Transcriptomic and Metabolomic Insights into Plant Hormone Modulation and Secondary Metabolite Accumulation in Basil Under Far-Red and Ultraviolet-A Light
Abstract
1. Introduction
2. Results
2.1. Effect of FR and UVA Treatments on the Growth Indicators of Basil
2.2. Effects of FR and UVA Treatments on Chlorophyll and Carotenoid Content of Basil
2.3. Effects of FR and UVA Treatments on the Transcriptome of Basil
2.3.1. Basil Transcriptome Sequencing Results
2.3.2. The Gene Expression and Differentially Expressed Gene (DEG) Analysis of Basil Under FR and UVA Treatments
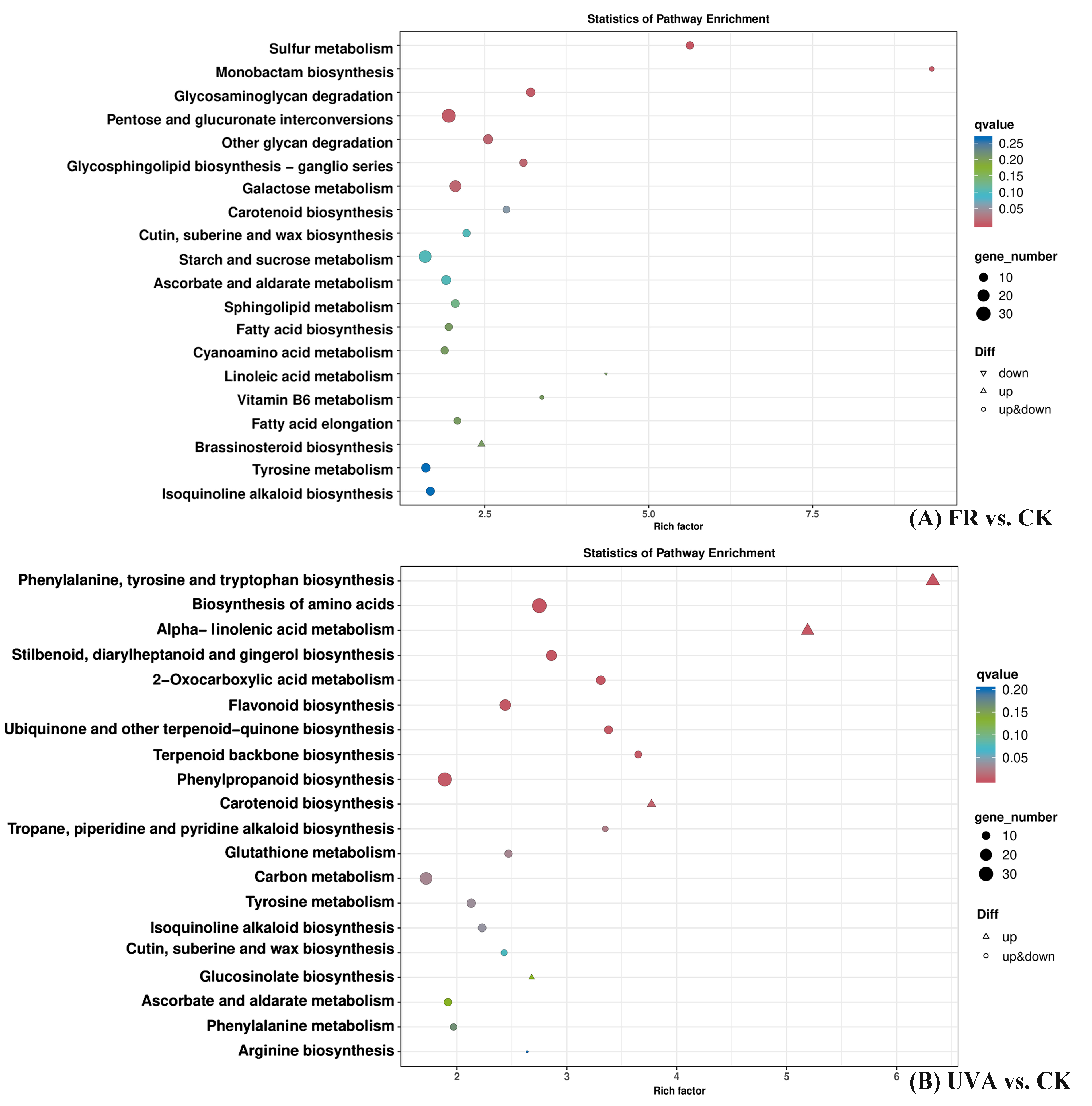
2.3.3. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis of Differentially Expressed Genes (DEGs) in Basil Under FR and UVA Treatments
2.4. The Effects of FR and UVA Treatments on the Metabolome of Basil
2.4.1. Metabolite Profiling and Differentially Accumulated Metabolite (DAM) Analysis of Basil Under FR and UVA Treatments
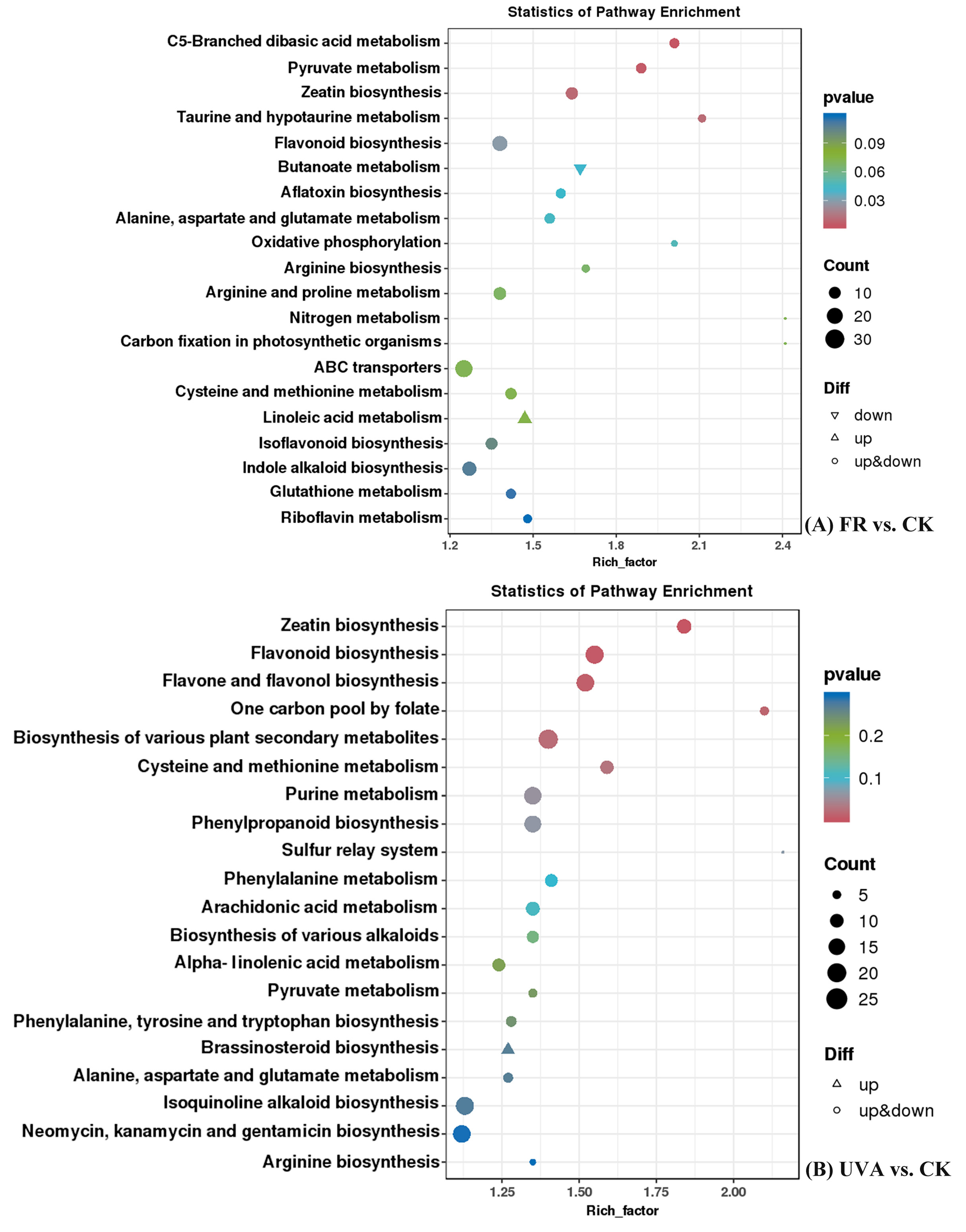
2.4.2. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis of Differentially Accumulated Metabolites (DAMs) in Basil Under FR and UVA Treatments
2.5. Combined Analysis of Transcriptome and Metabolites in Basil Under FR and UVA Treatments
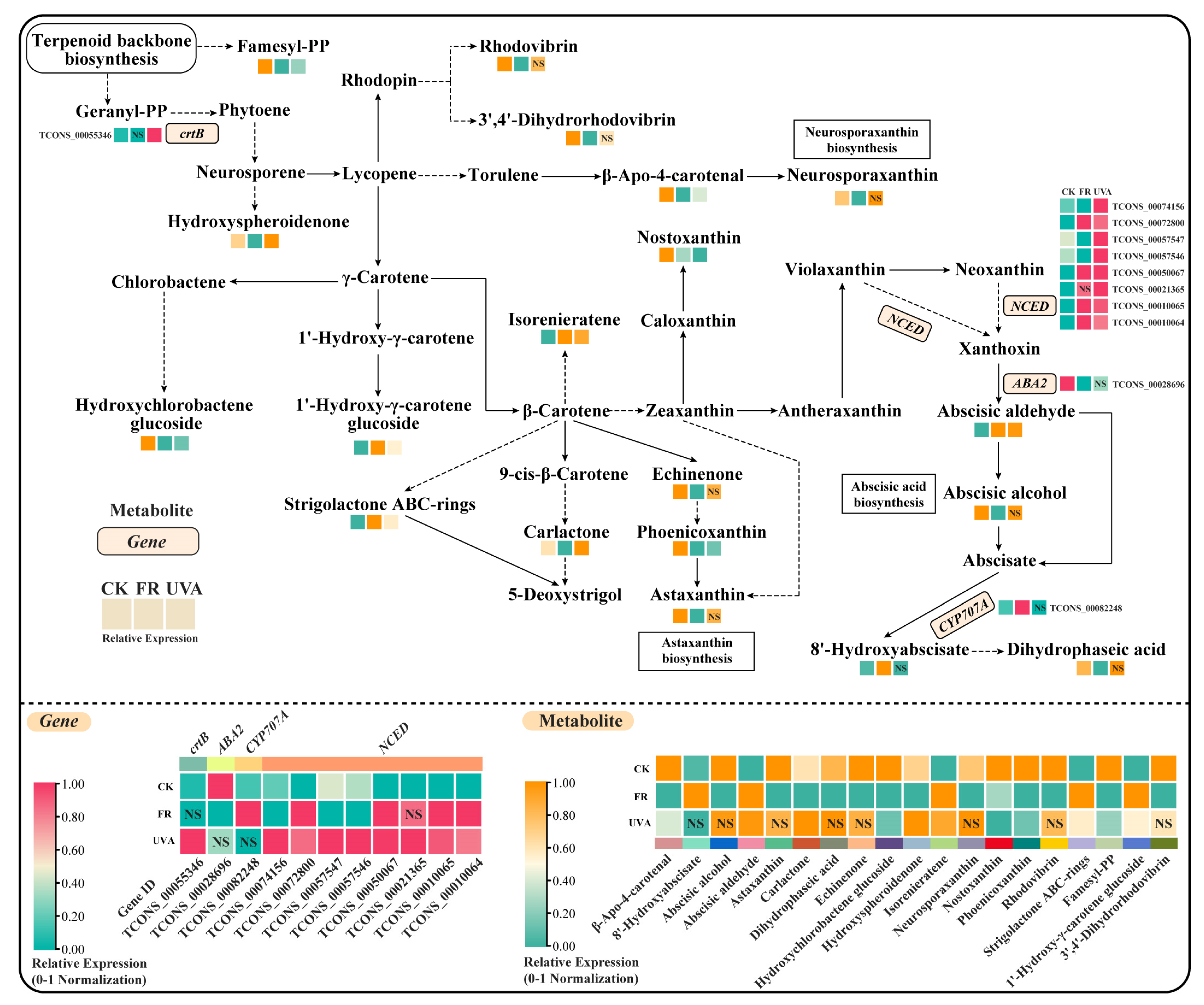
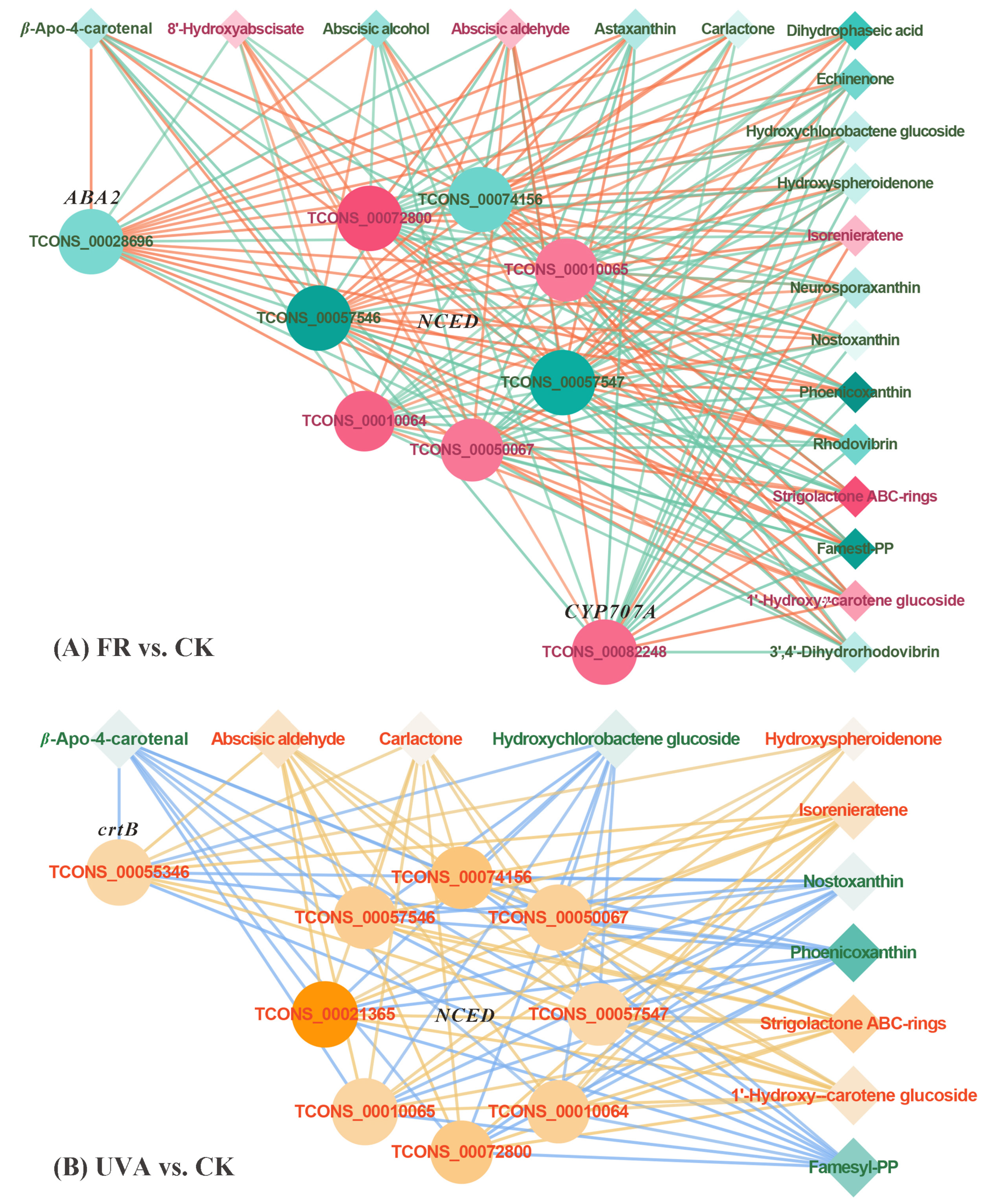
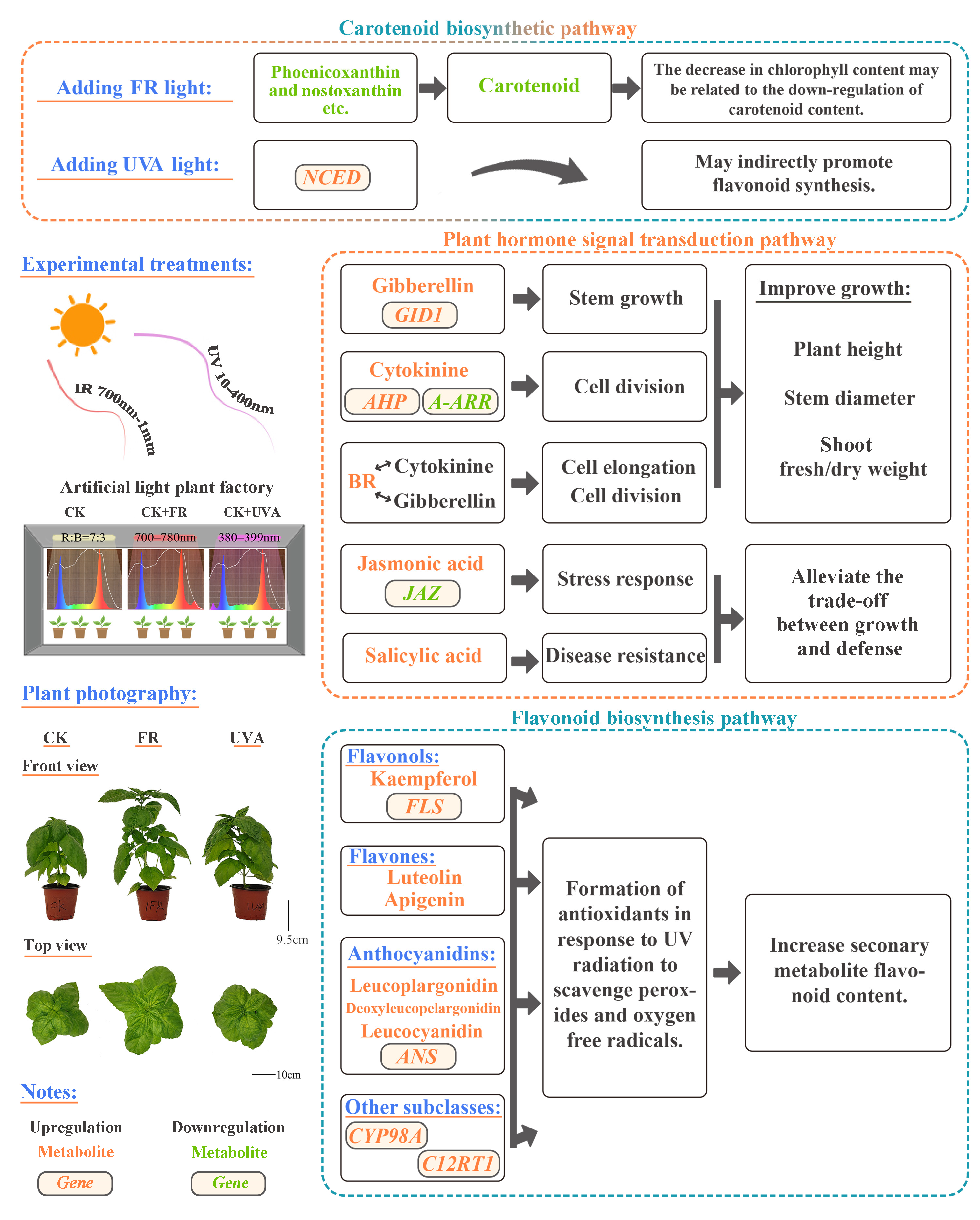
2.5.1. Combined Analysis of Transcriptome and Metabolites in Carotenoid Biosynthetic Pathway Under FR and UVA Treatments
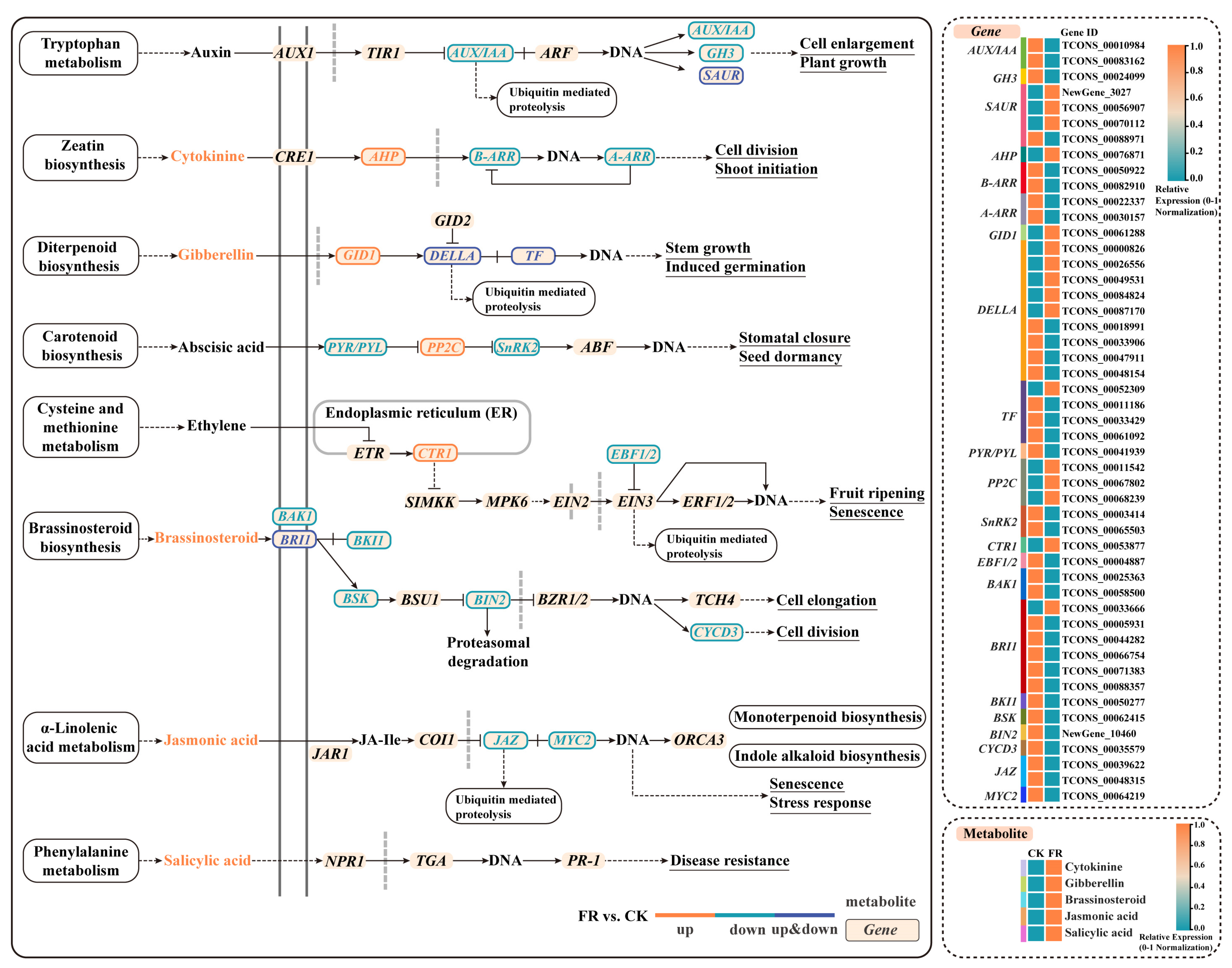
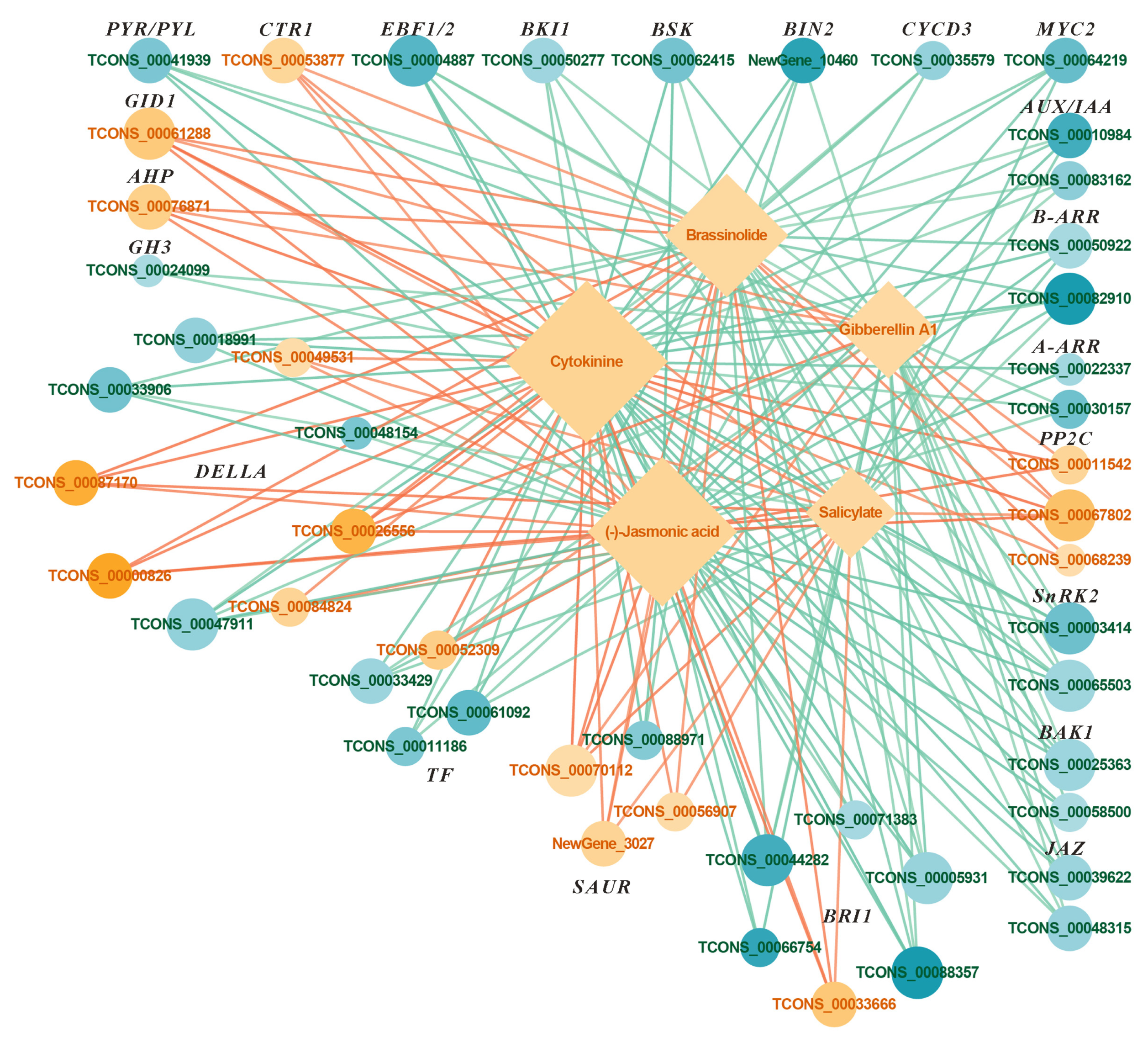
2.5.2. Combined Analysis of Transcriptome and Metabolites in Plant Hormone Signaling Pathways Under FR Treatment
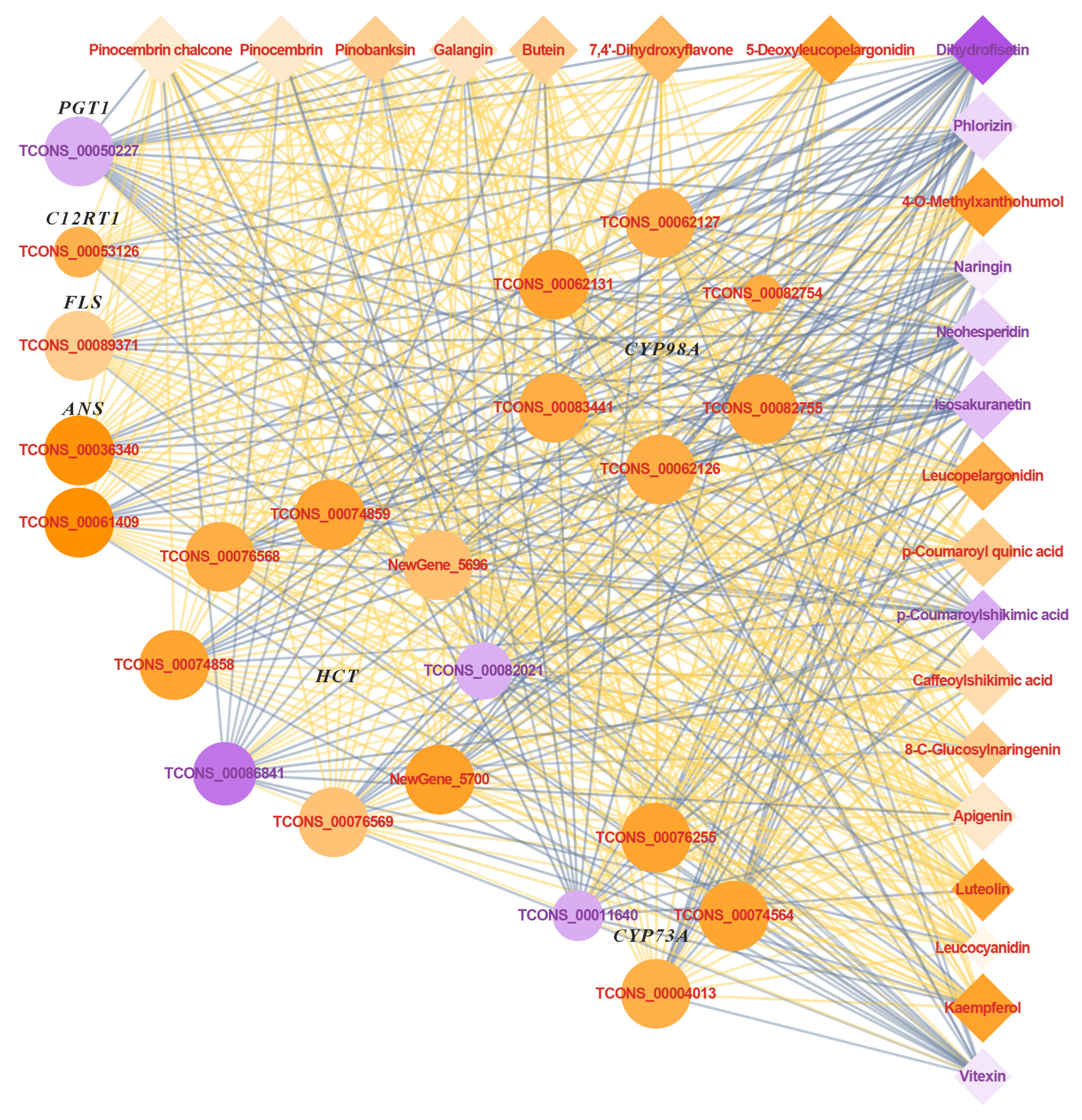
2.5.3. Combined Analysis of Transcriptome and Metabolites in Flavonoid Biosynthesis Pathway Under UVA Treatment
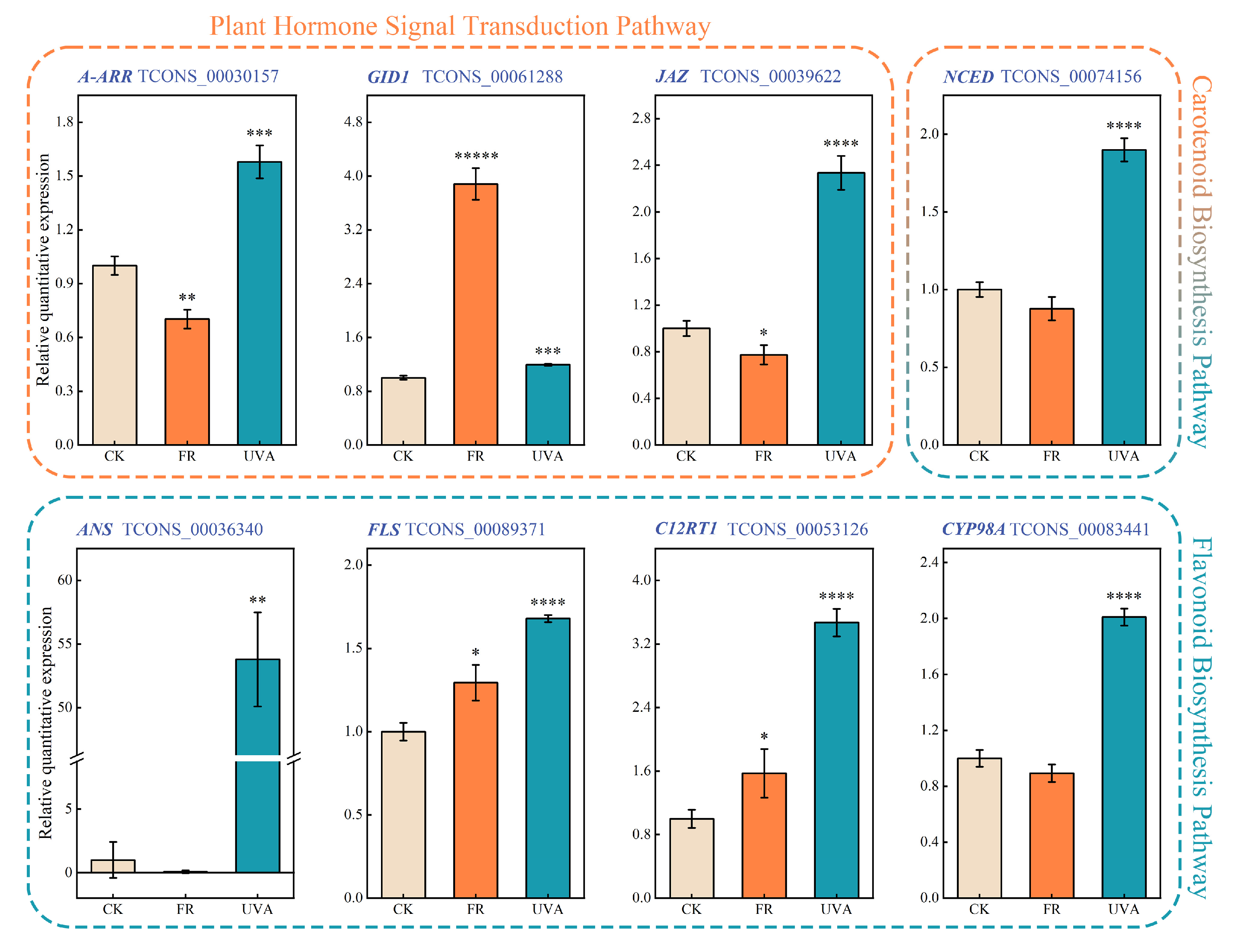
2.6. Validation of RNA-Seq-Based DEG Results via Real-Time Quantitative PCR (qRT-PCR)
3. Discussions
3.1. Effects of FR and UVA Treatments on the Growth of Basil
3.2. Effects of FR and UVA Treatments on Chlorophyll Content of Basil
3.3. Effects of FR and UVA Treatments on Carotenoid Content and the Carotenoid Biosynthesis Pathway in Basil
3.4. Effects of FR Treatment on the Hormone Signaling Transduction Pathway in Basil
3.5. Effects of UVA Treatment on the Flavonoid Biosynthetic Pathway in Basil
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Experimental Treatments
4.3. Project Measurement
4.3.1. Measurement of Growth Indicators
4.3.2. Determination of Chlorophyll and Carotenoid Contents
4.3.3. Transcriptome Sequencing and Data Analysis
- (1)
- RNA extraction: Total RNA from plants was extracted using the RNA prep Pure Plant Kit (Tiangen, Beijing, China). RNA concentration and purity were measured using a NanoDrop 2000 (Thermo Fisher Scientific, Wilmington, DE, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit with the Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA).
- (2)
- RNA quantification and qualification: RNA concentration and purity were measured using NanoDrop 2000. RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system.
- (3)
- Library preparation for transcriptome sequencing: A total amount of 1 μg RNA per sample was used as the input material for the RNA sample preparations. Sequencing libraries were generated using Hieff NGS Ultima Dual-mode mRNA Library Prep Kit for Illumina (Yeasen Biotechnology (Shanghai) Co., Ltd., Shanghai, China) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample.
- (4)
- Sequencing: The libraries were sequenced on an Illumina NovaSeq platform to generate 150 bp paired-end reads, according to the manufacturer’s instructions.
- (5)
- Data analysis: The raw reads were further processed with a bioinformatic pipeline tool—the BMKCloud (https://www.biocloud.net (accessed on 2 November 2023)) online platform.
4.3.4. Metabolome Sequencing and Data Analysis
- (1)
- Metabolite Extraction. The LC/MS system for metabolomics analysis comprises a Waters Acquity I-Class PLUS ultra-high performance liquid tandem Waters Xevo G2-XS QTof (Waters, Milford, MA, USA) high-resolution mass spectrometer. The column used is purchased from Waters Acquity UPLC HSS T3 column (1.8 μm 2.1 × 100 mm).
- (2)
- LC-MS/MS Analysis. The Waters Xevo G2-XS QTof high-resolution mass spectrometer can be used to collect primary and secondary mass spectrometry data in the MSe mode under the control of the acquisition software program (MassLynx v 4.2, Waters, Milford, MA, USA).
- (3)
- Data Preprocessing and Annotation. The raw data collected using MassLynx v 4.2 (Waters, Milford, MA, USA) is processed using the Progenesis QI software v 3.0 (Waters, Newcastle upon Tyne, UK) for peak extraction, peak alignment, and other data processing operations, based on the Progenesis QI software online METLIN database and Biomark’s self-built library (https://www.biocloud.net (accessed on 5 November 2023)) for identification, and at the same time, the theoretical fragment identification and mass deviation are all within 100ppm [80].
- (4)
- Data Analysis. The identified compounds are searched for classification and pathway information in the KEGG (http://www.genome.jp/kegg/ (accessed on 11 November 2023)), HMDB (https://hmdb.ca/ (accessed on 13 November 2023)), and lipidmaps (https://lipidmaps.org/ (accessed on 16 November 2023)) databases. According to the grouping information, the difference multiples were calculated and compared, and a t-test was used to calculate the difference in significance of each compound’s p value. The VIP value of the model was calculated using multiple cross-validation. The method of combining the difference multiple, the p value, and the VIP value of the OPLS-DA model was adopted to screen the differential metabolites. The screening criteria are a fold change > 1, p-value < 0.05, and VIP > 1. The differential metabolites of the KEGG pathway enrichment significance were calculated using the hypergeometric distribution test [81]. The raw data were further processed using the bioinformatics analysis platform BMKCloud (https://www.biocloud.net (accessed on 21 November 2023)) to obtain the analytical results related to differentially accumulated metabolites.
4.3.5. Correlation Network Diagram of Differentially Expressed Genes (DEGs) and Differentially Accumulated Metabolites (DAMs)
4.3.6. Quantitative Real-Time PCR (qRT-PCR) Validation
4.4. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Güez, C.M.; de Souza, R.O.; Fischer, P.; de Moura Leão, M.F.; Duarte, J.A.; Boligon, A.A.; Athayde, M.L.; Zuravski, L.; de Oliveira, L.F.S.; Machado, M.M. Evaluation of Basil Extract (Ocimum basilicum L.) on Oxidative, Anti-Genotoxic and Anti-Inflammatory Effects in Human Leukocytes Cell Cultures Exposed to Challenging Agents. Braz. J. Pharm. Sci. 2017, 53, e15098. [Google Scholar] [CrossRef]
- Zhang, H.; Tsao, R. Dietary Polyphenols, Oxidative Stress and Antioxidant and Anti-Inflammatory Effects. Curr. Opin. Food Sci. 2016, 8, 33–42. [Google Scholar] [CrossRef]
- Arranz, E.; Jaime, L.; López de las Hazas, M.C.; Reglero, G.; Santoyo, S. Supercritical Fluid Extraction as an Alternative Process to Obtain Essential Oils with Anti-Inflammatory Properties from Marjoram and Sweet Basil. Ind. Crops Prod. 2015, 67, 121–129. [Google Scholar] [CrossRef]
- Viyoch, J.; Pisutthanan, N.; Faikreua, A.; Nupangta, K.; Wangtorpol, K.; Ngokkuen, J. Evaluation of in Vitro Antimicrobial Activity of Thai Basil Oils and Their Micro-emulsion Formulas against Propionibacterium acnes. Int. J. Cosmet. Sci. 2006, 28, 125–133. [Google Scholar] [CrossRef]
- Zhan, Y.; An, X.; Wang, S.; Sun, M.; Zhou, H. Basil Polysaccharides: A Review on Extraction, Bioactivities and Pharmacological Applications. Bioorganic Med. Chem. 2020, 28, 115179. [Google Scholar] [CrossRef] [PubMed]
- Dhama, K.; Sharun, K.; Gugjoo, M.B.; Tiwari, R.; Alagawany, M.; Iqbal Yatoo, M.; Thakur, P.; Iqbal, H.M.N.; Chaicumpa, W.; Michalak, I.; et al. A Comprehensive Review on Chemical Profile and Pharmacological Activities of Ocimum basilicum. Food Rev. Int. 2023, 39, 119–147. [Google Scholar] [CrossRef]
- D’Aquino, L.; Lanza, B.; Gambale, E.; Sighicelli, M.; Menegoni, P.; Modarelli, G.C.; Rimauro, J.; Chianese, E.; Nenna, G.; Fasolino, T.; et al. Growth and Metabolism of Basil Grown in a New-Concept Microcosm under Different Lighting Conditions. Sci. Hortic. 2022, 299, 111035. [Google Scholar] [CrossRef]
- Fankhauser, C.; Chory, J. Light Control of Plant Development. Annu. Rev. Cell Dev. Biol. 1997, 13, 203–229. [Google Scholar] [CrossRef]
- García-López, I.; Vélez-Ramírez, A.I.; Gillmor, C.S.; Fernandez-Valverde, S.L. lncRNAs involved in the Shade Avoidance Syndrome (SAS) in Arabidopsis thaliana. BMC Genom. 2024, 25, 802. [Google Scholar] [CrossRef]
- Liu, Y.; Jafari, F.; Wang, H. Integration of Light and Hormone Signaling Pathways in the Regulation of Plant Shade Avoidance Syndrome. aBIOTECH 2021, 2, 131–145. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, H.; Gao, M.; He, R.; Liu, X.; Su, W.; Liu, H. Far-Red-Light-Induced Morphology Changes, Phytohormone, and Transcriptome Reprogramming of Chinese Kale (Brassica alboglabra Bailey). Int. J. Mol. Sci. 2023, 24, 5563. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Y.; Yang, Q.; Li, T. Overhead Supplemental Far-Red Light Stimulates Tomato Growth under Intra-Canopy Lighting with LEDs. J. Integr. Agric. 2019, 18, 62–69. [Google Scholar] [CrossRef]
- de Lucas, M.; Prat, S. PIFs Get BRright: PHYTOCHROME INTERACTING FACTORs as Integrators of Light and Hormonal Signals. New Phytol. 2014, 202, 1126–1141. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.; Xu, S.-L.; Tepperman, J.M.; Stanley, D.J.; Maltby, D.A.; Gross, J.D.; Burlingame, A.L.; Wang, Z.-Y.; Quail, P.H. A Mutually Assured Destruction Mechanism Attenuates Light Signaling in Arabidopsis. Science 2014, 344, 1160–1164. [Google Scholar] [CrossRef]
- Leivar, P.; Tepperman, J.M.; Cohn, M.M.; Monte, E.; Al-Sady, B.; Erickson, E.; Quail, P.H. Dynamic Antagonism between Phytochromes and PIF Family Basic Helix-Loop-Helix Factors Induces Selective Reciprocal Responses to Light and Shade in a Rapidly Responsive Transcriptional Network in Arabidopsis. Plant Cell 2012, 24, 1398–1419. [Google Scholar] [CrossRef]
- Bai, M.-Y.; Shang, J.-X.; Oh, E.; Fan, M.; Bai, Y.; Zentella, R.; Sun, T.; Wang, Z.-Y. Brassinosteroid, Gibberellin and Phytochrome Impinge on a Common Transcription Module in Arabidopsis. Nat. Cell Biol. 2012, 14, 810–817. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, Q.; Jiang, Y.; Yang, C.; Wang, Q.; Li, L. Shade-Induced Nuclear Localization of PIF7 Is Regulated by Phosphorylation and 14-3-3 Proteins in Arabidopsis. eLife 2018, 7, e31636. [Google Scholar] [CrossRef]
- Casal, J.J. Photoreceptor Signaling Networks in Plant Responses to Shade. Annu. Rev. Plant Biol. 2013, 64, 403–427. [Google Scholar] [CrossRef]
- Su, H.; Abernathy, S.D.; White, R.H.; Finlayson, S.A. Photosynthetic Photon Flux Density and Phytochrome B Interact to Regulate Branching in Arabidopsis. Plant Cell Environ. 2011, 34, 1986–1998. [Google Scholar] [CrossRef]
- Tucker, D.J.; Mansfield, T.A. Effects of Light Quality on Apical Dominance in Xanthium strumarium and the Associated Changes in Endogenous Levels of Abscisic Acid and Cytokinins. Planta 1971, 102, 140–151. [Google Scholar] [CrossRef]
- Dong, H.; Wang, J.; Song, X.; Hu, C.; Zhu, C.; Sun, T.; Zhou, Z.; Hu, Z.; Xia, X.; Zhou, J.; et al. HY5 Functions as a Systemic Signal by Integrating BRC1-Dependent Hormone Signaling in Tomato Bud Outgrowth. Proc. Natl. Acad. Sci. USA 2023, 120, e2301879120. [Google Scholar] [CrossRef] [PubMed]
- Casati, P.; Walbot, V. Gene Expression Profiling in Response to Ultraviolet Radiation in Maize Genotypes with Varying Flavonoid Content. Plant Physiol. 2003, 132, 1739–1754. [Google Scholar] [CrossRef] [PubMed]
- Kolb, C.A.; Käser, M.A.; Kopecký, J.; Zotz, G.; Riederer, M.; Pfündel, E.E. Effects of Natural Intensities of Visible and Ultraviolet Radiation on Epidermal Ultraviolet Screening and Photosynthesis in Grape Leaves. Plant Physiol. 2001, 127, 863–875. [Google Scholar] [CrossRef] [PubMed]
- Guidi, L.; Degl’Innocenti, E.; Remorini, D.; Biricolti, S.; Fini, A.; Ferrini, F.; Nicese, F.P.; Tattini, M. The Impact of UV-Radiation on the Physiology and Biochemistry of Ligustrum Vulgare Exposed to Different Visible-Light Irradiance. Environ. Exp. Bot. 2011, 70, 88–95. [Google Scholar] [CrossRef]
- Schreiner, M.; Mewis, I.; Huyskens-Keil, S.; Jansen, M.A.K.; Zrenner, R.; Winkler, J.B.; O’Brien, N.; Krumbein, A. UV-B-Induced Secondary Plant Metabolites—Potential Benefits for Plant and Human Health. Crit. Rev. Plant Sci. 2012, 31, 229–240. [Google Scholar] [CrossRef]
- Zhao, Q.; Wang, Z.; Wang, G.; Cao, F.; Yang, X.; Zhao, H.; Zhai, J. Effects of UVA on Flavonol Accumulation in Ginkgo biloba. Forests 2024, 15, 909. [Google Scholar] [CrossRef]
- Hashim, M.; Ahmad, B.; Drouet, S.; Hano, C.; Abbasi, B.H.; Anjum, S. Comparative Effects of Different Light Sources on the Production of Key Secondary Metabolites in Plants In Vitro Cultures. Plants 2021, 10, 1521. [Google Scholar] [CrossRef]
- Neugart, S.; Schreiner, M. UVB and UVA as Eustressors in Horticultural and Agricultural Crops. Sci. Hortic. 2018, 234, 370–381. [Google Scholar] [CrossRef]
- Verdaguer, D.; Jansen, M.A.K.; Llorens, L.; Morales, L.O.; Neugart, S. UV-A Radiation Effects on Higher Plants: Exploring the Known Unknown. Plant Sci. 2017, 255, 72–81. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, H.; Mei, Y.; Li, Q.; Bai, Y.; Yu, H.; Xu, X.; Ma, J.; Wu, Y.; Yang, Z. Integrated Transcriptome and Metabolome Analysis Reveals an Essential Role for Auxin in Hypocotyl Elongation during End-of-Day Far-Red Treatment of Cucurbita moschata (Duch. Ex Lam.). Agronomy 2021, 11, 853. [Google Scholar] [CrossRef]
- Ahmed, M.; Eun, J.B. Flavonoids in fruits and vegetables after thermal and nonthermal processing: A review. Crit. Rev. Food Sci. Nutr. 2017, 58, 3159–3188. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Xu, J.; Wang, W.; Rajashekar, C.B. The Effect of Supplemental Blue, Red and Far-Red Light on the Growth and the Nutritional Quality of Red and Green Leaf Lettuce. Am. J. Plant Sci. 2019, 10, 2219–2235. [Google Scholar] [CrossRef]
- Franklin, K.A.; Whitelam, G.C. Phytochromes and Shade-Avoidance Responses in Plants. Ann. Bot. 2005, 96, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Bernal, M.; Verdaguer, D.; Badosa, J.; Abadía, A.; Llusià, J.; Peñuelas, J.; Núñez-Olivera, E.; Llorens, L. Effects of Enhanced UV Radiation and Water Availability on Performance, Biomass Production and Photoprotective Mechanisms of Laurus Nobilis Seedlings. Environ. Exp. Bot. 2015, 109, 264–275. [Google Scholar] [CrossRef]
- Gao, M.; Li, Y.; Jiang, H.; He, R.; Shi, R.; Song, S.; Liu, H. UVA-Radiation Exposure of Different Durations Promoted the Growth, Phytochemicals and Glucosinolate Biosynthesis of Chinese Kale. Int. J. Mol. Sci. 2022, 23, 7619. [Google Scholar] [CrossRef]
- Dou, H.; Niu, G.; Gu, M.; Masabni, J. Far-red Light Effectively Accelerates the Accumulation of Biomass, Leaf Blade Anthocyanin, Total Phenolics, and Flavonoids in Basil Plant. China Veg. 2022, 5, 81–88. [Google Scholar]
- Demotes-Mainard, S.; Péron, T.; Corot, A.; Bertheloot, J.; Gourrierec, J.L.; Pelleschi-Travier, S.; Crespel, L.; Morel, P.; Huché-Thélier, L.; Boumaza, R.; et al. Plant Responses to Red and Far-Red Lights, Applications in Horticulture. Environ. Exp. Bot. 2016, 121, 4–21. [Google Scholar] [CrossRef]
- Štroch, M.; Materová, Z.; Vrábl, D.; Karlický, V.; Šigut, L.; Nezval, J.; Špunda, V. Protective Effect of UV-A Radiation during Acclimation of the Photosynthetic Apparatus to UV-B Treatment. Plant Physiol. Biochem. 2015, 96, 90–96. [Google Scholar] [CrossRef]
- Hu, Y.; He, R.; Ju, J.; Zhang, S.; He, X.; Li, Y.; Liu, X.; Liu, H. Effects of Substituting B with FR and UVA at Different Growth Stages on the Growth and Quality of Lettuce. Agronomy 2023, 13, 2547. [Google Scholar] [CrossRef]
- Chen, Y.; Li, T.; Yang, Q.; Zhang, Y.; Zou, J.; Bian, Z.; Wen, X. UVA Radiation Is Beneficial for Yield and Quality of Indoor Cultivated Lettuce. Front. Plant Sci. 2019, 10, 1563. [Google Scholar] [CrossRef]
- Kim, D.; Son, J.E. Adding Far-Red to Red, Blue Supplemental Light-Emitting Diode Interlighting Improved Sweet Pepper Yield but Attenuated Carotenoid Content. Front. Plant Sci. 2022, 13, 938199. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Kubota, C. Effects of Supplemental Light Quality on Growth and Phytochemicals of Baby Leaf Lettuce. Environ. Exp. Bot. 2009, 67, 59–64. [Google Scholar] [CrossRef]
- Kong, Y.; Nemali, K. Blue and Far-Red Light Affect Area and Number of Individual Leaves to Influence Vegetative Growth and Pigment Synthesis in Lettuce. Front. Plant Sci. 2021, 12, 667407. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, C.R.; Britz, S.J. Effect of Supplemental Ultraviolet Radiation on the Carotenoid and Chlorophyll Composition of Green House-Grown Leaf Lettuce (Lactuca sativa L.) Cultivars. J. Food Compos. Anal. 2006, 19, 637–644. [Google Scholar] [CrossRef]
- Guidi, L.; Brunetti, C.; Fini, A.; Agati, G.; Ferrini, F.; Gori, A.; Tattini, M. UV Radiation Promotes Flavonoid Biosynthesis, While Negatively Affecting the Biosynthesis and the de-Epoxidation of Xanthophylls: Consequence for Photoprotection? Environ. Exp. Bot. 2016, 127, 14–25. [Google Scholar] [CrossRef]
- Klem, K.; Holub, P.; Štroch, M.; Nezval, J.; Špunda, V.; Tříska, J.; Jansen, M.A.K.; Robson, T.M.; Urban, O. Ultraviolet and Photosynthetically Active Radiation Can Both Induce Photoprotective Capacity Allowing Barley to Overcome High Radiation Stress. Plant Physiol. Biochem. 2015, 93, 74–83. [Google Scholar] [CrossRef]
- Semenova, N.A.; Smirnov, A.A.; Ivanitskikh, A.S.; Izmailov, A.Y.; Dorokhov, A.S.; Proshkin, Y.A.; Yanykin, D.V.; Sarimov, R.R.; Gudkov, S.V.; Chilingaryan, N.O. Impact of Ultraviolet Radiation on the Pigment Content and Essential Oil Accumulation in Sweet Basil (Ocimum basilicum L.). Appl. Sci. 2022, 12, 7190. [Google Scholar] [CrossRef]
- Viršilė, A.; Laužikė, K.; Sutulienė, R.; Brazaitytė, A.; Kudirka, G.; Samuolienė, G. Distinct Impacts of UV-A Light Wavelengths on Nutraceutical and Mineral Contents in Green and Purple Basil Cultivated in a Controlled Environment. Horticulturae 2023, 9, 1168. [Google Scholar] [CrossRef]
- Zhang, X.; Zheng, X.; Yang, R.; Wang, Q.; Bai, L.; Gong, D.; Han, Y.; Prusky, D.; Bi, Y. UV-C Radiation Promoted Flavonoid Synthesis during Early Healing in Potato Tuber Wounds with the Possible Involvement of ABA and Related Transcription Factor Regulation. Postharvest Biol. Technol. 2024, 209, 112683. [Google Scholar] [CrossRef]
- Reid, J.B.; Hasan, O.; Ross, J.J. Internode Length in Pisum. Gibberellins and the Response to Far-Red-Rich Light. J. Plant Physiol. 1990, 137, 46–52. [Google Scholar] [CrossRef]
- Murase, K.; Hirano, Y.; Sun, T.; Hakoshima, T. Gibberellin-Induced DELLA Recognition by the Gibberellin Receptor GID1. Nature 2008, 456, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Vriezen, W.H.; Van Der Straeten, D.; Harberd, N.P. Ethylene Regulates Arabidopsis Development via the Modulation of DELLA Protein Growth Repressor Function. Plant Cell 2003, 15, 2816–2825. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Harberd, N.P. Auxin Promotes Arabidopsis Root Growth by Modulating Gibberellin Response. Nature 2003, 421, 740–743. [Google Scholar] [CrossRef]
- Thomas, T.; Hare, P.; van Staden, J. Phytochrome and Cytokinin Responses. Plant Growth Regul. 1997, 23, 105–122. [Google Scholar] [CrossRef]
- Halliday, K.J.; Fankhauser, C. Phytochrome-hormonal Signalling Networks. New Phytol. 2003, 157, 449–463. [Google Scholar] [CrossRef]
- Valio, I.F.M.; Schwabe, W.W. Correlative Growth in Seedlings of Phaseolus vulgaris L.: Inhibition of Stem Growth by the Primary Leaves. Ann. Bot. 1978, 42, 263–268. [Google Scholar] [CrossRef]
- Leite, V.M.; Rosolem, C.A.; Rodrigues, J.D. Gibberellin and Cytokinin Effects on Soybean Growth. Sci. Agric. 2003, 60, 537–541. [Google Scholar] [CrossRef]
- Smets, R.; Le, J.; Prinsen, E.; Verbelen, J.-P.; Van Onckelen, H.A. Cytokinin-Induced Hypocotyl Elongation in Light-Grown Arabidopsis Plants with Inhibited Ethylene Action or Indole-3-Acetic Acid Transport. Planta 2005, 221, 39–47. [Google Scholar] [CrossRef]
- Zurek, D.M.; Rayle, D.L.; McMorris, T.C.; Clouse, S.D. Investigation of Gene Expression, Growth Kinetics, and Wall Extensibility during Brassinosteroid-Regulated Stem Elongation. Plant Physiol. 1994, 104, 505–513. [Google Scholar] [CrossRef]
- Zhang, H. The Mechanism of Brassinosteroid and HY5 Induced Gibberellin Regulating Leaf and Stem Elongation in Tomato under Low Proportion of Red Light and Far Red Light. Master’s Thesis, Zhejiang University, Hangzhou, China, 2019; p. 79. [Google Scholar]
- Li, Z.; He, Y. Roles of Brassinosteroids in Plant Reproduction. Int. J. Mol. Sci. 2020, 21, 872. [Google Scholar] [CrossRef]
- Yuldashev, R.; Avalbaev, A.; Bezrukova, M.; Vysotskaya, L.; Khripach, V.; Shakirova, F. Cytokinin Oxidase Is Involved in the Regulation of Cytokinin Content by 24-Epibrassinolide in Wheat Seedlings. Plant Physiol. Biochem. 2012, 55, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ballaré, C.L. Light Regulation of Plant Defense. Annu. Rev. Plant Biol. 2014, 65, 335–363. [Google Scholar] [CrossRef] [PubMed]
- Cai, K.; Dong, T.; Xu, T. The physiological roles and resistance control in stress environment of jasmonates. Ecol. Environ. Sci. 2006, 15, 397–404. [Google Scholar]
- Wang, L.; Zhan, J.; Huang, W. Salicylic Acid and Response to Stress in Plants. Plant Physiol. J. 2002, 38, 619–624. [Google Scholar]
- Moreno, J.E.; Tao, Y.; Chory, J.; Ballaré, C.L. Ecological Modulation of Plant Defense via Phytochrome Control of Jasmonate Sensitivity. Proc. Natl. Acad. Sci. USA 2009, 106, 4935–4940. [Google Scholar] [CrossRef]
- Hou, X.; Lee, L.Y.C.; Xia, K.; Yan, Y.; Yu, H. DELLAs Modulate Jasmonate Signaling via Competitive Binding to JAZs. Dev. Cell 2010, 19, 884–894. [Google Scholar] [CrossRef]
- Russo, D. Flavonoids and the Structure-Antioxidant Activity Relationship. J. Pharmacogn. Nat. Prod. 2018, 4, e109. [Google Scholar] [CrossRef]
- Emiliani, J.; Grotewold, E.; Ferreyra, M.L.F.; Casati, P. Flavonols Protect Arabidopsis Plants against UV-B Deleterious Effects. Mol. Plant 2013, 6, 1376–1379. [Google Scholar] [CrossRef]
- Ryan, K.G.; Markham, K.R.; Bloor, S.J.; Bradley, J.M.; Mitchell, K.A.; Jordan, B.R. UVB Radiation Induced Increase in Quercetin: Kaempferol Ratio in Wild-Type and Transgenic Lines of Petunia. Photochem. Photobiol. 2008, 68, 323–330. [Google Scholar] [CrossRef]
- Agati, G.; Biricolti, S.; Guidi, L.; Ferrini, F.; Fini, A.; Tattini, M. The Biosynthesis of Flavonoids Is Enhanced Similarly by UV Radiation and Root Zone Salinity in L. Vulgare Leaves. J. Plant Physiol. 2011, 168, 204–212. [Google Scholar] [CrossRef]
- Righini, S.; Rodriguez, E.J.; Berosich, C.; Grotewold, E.; Casati, P.; Ferreyra, M.L.F. Apigenin Produced by Maize Flavone Synthase I and II Protects Plants against UV-B-induced Damage. Plant Cell Environ. 2019, 42, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wang, M.-H. Ultraviolet A-Specific Induction of Anthocyanin Biosynthesis and PAL Expression in Tomato (Solanum lycopersicum L.). Plant Growth Regul. 2010, 62, 1–8. [Google Scholar] [CrossRef]
- Castagna, A.; Dall’Asta, C.; Chiavaro, E.; Galaverna, G.; Ranieri, A. Effect of Post-Harvest UV-B Irradiation on Polyphenol Profile and Antioxidant Activity in Flesh and Peel of Tomato Fruits. Food Bioprocess Technol. 2014, 7, 2241–2250. [Google Scholar] [CrossRef]
- Stapleton, A.E.; Walbot, V. Flavonoids Can Protect Maize DNA from the Induction of Ultraviolet Radiation Damage. Plant Physiol. 1994, 105, 881–889. [Google Scholar] [CrossRef]
- Koes, R.E.; Spelt, C.E.; Mol, J.N.M. The Chalcone Synthase Multigene Family of Petunia hybrida (V30): Differential, Light-Regulated Expression during Flower Development and UV Light Induction. Plant Mol. Biol. 1989, 12, 213–225. [Google Scholar] [CrossRef]
- Shui, X.; Chen, T.; Qian, M.; Peng, J.; Du, J.; Zhou, K.; Liu, F. The Antioxidant Response Mechanism of Flavonoids in ‘Tainong 1’ Mango Pulp under Enhanced UV-B Radiation. Cogent Food Agric. 2024, 10, 2301273. [Google Scholar] [CrossRef]
- Liu, J.; Jiang, H.; Huang, Y.; Zhong, L.; Xu, Q.; Yang, Q.; Liu, S.; Wei, X.; Liang, Y.; Chai, S. Combined Analysis of Metagenome and Transcriptome Revealed the Adaptive Mechanism of Different Golden Camellia Species in Karst Regions. Front. Plant Sci. 2023, 14, 1180472. [Google Scholar] [CrossRef]
- Ding, X.; Miao, C.; Li, R.; He, L.; Zhang, H.; Jin, H.; Cui, J.; Wang, H.; Zhang, Y.; Lu, P.; et al. Artificial Light for Improving Tomato Recovery Following Grafting: Transcriptome and Physiological Analyses. Int. J. Mol. Sci. 2023, 24, 15928. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


| Treatment | Chlorophyll a (mg g−1) | Chlorophyll b (mg g−1) | Chlorophyll (mg g−1) | Carotenoid (mg g−1) |
|---|---|---|---|---|
| CK | 0.907 ± 0.003 a | 0.323 ± 0.002 b | 1.23 ± 0.004 a | 0.17 ± 0.001 a |
| FR | 0.81 ± 0.023 b | 0.285 ± 0.007 c | 1.095 ± 0.029 b | 0.147 ± 0.005 b |
| UVA | 0.914 ± 0.017 a | 0.332 ± 0.004 a | 1.247 ± 0.02 a | 0.165 ± 0.004 a |
| Treatment | Photosynthetic Photon Flux (PPF) (μmol s−1) | Photosynthetic Photon Flux Density (PPFD) (μmol m−2 s−1) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R | B | G | FR | UVA | Total PPF | R | B | G | FR | UVA | Total PPFD | |
| CK | 529 | 225 | 94 | 0.06 | 0.05 | 848.1 | 125.4 | 67.9 | 28.4 | 5.57 | 0.13 | 221.6 |
| FR | 530 | 224 | 93 | 106 | 0.02 | 953 | 125.4 | 68 | 28.5 | 27.7 | 0.09 | 221.9 |
| UVA | 531 | 232 | 93 | 0.11 | 17 | 873.1 | 125.2 | 70.7 | 28.2 | 5.4 | 1.85 | 224.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, D.; Shen, H.; Yu, L.; Zhu, K.; Zhang, Y.; Wu, S.; Chang, L.; Ding, X.; Jiang, Y. Transcriptomic and Metabolomic Insights into Plant Hormone Modulation and Secondary Metabolite Accumulation in Basil Under Far-Red and Ultraviolet-A Light. Int. J. Mol. Sci. 2025, 26, 3758. https://doi.org/10.3390/ijms26083758
Li D, Shen H, Yu L, Zhu K, Zhang Y, Wu S, Chang L, Ding X, Jiang Y. Transcriptomic and Metabolomic Insights into Plant Hormone Modulation and Secondary Metabolite Accumulation in Basil Under Far-Red and Ultraviolet-A Light. International Journal of Molecular Sciences. 2025; 26(8):3758. https://doi.org/10.3390/ijms26083758
Chicago/Turabian StyleLi, Dandan, Haibin Shen, Lishu Yu, Kaili Zhu, Yongxue Zhang, Shaofang Wu, Liying Chang, Xiaotao Ding, and Yuping Jiang. 2025. "Transcriptomic and Metabolomic Insights into Plant Hormone Modulation and Secondary Metabolite Accumulation in Basil Under Far-Red and Ultraviolet-A Light" International Journal of Molecular Sciences 26, no. 8: 3758. https://doi.org/10.3390/ijms26083758
APA StyleLi, D., Shen, H., Yu, L., Zhu, K., Zhang, Y., Wu, S., Chang, L., Ding, X., & Jiang, Y. (2025). Transcriptomic and Metabolomic Insights into Plant Hormone Modulation and Secondary Metabolite Accumulation in Basil Under Far-Red and Ultraviolet-A Light. International Journal of Molecular Sciences, 26(8), 3758. https://doi.org/10.3390/ijms26083758






