Identification of QTL for Grain Traits and Plant Height Using the Recombinant Inbred Line Population Derived from the Cross of Zhongke 331 × Nongda 399
Abstract
1. Introduction
2. Results
2.1. Phenotypic Evaluation of the RIL Population
2.2. Correlation Between Grain Traits and Plant Height
2.3. Construction of the Genetic Linkage Map
2.4. QTL Mapping
2.4.1. Thousand-Grain Weight (TGW)
2.4.2. Grain Length (GL)
2.4.3. Grain Width (GW)
2.4.4. Plant Height (PH)
2.5. Co-Localization of QTL Controlling Different Traits
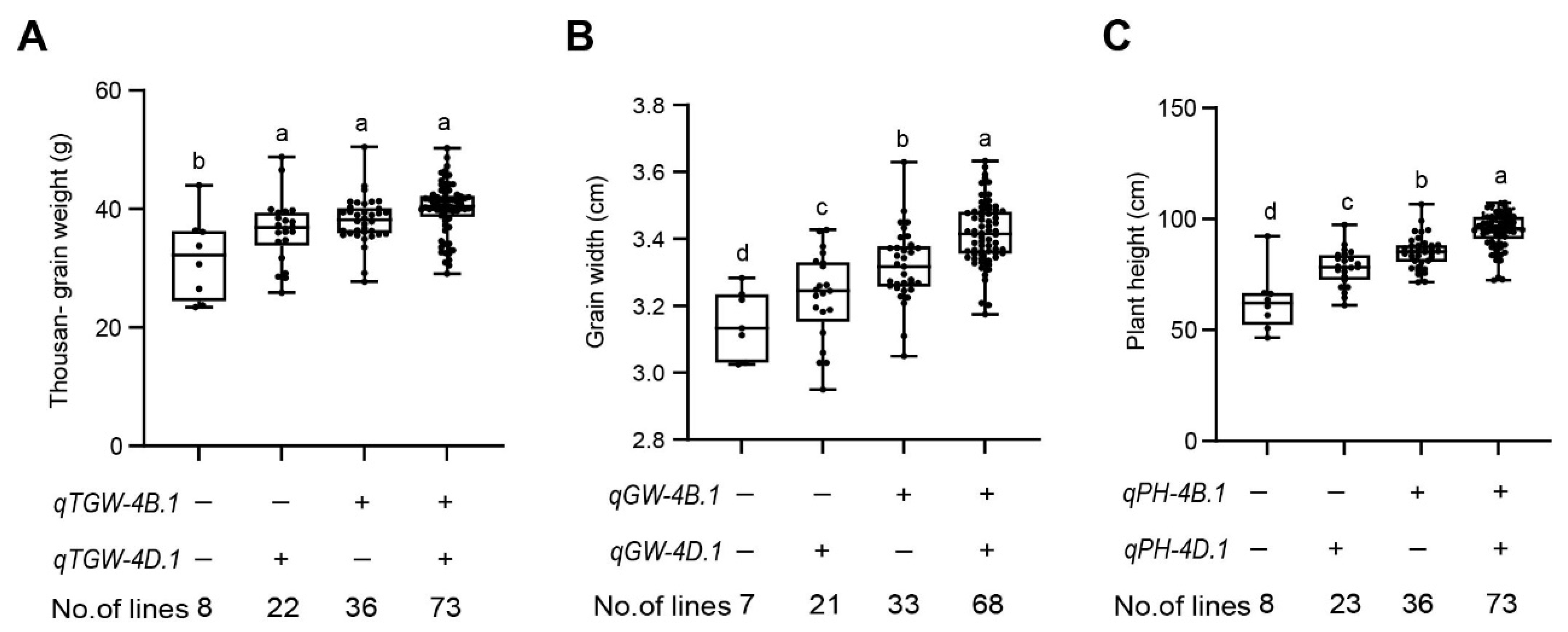
2.6. Analysis of Additive Effects of the Major QTL
2.7. Analysis of the Annotated Genes in the Target Genomic Interval of qTGW/GW/PH-4B.1 and qTGW/GW/PH-4D.1
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Field Experiments and Trait Phenotyping
4.3. Genetic Map Construction and QTL Analysis
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef] [PubMed]
- Molero, G.; Joynson, R.; Pinera-Chavez, F.J.; Gardiner, L.J.; Rivera-Amado, C.; Hall, A.; Reynolds, M.P. Elucidating the genetic basis of biomass accumulation and radiation use efficiency in spring wheat and its role in yield potential. Plant Biotechnol. J. 2019, 17, 1276–1288. [Google Scholar] [CrossRef] [PubMed]
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield trends are insufficient to double global crop production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.K.; Wei, W.; Cui, Q.; Xie, W. The prospects for China’s food security and imports: Will China starve the world via imports? J. Integr. Agric. 2017, 16, 2933–2944. [Google Scholar] [CrossRef]
- Lobell, D.B.; Schlenker, W.; Costa-Roberts, J. Climate trends and global crop production since 1980. Science 2011, 333, 616–620. [Google Scholar] [CrossRef]
- Cai, Y.A.; Zhou, X.A.; Wang, C.Y.; Liu, A.F.; Sun, Z.C.; Li, S.H.; Shi, X.Y.; Yang, S.; Guan, Y.X.; Cheng, J.J.; et al. Quantitative trait loci detection for three tiller-related traits and the effects on wheat (Triticum aestivum L.) yields. Theor. Appl. Genet. 2024, 137, 87. [Google Scholar] [CrossRef]
- Slafer, G.A. Genetic basis of yield as viewed from a crop physiologist’s perspective. Ann. Appl. Biol. 2003, 142, 117–128. [Google Scholar] [CrossRef]
- Gegas, V.C.; Nazari, A.; Griffiths, S.; Simmonds, J.; Fish, L.; Orford, S.; Sayers, L.; Doonan, J.H.; Snape, J.W. A genetic framework for grain size and shape variation in wheat. Plant Cell 2010, 22, 1046–1056. [Google Scholar] [CrossRef]
- Russo, M.A.; Ficco, D.B.M.; Laidò, G.; Marone, D.; Papa, R.; Blanco, A.; Gadaleta, A.; Vita, P.D.; Mastrangelo, A.M. A dense durum wheat × T. dicoccum linkage map based on SNP markers for the study of seed morphology. Mol. Breed. 2014, 34, 1579–1597. [Google Scholar] [CrossRef]
- Kumar, A.; Mantovani, E.E.; Seetan, R.; Soltani, A.; Echeverry-Solaarte, M.; Jain, S.; Simsek, S.; Doehlert, D.; Alamri, M.S.; Elias, E.M.; et al. Dissection of genetic factors underlying wheat kernel shape and size in an elite × nonadapted cross using a high-density SNP linkage map. Plant Genome 2016, 9, 1. [Google Scholar] [CrossRef]
- Wang, X.Q.; Dong, L.H.; Hu, J.M.; Pang, Y.L.; Hu, L.Q.; Xiao, G.L.; Ma, X.; Kong, X.Y.; Jia, J.Z.; Wang, H.W.; et al. Dissecting genetic loci affecting grain morphological traits to improve grain weight via nested association mapping. Theor. Appl. Genet. 2019, 132, 3115–3128. [Google Scholar] [CrossRef] [PubMed]
- Lizana, X.C.; Riegel, R.; Gomez, L.D.; Herrera, J.; Isla, A.; McQueen-Mason, S.J.; Calderini, D.F. Expansions expression is associated with grain size dynamics in wheat (Triticum aestivum L.). J. Exp. Bot. 2010, 61, 1147–1157. [Google Scholar] [CrossRef] [PubMed]
- Prashant, R.; Kadoo, N.; Desale, C.; Kore, P.; Dhaliwal, H.S.; Chhuneja, P.; Gupta, V. Kernel morphometric traits in hexaploid wheat (Triticum aestivum L.) are modulated by intricate QTL × QTL and genotype × environment interactions. J. Cereal Sci. 2012, 56, 432–439. [Google Scholar] [CrossRef]
- Wiersma, J.J.; Busch, R.H.; Fulcher, G.G.; Hareland, G.A. Recurrent selection for kernel weight in spring wheat. Crop Sci. 2001, 41, 568–572. [Google Scholar] [CrossRef]
- Breseghello, F.; Sorrells, M.E. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 2006, 172, 1165–1177. [Google Scholar] [CrossRef]
- Doerge, R.W. Mapping and analysis of quantitative trait loci in experimental populations. Nat. Rev. Genet. 2002, 3, 43–52. [Google Scholar] [CrossRef]
- Xu, D.G.; Wen, W.E.; Fu, L.P.; Li, F.J.; Li, J.H.; Xie, L.; Xia, X.C.; Ni, Z.F.; He, Z.H.; Cao, S. Genetic dissection of a major QTL for kernel weight spanning the Rht–B1 locus in bread wheat. Theor. Appl. Genet. 2019, 132, 3191–3200. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Liu, J.D.; Xia, X.C.; He, Z.H. TaGS-D1, an ortholog of rice OsGS3, is associated with grain weight and grain length in common wheat. Mol. Breed. 2014, 34, 1097–1107. [Google Scholar] [CrossRef]
- Brinton, J.; Simmonds, J.; Minter, F.; Leverington-Waite, M.; Snape, J.; Uauy, C. Increased pericarp cell length underlies a major quantitative trait locus for grain weight in hexaploid wheat. New Phytol. 2017, 215, 1026–1038. [Google Scholar] [CrossRef]
- Tian, X.L.; Wen, W.; Xie, L.; Fu, L.P.; Xu, D.G.; Fu, C.; Wang, D.S.; Chen, X.M.; Xia, X.C.; Chen, Q.J.; et al. Molecular mapping of reduced plant height gene Rht24 in bread wheat. Front. Plant Sci. 2017, 8, 1379. [Google Scholar] [CrossRef]
- Mo, Y.J.; Vanzetti, L.S.; Hale, I.; Spagnolo, E.J.; Guidobaldi, F.; Oboudi, J.A.; Odle, N.; Pearce, S.; Helguera, M.; Dubcovsky, J. Identification and characterization of Rht25, a locus on chromosome arm 6AS affecting wheat plant height, heading time, and spike development. Theor. Appl. Genet. 2018, 131, 2021–2035. [Google Scholar] [CrossRef] [PubMed]
- Meng, D.Y.; Batool, A.; Xuan, Y.Z.; Pan, R.Q.; Zhang, N.; Zhang, W.; Zhi, L.Y.; Ren, X.L.; Li, W.Q.; Li, J.J.; et al. Fine mapping and validation of a stable QTL for thousand-kernel weight in wheat (Triticum aestivum L.). Crop J. 2023, 11, 1491–1500. [Google Scholar] [CrossRef]
- Zhai, H.J.; Feng, Z.Y.; Li, J.; Liu, X.Y.; Xiao, S.H.; Ni, Z.F.; Sun, Q.X. QTL analysis of spike morphological traits and plant height in winter wheat (Triticum aestivum L.) using a high-density SNP and SSR-based linkage map. Front. Plant Sci. 2016, 7, 16–17. [Google Scholar] [CrossRef]
- Liao, S.M.; Xu, Z.B.; Fan, X.L.; Zhou, Q.; Liu, X.F.; Jiang, C.; Chen, L.G.; Lin, D.; Feng, B.; Wang, T. Genetic dissection and validation of a major QTL for grain weight on chromosome 3B in bread wheat (Triticum aestivum L.). J. Integr. Agric. 2024, 23, 77–92. [Google Scholar] [CrossRef]
- Liu, L.H.; Qu, P.P.; Zhou, Y.; Li, H.B.; Liu, Y.N.; Zhang, M.M.; Zhang, L.P.; Zhao, C.P.; Zhang, S.Q.; Pang, B.S. Consensus linkage map construction and QTL mapping for eight yield-related traits in wheat using the BAAFS Wheat 90K SNP array. J. Integr. Agric. 2024, 23, 3641–3656. [Google Scholar] [CrossRef]
- Li, Y.H.; Hu, J.H.; Qu, Y.F.; Qiu, D.; Lin, H.L.; Du, J.Y.; Hou, L.; Ma, L.; Wu, Q.H.; Zhou, Y.; et al. Alleles on locus chromosome 4B from different parents confer tiller number and the yield-associated traits in wheat. BMC Plant Biol. 2024, 24, 454. [Google Scholar] [CrossRef]
- Guan, P.F.; Lu, L.H.; Jia, L.J.; Kabir, M.R.; Zhang, J.B.; Lan, T.Y.; Zhao, Y.; Xin, M.M.; Hu, Z.R.; Yao, Y.Y.; et al. Global QTL analysis identifies genomic regions on chromosomes 4A and 4B harboring stable loci for yield-related traits across different environments in wheat (Triticum aestivum L.). Front. Plant Sci. 2018, 9, 529. [Google Scholar] [CrossRef]
- Chen, Z.Y.; Cheng, X.J.; Chai, L.L.; Wang, Z.H.; Bian, R.L.; Li, J.; Zhao, A.J.; Xin, M.M.; Guo, W.L.; Hu, Z.R.; et al. Dissection of genetic factors underlying grain size and fine mapping of QTgw.cau-7D in common wheat (Triticum aestivum L.). Theor. Appl. Genet. 2020, 133, 149–162. [Google Scholar] [CrossRef]
- Sun, X.Y.; Wu, K.; Zhao, Y.; Kong, F.M.; Han, G.Z.; Jiang, H.M.; Huang, X.J.; Li, R.J.; Wang, H.G.; Li, S.S. QTL analysis of kernel shape and weight using recombinant inbred lines in wheat. Euphytica 2008, 165, 615–624. [Google Scholar] [CrossRef]
- Zhai, H.J.; Feng, Z.Y.; Du, X.F.; Song, Y.; Liu, X.Y.; Qi, Z.Q.; Song, L.; Li, J.; Li, L.H.; Peng, H.R.; et al. A novel allele of TaGW2-A1 is located in a finely mapped QTL that increases grain weight but decreases grain number in wheat (Triticum aestivum L.). Theor. Appl. Genet. 2018, 131, 539–553. [Google Scholar] [CrossRef]
- Qu, X.R.; Li, C.; Liu, H.; Liu, J.J.; Luo, W.; Xu, Q.; Tang, H.P.; Mu, Y.; Deng, M.; Pu, Z.E.; et al. Quick mapping and characterization of a co-located kernel length and thousand-kernel weight-related QTL in wheat. Theor. Appl. Genet. 2022, 135, 2849–2860. [Google Scholar] [CrossRef]
- Liu, G.; Jia, L.J.; Lu, L.H.; Qin, D.D.; Zhang, J.P.; Guan, P.F.; Ni, Z.F.; Yao, Y.Y.; Sun, Q.X.; Peng, H.R. Mapping QTLs of yield-related traits using RIL population derived from common wheat and Tibetan semi-wild wheat. Theor. Appl. Genet. 2014, 127, 2415–2432. [Google Scholar] [CrossRef]
- Gao, F.M.; Wen, W.E.; Liu, J.D.; Rashed, A.; Yin, G.H.; Xia, X.C.; Wu, X.X.; He, Z.H. Genome-wide linkage mapping of QTL for yield components, plant height and yield-related physiological traits in the Chinese wheat cross Zhou 8425B/Chinese Spring. Front Plant Sci. 2015, 18, 6. [Google Scholar] [CrossRef]
- Zhang, L.Y.; Liu, D.C.; Guo, X.L.; Yang, W.L.; Sun, J.Z.; Wang, D.W.; Zhang, A.M. Genomic distribution of quantitative trait loci for yield and yield-related traits in common wheat. J. Integr. Plant Biol. 2010, 52, 996–1007. [Google Scholar] [CrossRef]
- Xin, F.; Zhu, T.; Wei, S.W.; Han, Y.C.; Zhao, Y.; Zhang, D.Z.; Ma, L.J.; Ding, Q. QTL mapping of kernel traits and validation of a major QTL for kernel length-width ratio using SNP and bulked segregant analysis in wheat. Sci. Rep. 2020, 10, 25. [Google Scholar] [CrossRef]
- Peng, J.R.; Richards, D.E.; Hartley, N.M.; Murphy, G.P.; Devos, K.M.; Flintham, J.E.; Beales, J.; Fish, L.J.; Worland, A.J.; Pelica, F.; et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 1999, 400, 256–261. [Google Scholar] [CrossRef]
- Chu, C.G.; Xu, S.S.; Friesen, T.L.; Faris, J.D. Whole genome mapping in a wheat doubled haploid population using SSRs and TRAPs and the identification of QTL for agronomic traits. Mol. Breed. 2008, 22, 251–266. [Google Scholar] [CrossRef]
- Li, W.Q.; Han, M.M.; Pang, D.W.; Chen, J.; Wang, Y.Y.; Dong, H.H.; Chang, Y.L.; Jin, M.; Luo, Y.L.; Li, Y.; et al. Characteristics of lodging resistance of high-yield winter wheat as affected by nitrogen rate and irrigation managements. J. Integr. Agric. 2022, 21, 1290–1309. [Google Scholar] [CrossRef]
- Gao, Z.X.; Wang, X.Y.; Tian, G.Y.; Zhao, Y.K.; Li, C.H.; Cao, Q.; Han, R.; Shi, Z.L.; He, M.Q. Plant height and its relationship with yield in wheat under different irrigation regime. Irrig. Sci. 2020, 38, 365–371. [Google Scholar] [CrossRef]
- Wen, S.Z.; Zhang, M.H.; Tu, K.L.; Fan, C.F.; Tian, S.; Bi, C.; Chen, Z.L.; Zhao, H.H.; Wei, C.X.; Shi, X.T.; et al. A major quantitative trait loci cluster controlling three components of yield and plant height identified on chromosome 4B of common wheat. Front. Plant Sci. 2022, 12, 799520. [Google Scholar] [CrossRef]
- Cheng, X.J.; Chai, L.L.; Chen, Z.Y.; Xu, L.; Zhai, H.J.; Zhao, A.J.; Peng, H.R.; Yao, Y.Y.; You, M.S.; Sun, Q.X.; et al. Identification and characterization of a high kernel weight mutant induced by gamma radiation in wheat (Triticum aestivum L.). BMC Genet. 2015, 16, 127. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- DePristo, M.A.; Banks, E.; Popli, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; Angel, G.D.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.H.; Zhang, L.Y.; Wang, J.K. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]

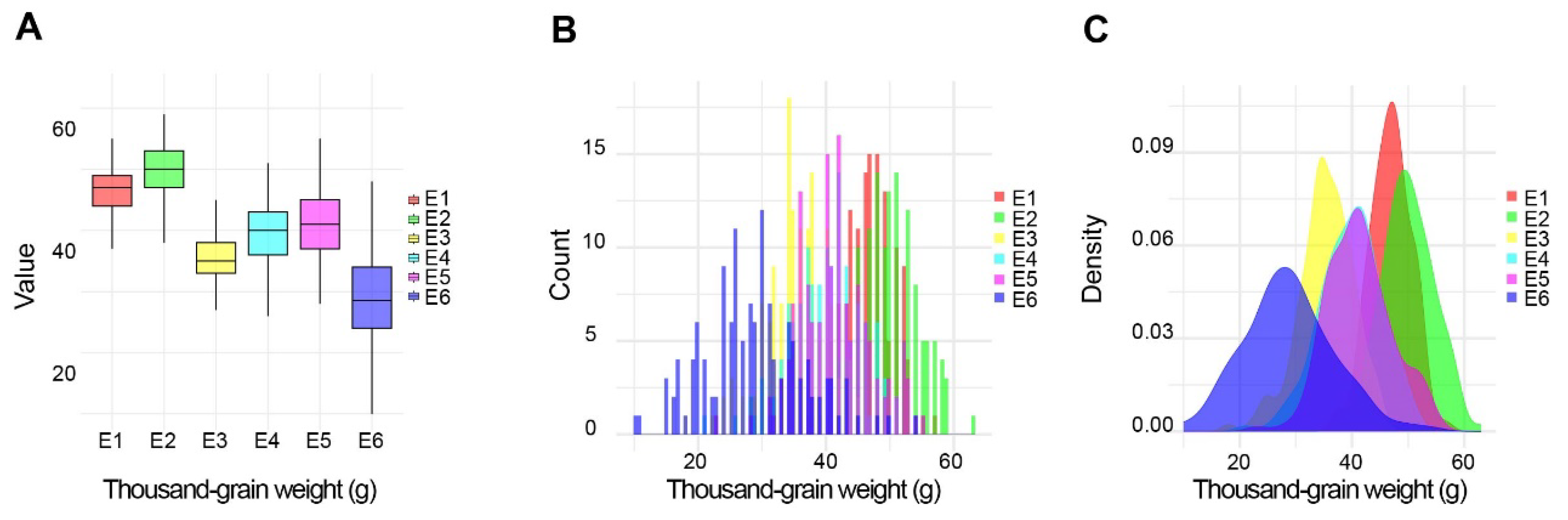

| ZK331 | ND399 | Min. | Max. | Average | SD | Skewness | Kurtosis | h2 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| TGW (g) | E1 | 53.4 | 45.6 | 36.5 | 57.1 | 46.9 | 3.60 | −0.11 | 0.16 | 77.5 |
| E2 | 52.8 | 46.3 | 37.7 | 63.0 | 49.9 | 4.58 | 0.04 | −0.09 | ||
| E3 | 37.2 | 35.4 | 18.0 | 45.3 | 35.3 | 4.82 | −0.54 | 0.78 | ||
| E4 | 36.5 | 34.2 | 21.0 | 51.0 | 39.6 | 5.41 | −0.36 | 0.32 | ||
| E5 | 43.7 | 41.2 | 22.9 | 54.7 | 41.3 | 5.66 | 0.14 | 0.22 | ||
| E6 | 37.8 | 34.2 | 9.7 | 53.7 | 28.8 | 7.95 | 0.32 | 0.26 | ||
| GL (mm) | E1 | 6.7 | 6.3 | 6.0 | 7.6 | 6.8 | 0.31 | 0.21 | 0.18 | 93.2 |
| E2 | 7.1 | 6.7 | 6.2 | 7.9 | 6.9 | 0.29 | 0.55 | 0.87 | ||
| E3 | 6.8 | 6.2 | 6.1 | 7.5 | 6.6 | 0.29 | −0.66 | 0.39 | ||
| E4 | 6.8 | 6.2 | 6.1 | 7.9 | 6.8 | 0.30 | 0.67 | 1.18 | ||
| E5 | 6.9 | 6.2 | 5.7 | 7.3 | 6.4 | 0.30 | 0.48 | 0.49 | ||
| E6 | 6.8 | 6.3 | 5.5 | 7.4 | 6.4 | 0.39 | 0.31 | −0.15 | ||
| GW (mm) | E1 | 3.3 | 2.8 | 3.0 | 3.8 | 3.5 | 0.14 | −0.61 | 1.1 | 75.9 |
| E2 | 3.7 | 3.2 | 3.1 | 4.0 | 3.6 | 0.14 | −0.58 | 1.92 | ||
| E3 | 3.1 | 2.8 | 2.6 | 3.8 | 3.2 | 0.20 | −0.15 | 0.48 | ||
| E4 | 3.6 | 3.2 | 2.9 | 3.8 | 3.3 | 0.17 | −0.06 | 0.09 | ||
| E5 | 3.6 | 3.2 | 2.8 | 3.9 | 3.4 | 0.18 | −0.08 | 0.4 | ||
| E6 | 3.4 | 3.0 | 2.3 | 4.0 | 3.0 | 0.29 | 0.25 | 0.74 | ||
| PH (cm) | E2 | 72.0 | 68.0 | 40.3 | 99.7 | 74.3 | 12.57 | −0.61 | 0.47 | 90.5 |
| E3 | 71.7 | 67.8 | 41.7 | 103.3 | 83.1 | 11.87 | −0.86 | 0.58 | ||
| E4 | 76.5 | 71.2 | 47.0 | 129.3 | 96.7 | 17.23 | −0.66 | 0.54 | ||
| E5 | 78.2 | 73.6 | 54.0 | 120.7 | 92.7 | 10.95 | −0.55 | 1.18 | ||
| E6 | 68.3 | 64.9 | 50.0 | 120.3 | 93.8 | 13.98 | −0.4 | −0.07 |
| Trait | QTL | Environment | Position | Left Marker | Right Marker | LOD | PVE a | Add b | Previous Study |
|---|---|---|---|---|---|---|---|---|---|
| TGW | qTGW-1B.1 | E5 | 82 | 1B_637885015 | 1B_641922827 | 3.65 | 11.38 | 1.90 | |
| E6 | 85 | 1B_637885015 | 1B_641922827 | 4.40 | 14.44 | 2.98 | |||
| qTGW-2B.1 | E3 | 63 | 2B_39795004 | 2B_67981606 | 4.91 | 14.63 | 1.87 | ||
| E5 | 64 | 2B_67981922 | 2B_68424868 | 5.01 | 11.20 | 2.12 | |||
| qTGW-3B.1 | E3 | 59 | 3B_44543422 | 3B_50191187 | 3.81 | 11.86 | 1.71 | QTgw.cib-3B.1 [24] | |
| E5 | 57 | 3B_43599467 | 3B_44857581 | 3.18 | 10.99 | 1.85 | |||
| qTGW-4B.1 | E4 | 55 | 4B_28739948 | 4B_32174878 | 5.94 | 18.65 | 2.46 | QTGW.baafs-4B [25]; QTgw.caau-4B [26] | |
| E6 | 54 | 4B_28739948 | 4B_32174878 | 4.62 | 14.41 | 3.34 | QTgw.cau-4B.2 [27]; QTgw.cau-4B-1 [28] | ||
| qTGW-4D.1 | E1 | 40 | 4D_19090268 | 4D_48697668 | 2.64 | 8.82 | −1.42 | ||
| E2 | 37 | 4D_13215376 | 4D_19090268 | 2.75 | 7.93 | −1.73 | |||
| GL | qGL-1B.1 | E1 | 82 | 1B_637885015 | 1B_641922827 | 4.86 | 14.47 | 0.12 | QKl.sdau-1B.2 [29] |
| E2 | 80 | 1B_564852828 | 1B_637584804 | 7.15 | 21.14 | 0.13 | |||
| E3 | 83 | 1B_637885015 | 1B_641922827 | 8.54 | 24.27 | 0.15 | |||
| E4 | 80 | 1B_564852828 | 1B_637584804 | 5.75 | 20.07 | 0.13 | |||
| E5 | 83 | 1B_637885015 | 1B_641922827 | 6.81 | 19.39 | 0.14 | |||
| qGL-2B.1 | E3 | 59 | 2B_39795004 | 2B_67981606 | 3.49 | 8.89 | 0.11 | ||
| E6 | 63 | 2B_39795004 | 2B_67981606 | 4.78 | 8.69 | 0.15 | |||
| qGL-2B.2 | E2 | 161 | 2B_690211230 | 2B_694440260 | 2.67 | 10.20 | −0.09 | QGl.cau-2B.2 [30] | |
| E5 | 161 | 2B_690211230 | 2B_694440260 | 3.22 | 11.56 | −0.10 | |||
| qGL-2D.1 | E3 | 107 | 2D_531701660 | 2D_562710587 | 3.29 | 10.96 | 0.11 | QKL.sicau-SSY-2D [31] | |
| E4 | 104 | 2D_531701660 | 2D_562710587 | 2.93 | 10.96 | 0.11 | |||
| qGL-3A.1 | E2 | 258 | 3A_741822204 | 3A_746542290 | 3.11 | 9.70 | 0.10 | ||
| E3 | 268 | 3A_746542290 | 3A_747278706 | 3.04 | 8.37 | 0.09 | |||
| E4 | 268 | 3A_746542290 | 3A_747278706 | 2.74 | 8.73 | 0.09 | |||
| qGL-3B.1 | E3 | 45 | 3B_31983800 | 3B_34669808 | 5.22 | 15.86 | 0.12 | ||
| E4 | 44 | 3B_31983800 | 3B_34669808 | 3.21 | 9.71 | 0.09 | |||
| qGL-3B.2 | E3 | 188 | 3B_757269550 | 3B_759449813 | 2.84 | 7.74 | 0.08 | ||
| E6 | 188 | 3B_757269550 | 3B_759449813 | 19.92 | 11.10 | 0.35 | |||
| qGL-4A.1 | E2 | 50 | 4A_466204879 | 4A_468921463 | 3.05 | 11.66 | 0.09 | ||
| E4 | 50 | 4A_466204879 | 4A_468921463 | 4.14 | 13.38 | 0.11 | |||
| qGL-7A.1 | E3 | 148 | 7A_607579794 | 7A_617666459 | 12.10 | 17.27 | 0.12 | ||
| E6 | 148 | 7A_607579794 | 7A_617666459 | 8.73 | 16.25 | 0.17 | |||
| GW | qGW-3D.1 | E2 | 144 | 3D_538481562 | 3D_551980404 | 3.23 | 10.03 | 0.04 | |
| E4 | 142 | 3D_538481562 | 3D_551980404 | 3.78 | 11.34 | 0.06 | |||
| qGW-4B.1 | E1 | 55 | 4B_28739948 | 4B_32174878 | 2.75 | 8.45 | 0.04 | QGw.caau-4B [26] | |
| E2 | 51 | 4B_24900757 | 4B_28739948 | 4.07 | 13.67 | 0.05 | |||
| E3 | 45 | 4B_24900757 | 4B_28739948 | 4.07 | 13.56 | 0.08 | |||
| E4 | 47 | 4B_24900757 | 4B_28739948 | 3.54 | 11.63 | 0.07 | |||
| E6 | 47 | 4B_24900757 | 4B_28739948 | 3.62 | 12.59 | 0.11 | |||
| qGW-4D.1 | E5 | 36 | 4D_13215376 | 4D_19090268 | 3.18 | 9.41 | -0.07 | ||
| E6 | 45 | 4D_19090268 | 4D_48697668 | 3.08 | 9.98 | −0.13 | |||
| PH | qPH-1A.1 | E4 | 25 | 1A_6791455 | 1A_14475390 | 2.93 | 9.15 | 4.95 | |
| E5 | 27 | 1A_14475390 | 1A_23131762 | 2.65 | 9.36 | 3.17 | |||
| qPH-3D.1 | E5 | 143 | 3D_538481562 | 3D_551980404 | 3.19 | 9.29 | 3.61 | ||
| E6 | 143 | 3D_538481562 | 3D_551980404 | 5.41 | 15.70 | 5.00 | |||
| qPH-4B.1 | E2 | 53 | 4B_28739948 | 4B_32174878 | 5.67 | 16.81 | 4.73 | QPH.baafs-4B [25] | |
| E3 | 53 | 4B_28739948 | 4B_32174878 | 5.82 | 17.60 | 7.52 | QPh.cau-4B.2 [27] | ||
| E4 | 53 | 4B_28739948 | 4B_32174878 | 6.02 | 17.25 | 4.85 | QPh-4B1 [32] | ||
| E5 | 54 | 4B_28739948 | 4B_32174878 | 9.38 | 26.24 | 8.32 | QPH-caas-4BS.2 [33] | ||
| E6 | 53 | 4B_28739948 | 4B_32174878 | 15.81 | 21.48 | 6.99 | |||
| qPH-4D.1 | E2 | 40 | 4D_19090268 | 4D_48697668 | 11.37 | 29.91 | −8.07 | QTgw.cau-4D.1 [27] | |
| E3 | 37 | 4D_13215376 | 4D_19090268 | 16.42 | 40.51 | −9.31 | QPh-4D [32] | ||
| E4 | 37 | 4D_13215376 | 4D_48697668 | 13.41 | 39.36 | −11.53 | QPH.caas-4DS.1 [33] | ||
| E5 | 39 | 4D_19090268 | 4D_48697668 | 12.22 | 34.29 | −7.49 | |||
| E6 | 40 | 4D_19090268 | 4D_48697668 | 10.79 | 31.43 | −9.35 | |||
| qPH-5A.1 | E2 | 74 | 5A_532377854 | 5A_535830656 | 4.18 | 12.78 | 3.88 | ||
| E4 | 74 | 5A_532377854 | 5A_535830656 | 5.01 | 15.12 | 5.88 | |||
| E5 | 75 | 5A_535032560 | 5A_539540555 | 4.10 | 11.87 | 3.90 | |||
| E6 | 75 | 5A_535032560 | 5A_539540555 | 4.17 | 12.23 | 5.04 | |||
| qPH-7A.1 | E2 | 208 | 7A_701630471 | 7A_724015398 | 5.11 | 6.82 | 2.90 | ||
| E4 | 208 | 7A_701630471 | 7A_724015398 | 2.60 | 3.94 | 3.74 | |||
| E6 | 208 | 7A_701630471 | 7A_724015398 | 3.52 | 4.00 | 2.80 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Chen, Y.; Yang, Y.; Qiu, D.; Zhang, H.; Hu, J.; Guo, G.; Zhu, K.; Fu, H.; Li, H.; et al. Identification of QTL for Grain Traits and Plant Height Using the Recombinant Inbred Line Population Derived from the Cross of Zhongke 331 × Nongda 399. Int. J. Mol. Sci. 2025, 26, 3526. https://doi.org/10.3390/ijms26083526
Liu Y, Chen Y, Yang Y, Qiu D, Zhang H, Hu J, Guo G, Zhu K, Fu H, Li H, et al. Identification of QTL for Grain Traits and Plant Height Using the Recombinant Inbred Line Population Derived from the Cross of Zhongke 331 × Nongda 399. International Journal of Molecular Sciences. 2025; 26(8):3526. https://doi.org/10.3390/ijms26083526
Chicago/Turabian StyleLiu, Yi, Yongxing Chen, Yijun Yang, Dan Qiu, Huaizhi Zhang, Jinghuang Hu, Guanghao Guo, Keyu Zhu, Hongkui Fu, Hongjie Li, and et al. 2025. "Identification of QTL for Grain Traits and Plant Height Using the Recombinant Inbred Line Population Derived from the Cross of Zhongke 331 × Nongda 399" International Journal of Molecular Sciences 26, no. 8: 3526. https://doi.org/10.3390/ijms26083526
APA StyleLiu, Y., Chen, Y., Yang, Y., Qiu, D., Zhang, H., Hu, J., Guo, G., Zhu, K., Fu, H., Li, H., Liu, Z., Wang, R., & Wu, Q. (2025). Identification of QTL for Grain Traits and Plant Height Using the Recombinant Inbred Line Population Derived from the Cross of Zhongke 331 × Nongda 399. International Journal of Molecular Sciences, 26(8), 3526. https://doi.org/10.3390/ijms26083526







