Cis-Regulation of the CFTR Gene in Pancreatic Cells
Abstract
1. Introduction
2. Results
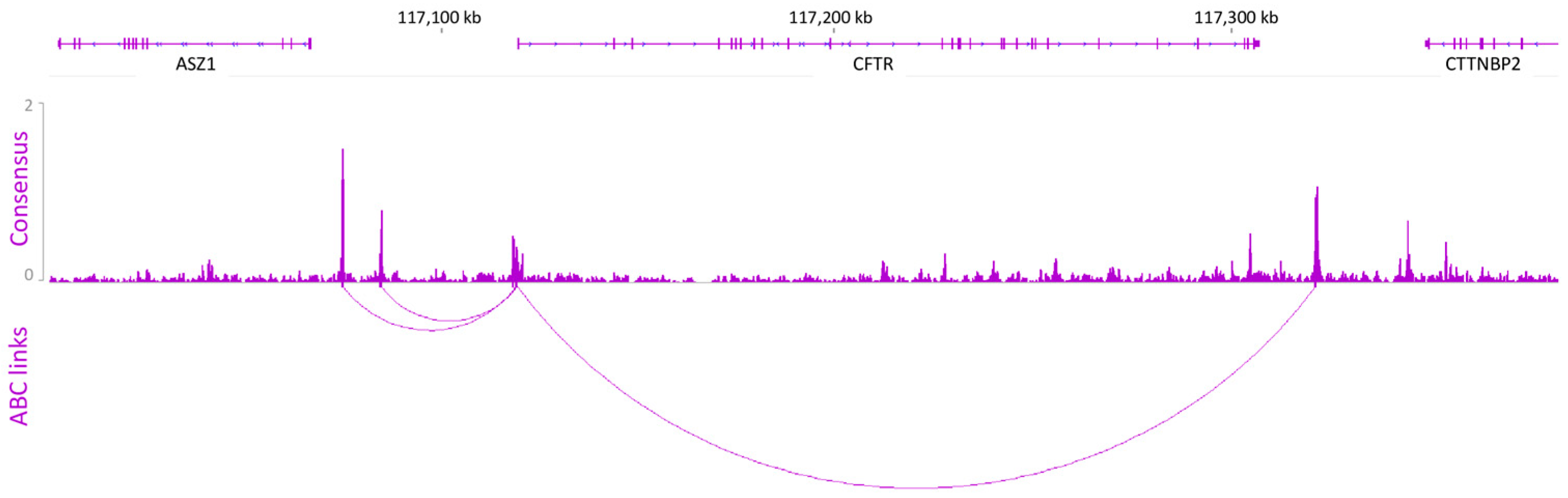
2.1. Prediction Model Identifying Putative Regulatory Elements
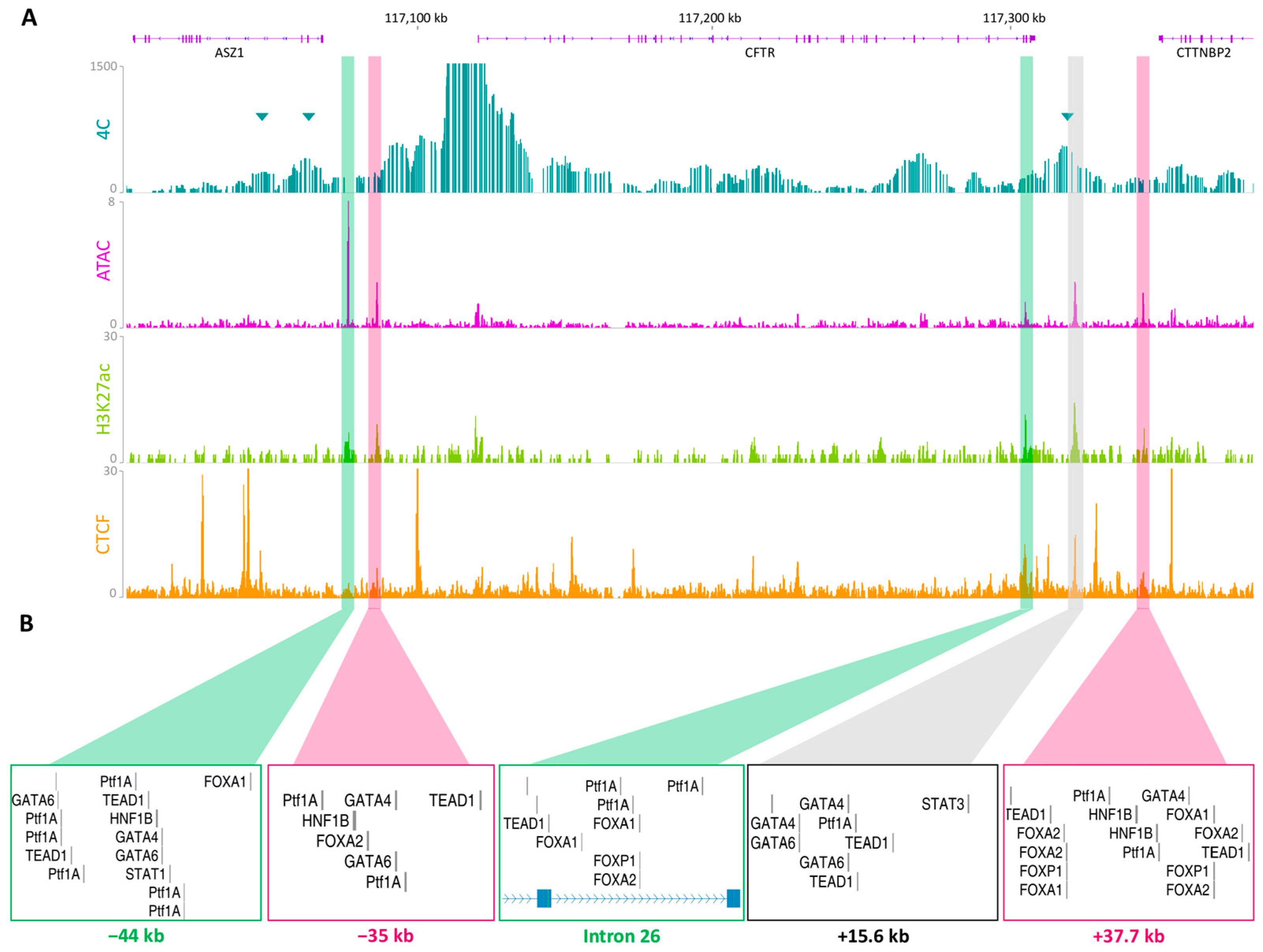
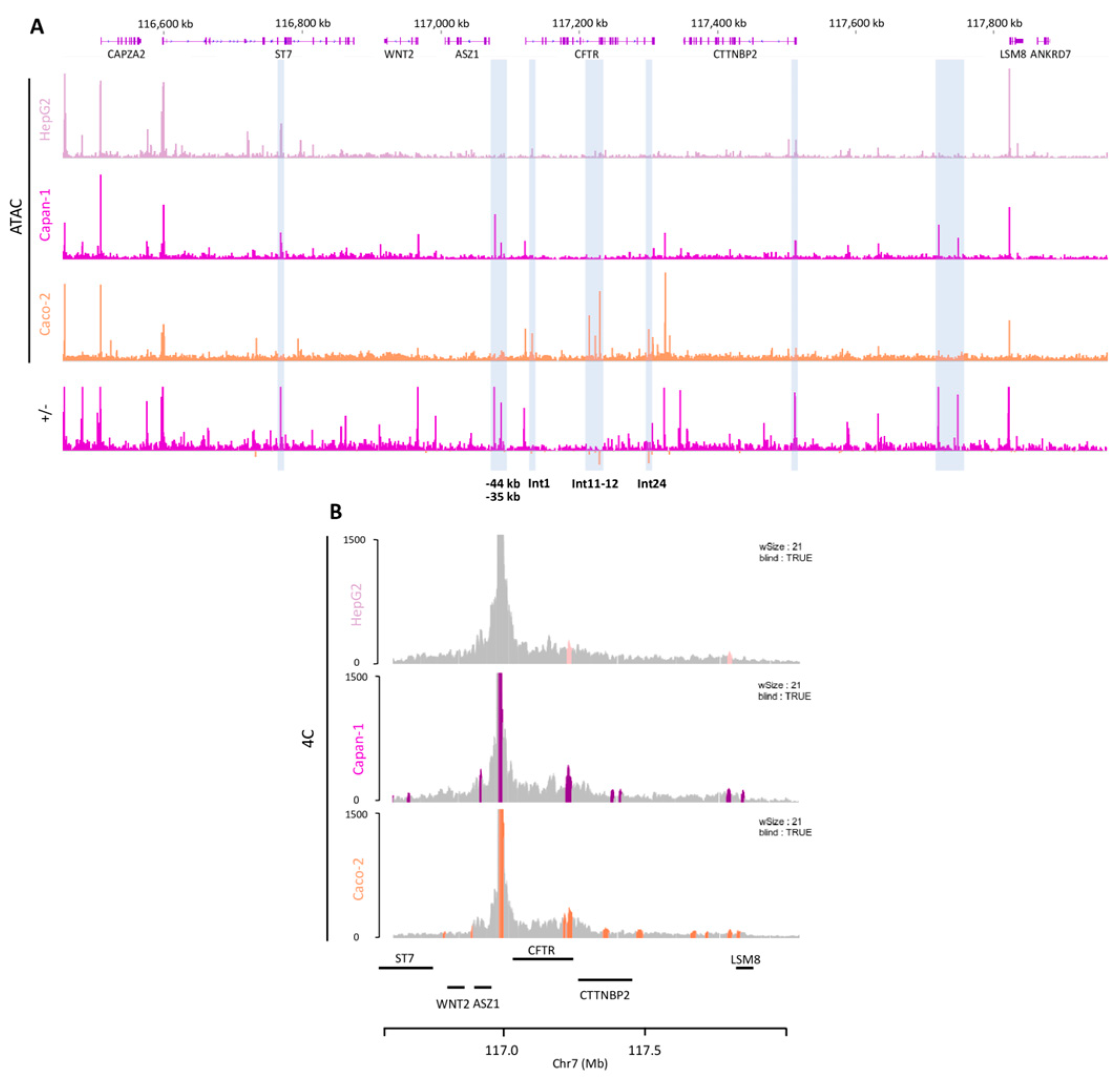
2.2. Chromatin Organization Confirms the Presence of Active Regulatory Regions
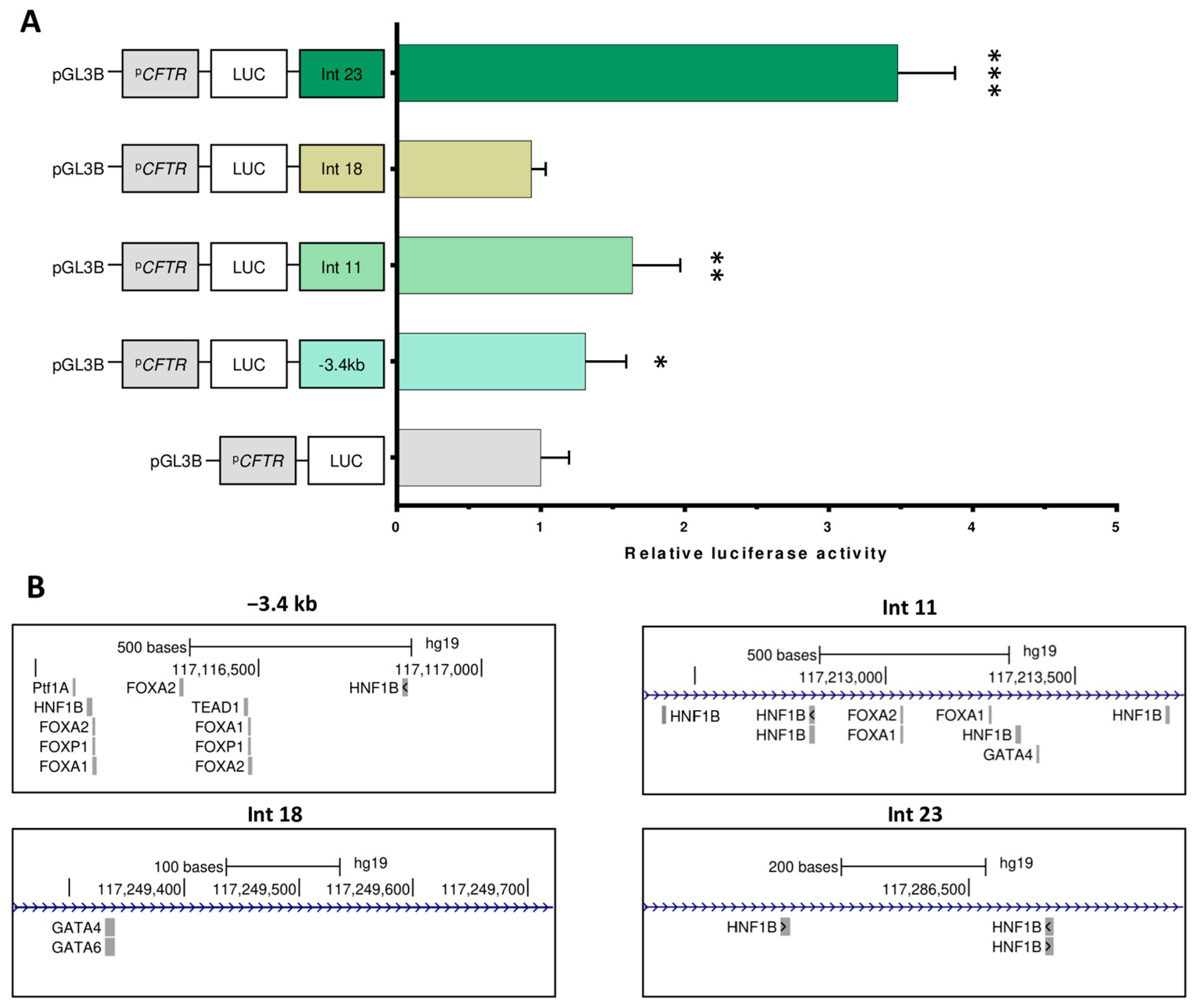
2.3. Luciferase Assays Characterize cCRE Activity
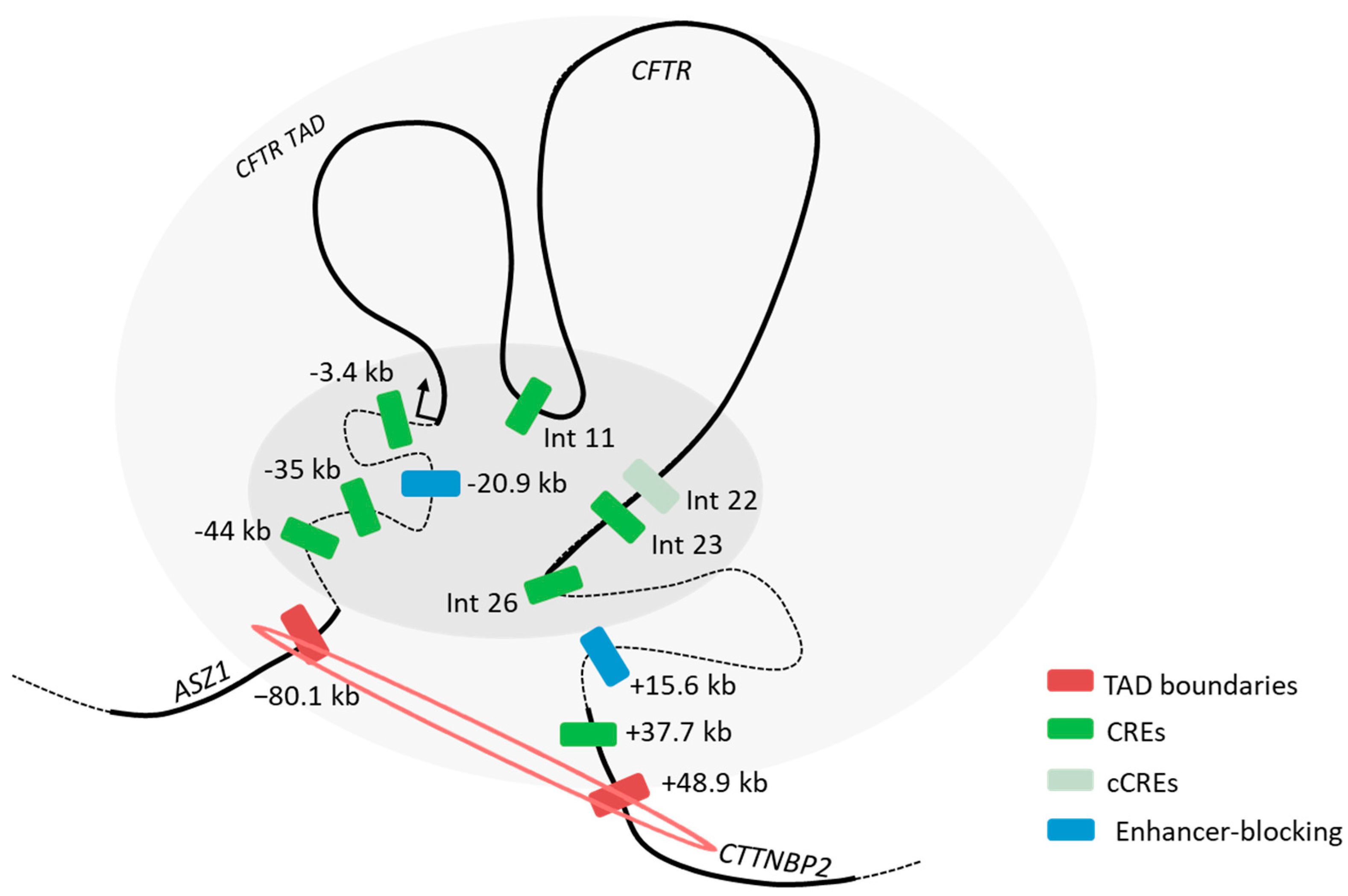
2.4. Endogenous Assay Validated the Enhancer Effect of the −44 kb Region
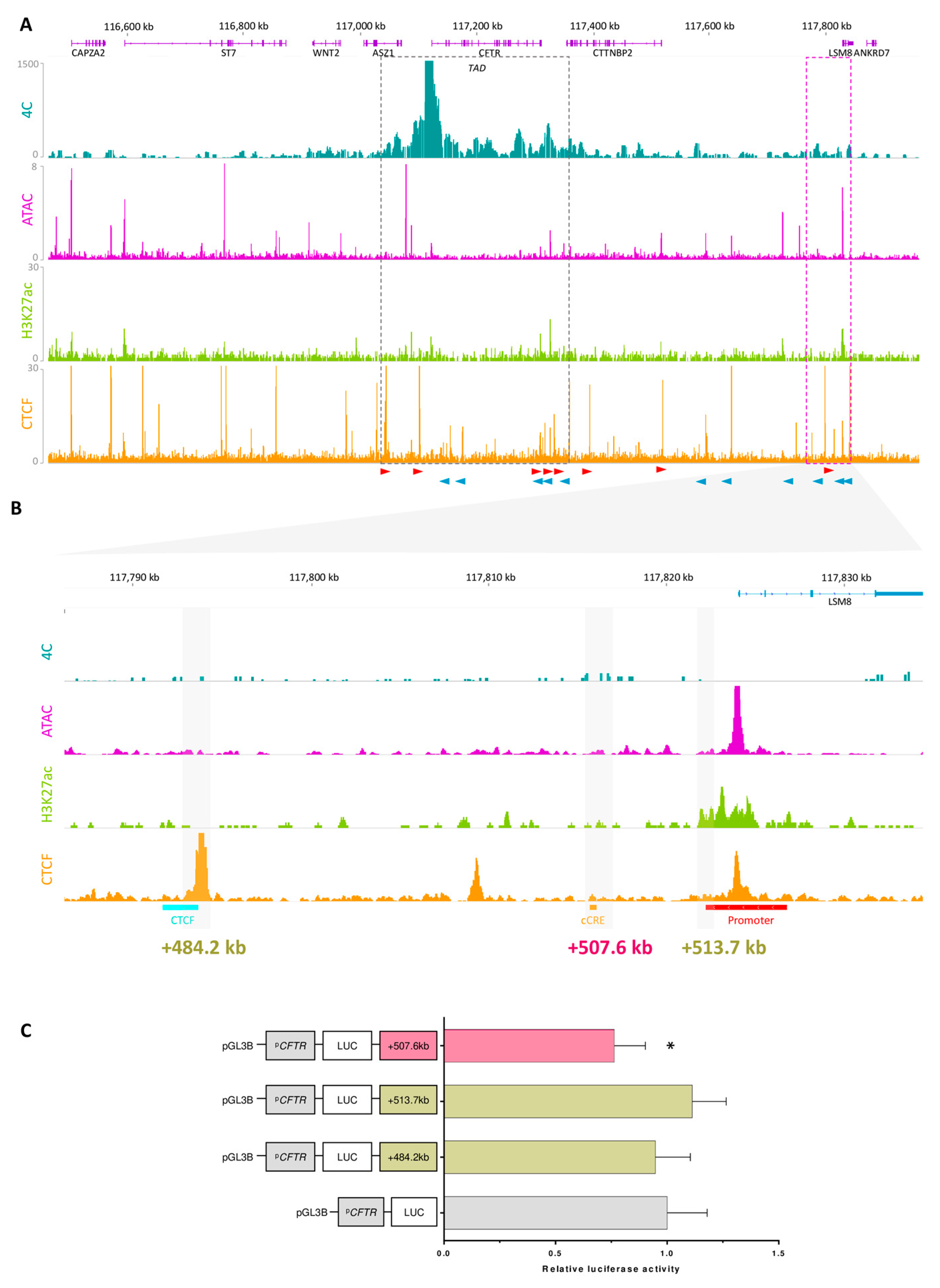
2.5. Exploring the Landscape Outside the TAD
2.6. Cell Type Specificity
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. ATAC-Seq
4.3. CUT&RUN-Seq
4.4. 4C-Seq
4.5. ABC Model
4.6. Plasmid Construction
4.7. Reporter Gene Assay
4.8. CRISPR/Cas9 Deletion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 4C | circular chromosome conformation capture |
| ABC | activity-by-contact |
| ATAC | assay for transposase-accessible chromatin |
| CBAVD | congenital bilateral absence of vas deferens |
| CF | cystic fibrosis |
| CFTR | cystic fibrosis transmembrane conductance regulator |
| CFTR-RD | CFTR-related diseases |
| cCRE | candidate cis-regulatory elements |
| CRE | cis-regulatory elements |
| CRISPR | clustered regularly interspaced short palindromic repeats |
| CTCF | CCCTC-binding factor |
| DHS | DNase I hypersensitive site |
| CUT&RUN | cleavage under targets and release using nuclease |
| gRNA | guide RNA |
| OCR | open chromatin region |
| PTM | post-translational modifications |
| TAD | topologically associating domain |
| TF | transcription factor |
| TSS | transcription start site |
References
- The ENCODE Project Consortium; Abascal, F.; Acosta, R.; Addleman, N.J.; Adrian, J.; Afzal, V.; Aken, B.; Ai, R.; Akiyama, J.A.; Jammal, O.A.; et al. Perspectives on ENCODE. Nature 2020, 583, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.E.; Purcaro, M.J.; Pratt, H.E.; Epstein, C.B.; Shoresh, N.; Adrian, J.; Kawli, T.; Davis, C.A.; Dobin, A.; Kaul, R.; et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 2020, 583, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Gasperini, M.; Tome, J.M.; Shendure, J. Towards a comprehensive catalogue of validated and target-linked human enhancers. Nat. Rev. Genet. 2020, 21, 292–310. [Google Scholar] [CrossRef]
- Lopez Soriano, V.; Dueñas Rey, A.; Mukherjee, R.; Genomics England Research Consortium; Coppieters, F.; Bauwens, M.; Willaert, A.; De Baere, E. Multi-omics analysis in human retina uncovers ultraconserved cis-regulatory elements at rare eye disease loci. Nat. Commun. 2024, 15, 1600. [Google Scholar] [CrossRef]
- Zaugg, J.B.; Sahlén, P.; Andersson, R.; Alberich-Jorda, M.; de Laat, W.; Deplancke, B.; Ferrer, J.; Mandrup, S.; Natoli, G.; Plewczynski, D.; et al. Current challenges in understanding the role of enhancers in disease. Nat. Struct. Mol. Biol. 2022, 29, 1148–1158. [Google Scholar] [CrossRef]
- The 100,000 Genomes Project Pilot Investigators. 100,000 Genomes Pilot on Rare-Disease Diagnosis in Health Care—Preliminary Report. N. Engl. J. Med. 2021, 385, 1868–1880. [Google Scholar] [CrossRef]
- Pang, B.; van Weerd, J.H.; Hamoen, F.L.; Snyder, M.P. Identification of non-coding silencer elements and their regulation of gene expression. Nat. Rev. Mol. Cell Biol. 2023, 24, 383–395. [Google Scholar] [CrossRef]
- El-Seedy, A.; Ladeveze, V. CFTR complex alleles and phenotypic variability in cystic fibrosis disease. Cell Mol. Biol. 2024, 70, 244–260. [Google Scholar] [CrossRef] [PubMed]
- Sermet-Gaudelus, I.; Girodon, E.; Vermeulen, F.; Solomon, G.M.; Melotti, P.; Graeber, S.Y.; Bronsveld, I.; Rowe, S.M.; Wilschanski, M.; Tümmler, B.; et al. ECFS standards of care on CFTR-related disorders: Diagnostic criteria of CFTR dysfunction. J. Cyst. Fibros. 2022, 21, 922–936. [Google Scholar] [CrossRef]
- Simmonds, N.J.; Southern, K.W.; De Wachter, E.; De Boeck, K.; Bodewes, F.; Mainz, J.G.; Middleton, P.G.; Schwarz, C.; Vloeberghs, V.; Wilschanski, M.; et al. ECFS standards of care on CFTR-related disorders: Identification and care of the disorders. J. Cyst. Fibros. 2024, 23, 590–602. [Google Scholar] [CrossRef]
- Gosalia, N.; Neems, D.; Kerschner, J.L.; Kosak, S.T.; Harris, A. Architectural proteins CTCF and cohesin have distinct roles in modulating the higher order structure and expression of the CFTR locus. Nucleic Acids Res. 2014, 42, 9612–9622. [Google Scholar] [CrossRef]
- Smith, E.M.; Lajoie, B.R.; Jain, G.; Dekker, J. Invariant TAD Boundaries Constrain Cell-Type-Specific Looping Interactions between Promoters and Distal Elements around the CFTR Locus. Am. J. Hum. Genet. 2016, 98, 185–201. [Google Scholar] [CrossRef]
- Smith, A.N.; Barth, M.L.; McDowell, T.L.; Moulin, D.S.; Nuthall, H.N.; Hollingsworth, M.A.; Harris, A. A regulatory element in intron 1 of the cystic fibrosis transmembrane conductance regulator gene. J. Biol. Chem. 1996, 271, 9947–9954. [Google Scholar] [CrossRef]
- Blotas, C.; Férec, C.; Moisan, S. Tissue-Specific Regulation of CFTR Gene Expression. Int. J. Mol. Sci. 2023, 24, 10678. [Google Scholar] [CrossRef]
- Becq, F.; Fanjul, M.; Merten, M.; Figarella, C.; Hollande, E.; Gola, M. Possible regulation of CFTR-chloride channels by membrane-bound phosphatases in pancreatic duct cells. FEBS Lett. 1993, 327, 337–342. [Google Scholar] [CrossRef]
- Fulco, C.P.; Nasser, J.; Jones, T.R.; Munson, G.; Bergman, D.T.; Subramanian, V.; Grossman, S.R.; Anyoha, R.; Doughty, B.R.; Patwardhan, T.A.; et al. Activity-by-contact model of enhancer–promoter regulation from thousands of CRISPR perturbations. Nat. Genet. 2019, 51, 1664–1669. [Google Scholar] [CrossRef]
- Nasser, J.; Bergman, D.T.; Fulco, C.P.; Guckelberger, P.; Doughty, B.R.; Patwardhan, T.A.; Jones, T.R.; Nguyen, T.H.; Ulirsch, J.C.; Lekschas, F.; et al. Genome-wide enhancer maps link risk variants to disease genes. Nature 2021, 593, 238–243. [Google Scholar] [CrossRef]
- Moisan, S.; Berlivet, S.; Ka, C.; Le Gac, G.; Dostie, J.; Férec, C. Analysis of long-range interactions in primary human cells identifies cooperative CFTR regulatory elements. Nucleic Acids Res. 2016, 44, 2564–2576. [Google Scholar] [CrossRef]
- Nuthall, H.N.; Moulin, D.S.; Huxley, C.; Harris, A. Analysis of DNase-I-hypersensitive sites at the 3’ end of the cystic fibrosis transmembrane conductance regulator gene (CFTR). Biochem. J. 1999, 341, 601–611. [Google Scholar] [CrossRef]
- Blackledge, N.P.; Carter, E.J.; Evans, J.R.; Lawson, V.; Rowntree, R.K.; Harris, A. CTCF mediates insulator function at the CFTR locus. Biochem. J. 2007, 408, 267–275. [Google Scholar] [CrossRef]
- Simonis, M.; Klous, P.; Splinter, E.; Moshkin, Y.; Willemsen, R.; de Wit, E.; van Steensel, B.; de Laat, W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture–on-chip (4C). Nat. Genet. 2006, 38, 1348–1354. [Google Scholar] [CrossRef]
- Krijger, P.H.L.; Geeven, G.; Bianchi, V.; Hilvering, C.R.E.; de Laat, W. 4C-seq from beginning to end: A detailed protocol for sample preparation and data analysis. Methods 2020, 170, 17–32. [Google Scholar] [CrossRef]
- Rauluseviciute, I.; Riudavets-Puig, R.; Blanc-Mathieu, R.; Castro-Mondragon, J.A.; Ferenc, K.; Kumar, V.; Lemma, R.B.; Lucas, J.; Chèneby, J.; Baranasic, D.; et al. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2024, 52, D174–D182. [Google Scholar] [CrossRef]
- Ott, C.; Blackledge, N.P.; Kerschner, J.L.; Leir, S.-H.; Crawford, G.E.; Cotton, C.U.; Harris, A. Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc. Natl. Acad. Sci. USA 2009, 106, 19934–19939. [Google Scholar] [CrossRef]
- Martinez-Ara, M.; Comoglio, F.; Steensel, B.V. Large-scale analysis of the integration of enhancer-enhancer signals by promoters. eLife 2024, 12, PR91994. [Google Scholar] [CrossRef]
- Osterwalder, M.; Barozzi, I.; Tissières, V.; Fukuda-Yuzawa, Y.; Mannion, B.J.; Afzal, S.Y.; Lee, E.A.; Zhu, Y.; Plajzer-Frick, I.; Pickle, C.S.; et al. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 2018, 554, 239–243. [Google Scholar] [CrossRef]
- Kim, S.; Wysocka, J. Deciphering the multi-scale, quantitative cis-regulatory code. Mol. Cell 2023, 83, 373–392. [Google Scholar] [CrossRef]
- van Mierlo, G.; Pushkarev, O.; Kribelbauer, J.F.; Deplancke, B. Chromatin modules and their implication in genomic organization and gene regulation. Trends Genet. 2023, 39, 140–153. [Google Scholar] [CrossRef]
- McCarthy, V.A.; Ott, C.J.; Phylactides, M.; Harris, A. Interaction of intestinal and pancreatic transcription factors in the regulation of CFTR gene expression. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2009, 1789, 709–718. [Google Scholar] [CrossRef]
- Zhang, Z.; Ott, C.J.; Lewandowska, M.A.; Leir, S.-H.; Harris, A. Molecular mechanisms controlling CFTR gene expression in the airway. J. Cell. Mol. Med. 2012, 16, 1321–1330. [Google Scholar] [CrossRef]
- Collobert, M.; Bocher, O.; Le Nabec, A.; Génin, E.; Férec, C.; Moisan, S. CFTR Cooperative Cis-Regulatory Elements in Intestinal Cells. Int. J. Mol. Sci. 2021, 22, 2599. [Google Scholar] [CrossRef] [PubMed]
- Hung, T.-C.; Kingsley, D.M.; Boettiger, A.N. Boundary stacking interactions enable cross-TAD enhancer–promoter communication during limb development. Nat. Genet. 2024, 56, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Tak, Y.E.; Horng, J.E.; Perry, N.T.; Schultz, H.T.; Iyer, S.; Yao, Q.; Zou, L.S.; Aryee, M.J.; Pinello, L.; Joung, J.K. Augmenting and directing long-range CRISPR-mediated activation in human cells. Nat. Methods 2021, 18, 1075–1081. [Google Scholar] [CrossRef]
- Gheldof, N.; Smith, E.M.; Tabuchi, T.M.; Koch, C.M.; Dunham, I.; Stamatoyannopoulos, J.A.; Dekker, J. Cell-type-specific long-range looping interactions identify distant regulatory elements of the CFTR gene. Nucleic Acids Res. 2010, 38, 4325–4336. [Google Scholar] [CrossRef] [PubMed]
- Rowntree, R.K.; Vassaux, G.; McDowell, T.L.; Howe, S.; McGuigan, A.; Phylactides, M.; Huxley, C.; Harris, A. An element in intron 1 of the CFTR gene augments intestinal expression in vivo. Hum. Mol. Genet. 2001, 10, 1455–1464. [Google Scholar] [CrossRef]
- Chambers, J.A.; Harris, A. Expression of the cystic fibrosis gene and the major pancreatic mucin gene, MUC1, in human ductal epithelial cells. J. Cell Sci. 1993, 105, 417–422. [Google Scholar] [CrossRef]
- Yang, J.H.; Hansen, A.S. Enhancer selectivity in space and time: From enhancer–promoter interactions to promoter activation. Nat. Rev. Mol. Cell Biol. 2024, 25, 574–591. [Google Scholar] [CrossRef]
- Mummey, H.M.; Elison, W.; Korgaonkar, K.; Elgamal, R.M.; Kudtarkar, P.; Griffin, E.; Benaglio, P.; Miller, M.; Jha, A.; Fox, J.E.M.; et al. Single cell multiome profiling of pancreatic islets reveals physiological changes in cell type-specific regulation associated with diabetes risk. bioRxiv 2024. [Google Scholar] [CrossRef]
- Hecker, D.; Behjati Ardakani, F.; Karollus, A.; Gagneur, J.; Schulz, M.H. The adapted Activity-By-Contact model for enhancer–gene assignment and its application to single-cell data. Bioinformatics 2023, 39, btad062. [Google Scholar] [CrossRef]
- Hoellinger, T.; Mestre, C.; Aschard, H.; Le Goff, W.; Foissac, S.; Faraut, T.; Djebali, S. Enhancer/gene relationships: Need for more reliable genome-wide reference sets. Front. Bioinform. 2023, 3, 1092853. [Google Scholar] [CrossRef]
- Ying, P.; Chen, C.; Lu, Z.; Chen, S.; Zhang, M.; Cai, Y.; Zhang, F.; Huang, J.; Fan, L.; Ning, C.; et al. Genome-wide enhancer-gene regulatory maps link causal variants to target genes underlying human cancer risk. Nat. Commun. 2023, 14, 5958. [Google Scholar] [CrossRef]
- Malfait, J.; Wan, J.; Spicuglia, S. Epromoters are new players in the regulatory landscape with potential pleiotropic roles. BioEssays 2023, 45, e2300012. [Google Scholar] [CrossRef]
- Smith, A.N.; Wardle, C.J.C.; Harris, A. Characterization of DNASE I hypersensitive sites in the 120kb 5’ to the CFTR gene. Biochem. Biophys. Res. Commun. 1995, 211, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Batut, P.J.; Bing, X.Y.; Sisco, Z.; Raimundo, J.; Levo, M.; Levine, M.S. Genome organization controls transcriptional dynamics during development. Science 2022, 375, 566–570. [Google Scholar] [CrossRef]
- Blayney, J.W.; Francis, H.; Rampasekova, A.; Camellato, B.; Mitchell, L.; Stolper, R.; Cornell, L.; Babbs, C.; Boeke, J.D.; Higgs, D.R.; et al. Super-enhancers include classical enhancers and facilitators to fully activate gene expression. Cell 2023, 186, 5826–5839.e18. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H.F.; Kotova, E.; Jayaram, S.; Pilz, A.; Romeike, M.; Lackner, A.; Penz, T.; Bock, C.; Leeb, M.; Halbritter, F.; et al. Temporal dissection of an enhancer cluster reveals distinct temporal and functional contributions of individual elements. Mol. Cell 2021, 81, 969–982.e13. [Google Scholar] [CrossRef]
- Thomas, H.F.; Feng, S.; Haslhofer, F.; Huber, M.; García Gallardo, M.; Loubiere, V.; Vanina, D.; Pitasi, M.; Stark, A.; Buecker, C. Enhancer cooperativity can compensate for loss of activity over large genomic distances. Mol. Cell 2025, 85, 362–375.e9. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Armand, E.J.; Yao, Z.; Liu, H.; Bartlett, A.; Behrens, M.M.; Li, Y.E.; Lucero, J.D.; Luo, C.; Nery, J.R.; et al. Robust enhancer-gene regulation identified by single-cell transcriptomes and epigenomes. Cell Genom. 2023, 3, 100342. [Google Scholar] [CrossRef]
- Merkenschlager, M.; Nora, E.P. CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annu. Rev. Genom. Hum. Genet. 2016, 17, 17–43. [Google Scholar] [CrossRef]
- Brosh, R.; Laurent, J.M.; Ordoñez, R.; Huang, E.; Hogan, M.S.; Hitchcock, A.M.; Mitchell, L.A.; Pinglay, S.; Cadley, J.A.; Luther, R.D.; et al. A versatile platform for locus-scale genome rewriting and verification. Proc. Natl. Acad. Sci. USA 2021, 118, e2023952118. [Google Scholar] [CrossRef]
- Xu, Y.; Nipper, M.H.; Dominguez, A.A.; Ye, Z.; Akanuma, N.; Lopez, K.; Deng, J.J.; Arenas, D.; Sanchez, A.; Sharkey, F.E.; et al. Reconstitution of human PDAC using primary cells reveals oncogenic transcriptomic features at tumor onset. Nat. Commun. 2024, 15, 818. [Google Scholar] [CrossRef]
- Corces, M.R.; Trevino, A.E.; Hamilton, E.G.; Greenside, P.G.; Sinnott-Armstrong, N.A.; Vesuna, S.; Satpathy, A.T.; Rubin, A.J.; Montine, K.S.; Wu, B.; et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 2017, 14, 959–962. [Google Scholar] [CrossRef] [PubMed]
- Skene, P.J.; Henikoff, S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 2017, 6, e21856. [Google Scholar] [CrossRef] [PubMed]
- Geeven, G.; Teunissen, H.; de Laat, W.; de Wit, E. peakC: A flexible, non-parametric peak calling package for 4C and Capture-C data. Nucleic Acids Res. 2018, 46, e91. [Google Scholar] [CrossRef]
- Ibrahimi, A.; Velde, G.V.; Reumers, V.; Toelen, J.; Thiry, I.; Vandeputte, C.; Vets, S.; Deroose, C.; Bormans, G.; Baekelandt, V.; et al. Highly Efficient Multicistronic Lentiviral Vectors with Peptide 2A Sequences. Hum. Gene Ther. 2009, 20, 845–860. [Google Scholar] [CrossRef]
- Mangeot, P.E.; Risson, V.; Fusil, F.; Marnef, A.; Laurent, E.; Blin, J.; Mournetas, V.; Massouridès, E.; Sohier, T.J.M.; Corbin, A.; et al. Genome editing in primary cells and in vivo using viral-derived Nanoblades loaded with Cas9-sgRNA ribonucleoproteins. Nat. Commun. 2019, 10, 45. [Google Scholar] [CrossRef]
- Banskota, S.; Raguram, A.; Suh, S.; Du, S.W.; Davis, J.R.; Choi, E.H.; Wang, X.; Nielsen, S.C.; Newby, G.A.; Randolph, P.B.; et al. Engineered virus-like particles for efficient in vivo delivery of therapeutic proteins. Cell 2022, 185, 250–265.e16. [Google Scholar] [CrossRef]
- Sturgill, D.; Wang, L.; Arda, H.E. PancrESS—A meta-analysis resource for understanding cell-type specific expression in the human pancreas. BMC Genom. 2024, 25, 76. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blotas, C.; Le Nabec, A.; Collobert, M.; Bulcaen, M.; Carlon, M.S.; Férec, C.; Moisan, S. Cis-Regulation of the CFTR Gene in Pancreatic Cells. Int. J. Mol. Sci. 2025, 26, 3788. https://doi.org/10.3390/ijms26083788
Blotas C, Le Nabec A, Collobert M, Bulcaen M, Carlon MS, Férec C, Moisan S. Cis-Regulation of the CFTR Gene in Pancreatic Cells. International Journal of Molecular Sciences. 2025; 26(8):3788. https://doi.org/10.3390/ijms26083788
Chicago/Turabian StyleBlotas, Clara, Anaïs Le Nabec, Mégane Collobert, Mattijs Bulcaen, Marianne S. Carlon, Claude Férec, and Stéphanie Moisan. 2025. "Cis-Regulation of the CFTR Gene in Pancreatic Cells" International Journal of Molecular Sciences 26, no. 8: 3788. https://doi.org/10.3390/ijms26083788
APA StyleBlotas, C., Le Nabec, A., Collobert, M., Bulcaen, M., Carlon, M. S., Férec, C., & Moisan, S. (2025). Cis-Regulation of the CFTR Gene in Pancreatic Cells. International Journal of Molecular Sciences, 26(8), 3788. https://doi.org/10.3390/ijms26083788






