (b) an effect estimate and its precision (e.g., confidence/credible interval), ideally using structured tables or plots. | Lines 176 to 207 and Section 3.2 |
| Results of syntheses | 20a | For each synthesis, briefly summarise the characteristics and risk of bias among contributing studies. | Lines 167 to 175 and Figure A1B |
| 20b | Present results of all statistical syntheses conducted. If meta-analysis was performed, present for each the summary estimate and its precision (e.g., confidence/credible interval) and measures of statistical heterogeneity. If comparing groups, describe the direction of the effect. | Lines 113 to 124 and Figure A1A | |
| 20c | Present results of all investigations of possible causes of heterogeneity among study results. | Lines 167 to 173 and Figure A1A | |
| 20d | Present results of all sensitivity analyses conducted to assess the robustness of the synthesized results. | Lines 167 to 173 Figure A1A | |
| Reporting biases | 21 | Present assessments of risk of bias due to missing results (arising from reporting biases) for each synthesis assessed. | Lines 173 to 175 Figure A1B |
| Certainty of evidence | 22 | Present assessments of certainty (or confidence) in the body of evidence for each outcome assessed. | Lines 176 to 207 Section 2.3 |
| Discussion | |||
| Discussion | 23a | Provide a general interpretation of the results in the context of other evidence. | Lines 230 to 243 |
| 23b | Discuss any limitations of the evidence included in the review. | Lines 192 to 194 | |
| 23c | Discuss any limitations of the review processes used. | Lines 258 to 263 and 281 to 283. | |
| 23d | Discuss implications of the results for practice, policy, and future research. | Lines 272 to 278 | |
| Other Information | |||
| Registration and protocol | 24a | Provide registration information for the review, including the registered name and registration number (or state that the review was not registered). | not registered |
| 24b | Indicate where the review protocol can be accessed, or state that a protocol was not prepared. | not registered | |
| 24c | Describe and explain any amendments to information provided at registration or in the protocol. | not registered | |
| Support | 25 | Describe sources of financial or non-financial support for the review, and the role of the funders or sponsors in the review. | Lines 340 to 342 |
| Competing interests | 26 | Declare any competing interests of review authors. | Line 351 to 352 |
| Availability of data, code and other materials | 27 | Report which of the following are publicly available and where they can be found: template data collection forms; data extracted from included studies; data used for all analyses; analytic code; any other materials used in the review. | Upon request of the reviewer |
| No. | Study | n | Age (wks) | Muscle Type | Muscle Atrophy Protocol | Time Course (Days) | E3 Ligases Modulation (Times) | CSA (%) | |
|---|---|---|---|---|---|---|---|---|---|
| MAFbx (Fbxo32) | MuRF1 (Trim63) | ||||||||
| 1 | Polge et al. [23] (DEX 9) | 4 | 20 | TA | Dexamethasone treatment | 9 | ↑ 2.7× | ↑ 2.4× | ↓ 26.8 |
| Polge et al. [23] (DEX 14) | 4 | 14 | ↑ 2.6× | ↑ 1.8× | ↓ 22.4 | ||||
| 2 | Chen et al. [26] | 6 | 4 | GAS | Cancer cachexia | 21 | ↑ 13.6× | ↑ 9.7× | ↓ 54.4 |
| 3 | Chen et al. [29] | 3 | 6–8 | GAS | Cancer cachexia | 15 | ↑ 3.9× | ↑ 3.6× | ↓ 58.5 |
| 4 | Cunha et al. [38] | 6 | 28 | PLA | Heart failure | 210 | ↑ 1.6× | ↑ 1.3× | ↓ 29.8 |
| 5 | de Theije et al. [32] | 8 | 20 | GAS | Hypoxia | 21 | ↑ 3.3× | ↑ 2.3× | ↓ 14.9 |
| 6 | Files et al. [36] | 5 | 8 | EDL | Lung injury | 3 | ↑ 16.3× | ↑ 12.5× | ↓ 23.8 |
| 7 | Giordano et al. [31] | 5 | 8 | DIA | Hypoxia | 4 | ↑ 1.2× | ↑ 1.1× | ↓ 28.9 |
| 8 | Haegens et al. [39] | 6 | 20 | DIA | Lung inflammation | 1 | ↑ 2.6× | ↑ 5.4× | ↓ 17.4 |
| 9 | Hillege et al. [13] (Tgfbr1KO) | 6 | 6 | TA | Cardiotoxin-induced injury | 4 | ↑ 1× | ↑ 1× | ↓ 30.6 |
| Hillege et al. [13] (Acvr1bKO) | 6 | ↑ 1× | ↑ 1× | ↓ 22.0 | |||||
| Hillege et al. [13] (Acvr:TgfbKO) | 6 | ↑ 3.3× | ↑ 1.5× | ↑ 25.7 | |||||
| 10 | Hirata et al. [30] | 4 | 10 | TA | Diabetes mellitus | 21 | ↑ 1.3× | ↑ 1.3× | ↓ 38.8 |
| 11 | Hwee et al. [24] | 5 | 26 to 100 | GAS | Denervation | 7–14 | ↑ 1.8× | ↑ 0.04× | ↑ 39.6 |
| 12 | Ibebunjo et al. [41] | 6 | 16 | TRI | Orchidectomy | 28 | ↑ 1× | ↑ 2.3× | ↓ 13.2 |
| 13 | Jaitovich et al. [35] | 4 | 14–16 | SOL | Denervation and dexamethasone treatment | 21 | - | ↑ 4.6× | ↓ 41.2 |
| 14 | Kawanishi et al. [33] | 6 | 10 | GAS | Immobilization | 14 | ↑ 6.3× | ↑ 4.5× | ↓ 40 |
| 15 | Keeble et al. [19] (6 h) | 6 | 14–21 | QUA | Anterior cruciate ligament tear injury | 0.25 | ↑ 0.9× | ↑ 0.9× | ↑ 4.5 |
| Keeble et al. [19] (1 d) | 6 | 1 | ↑ 1.6× | ↑ 1.6× | ↑ 5.9 | ||||
| Keeble et al. [19] (3 d) | 6 | 3 | ↑ 2× | ↑ 3.3× | ↓ 10.3 | ||||
| Keeble et al. [19] (7 d) | 6 | 7 | ↑ 0.9× | ↑ 1.2× | ↓ 8.8 | ||||
| 16 | Lee et al. [18] (TC) | 4 | 6 | QUA | Cancer cachexia | 15 | ↑ 5× | ↑ 7.5× | ↓ 18 |
| Lee et al. [18] (A5) | 4 | ↑ 1.8× | ↑ 2.5× | ↓ 5.1 | |||||
| Lee et al. [18] (A10) | 4 | ↑ 1.3× | ↑ 1.7× | ↑ 18.8 | |||||
| 17 | Li et al. [16] (TNF) | 8 | 10 to 77 | TA | Aging | 5 | ↑ 2.1× | ↑ 4.2× | ↓ 60.6 |
| Li et al. [16] (TRAF) | 8 | ↑ 0.9× | ↑ 2.7× | ↓ 8.7 | |||||
| Li et al. [16] (TNF:TRAF) | 8 | ↑ 0.7× | ↑ 1.6× | ↓ 30.3 | |||||
| 18 | Moarbes et al. [28] (24 h) | 8 | 8 | TA | Burn injury | 1 | ↑ 15.8× | ↑ 24× | ↓ 44.8 |
| Moarbes et al. [28] (48 h) | 8 | 2 | ↑ 7.3× | ↑ 8.3× | ↓ 37.4 | ||||
| Moarbes et al. [28] (96 h) | 8 | 4 | ↑ 25× | ↑ 14.5× | ↓ 31.3 | ||||
| 19 | Niida et al. [14] (CKD) | 5 | 8 | GAS | Chronic kidney disease | 42 | ↑ 2.6× | ↑ 3.3× | ↓ 43.7 |
| Niida et al. [14] (CKD + 4PBA) | 5 | ↑ 1.4× | ↑ 1.7× | ↓ 23.4 | |||||
| 20 | Onda et al. [40] | 7 | 8 | SOL | Immobilization | 14 | ↑ 3× | ↑ 2.4× | ↓ 44 |
| 21 | Ono et al. [34] | 10 | 8 to 10 | GAS | Diabetes mellitus | 14 | ↑ 5.3× | ↑ 3.3× | ↓ 40.4 |
| 22 | Ozaki et al. [20] | 4 | 8 to 10 | GAS | Dexamethasone treatment | 14 | ↑ 2.9× | ↑ 2.8× | ↓ 22.6 |
| 23 | Rupert et al. [21] (KPC) | 4 | 8 | QUA | Cancer cachexia | 17 | ↑ 15.6× | ↑ 17.8× | ↓ 32.2 |
| Rupert et al. [21] (KPC IL) | 4 | ↑ 3.2× | ↑ 1.3× | ↑ 4.8 | |||||
| 24 | Saul et al. [27] | 8 | 10 | QUA | Colitis | 14 | ↑ 6.8× | ↑ 5.2× | ↓ 25.9 |
| 25 | Sun et al. [22] | 9 | 29 | TA | Dexamethasone treatment | 14 | ↑ 3.1× | ↑ 3.8× | ↓ 27.1 |
| 26 | Xu et al. [15] (IM(gyy)) | 6 | 10 | TA | Immobilization | 14 | ↑ 6.3× | ↑ 4.6× | ↓ 44 |
| Xu et al. [15] (IM(nahs)) | 6 | ↑ 5.8× | ↑ 4.2× | ↓ 43.6 | |||||
| Xu et al. [15] (IM + GYY) | 6 | ↑ 2.9× | ↑ 2× | ↓ 16.9 | |||||
| Xu et al. [15] (IM + NaHS) | 6 | ↑ 2.2× | ↑ 2.2× | ↓ 17.8 | |||||
| 27 | Zhang et al. [37] | 8 | 6 to 8 | GAS | Cancer cachexia | 19 | ↑ 9.3× | ↑ 7× | ↓ 49.1 |
| 28 | Zheng et al. [17] | 12 | GAS | Angiotensin infusion | ↑ 1.5× | ↑ 1.6× | ↓ 9.1 | ||
| 29 | Zheng et al. [25] (Day 1) | 5 | 8–10 | TA | Sepsis | 1 | ↑ 4.6× | ↑ 4.7× | ↓ 10.5 |
| Zheng et al. [25] (Day 3) | 5 | 3 | ↑ 3.8× | ↑ 3.4× | ↓ 44.8 | ||||
| Zheng et al. [25] (Day 5) | 5 | 7 | ↑ 1.5× | ↑ 0.9× | ↓ 46.2 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Souza, A.L.G.d.; Alves, A.L.R.; Martinez, C.G.; Sousa, J.C.d.; Kurtenbach, E. Biomarkers of Skeletal Muscle Atrophy Based on Atrogenes Evaluation: A Systematic Review and Meta-Analysis Study. Int. J. Mol. Sci. 2025, 26, 3516. https://doi.org/10.3390/ijms26083516
Souza ALGd, Alves ALR, Martinez CG, Sousa JCd, Kurtenbach E. Biomarkers of Skeletal Muscle Atrophy Based on Atrogenes Evaluation: A Systematic Review and Meta-Analysis Study. International Journal of Molecular Sciences. 2025; 26(8):3516. https://doi.org/10.3390/ijms26083516
Chicago/Turabian StyleSouza, André Luiz Gouvêa de, Anna Luisa Rosa Alves, Camila Guerra Martinez, Júlia Costa de Sousa, and Eleonora Kurtenbach. 2025. "Biomarkers of Skeletal Muscle Atrophy Based on Atrogenes Evaluation: A Systematic Review and Meta-Analysis Study" International Journal of Molecular Sciences 26, no. 8: 3516. https://doi.org/10.3390/ijms26083516
APA StyleSouza, A. L. G. d., Alves, A. L. R., Martinez, C. G., Sousa, J. C. d., & Kurtenbach, E. (2025). Biomarkers of Skeletal Muscle Atrophy Based on Atrogenes Evaluation: A Systematic Review and Meta-Analysis Study. International Journal of Molecular Sciences, 26(8), 3516. https://doi.org/10.3390/ijms26083516






