Exosomal microRNAs as Early Transition Biomarkers from Recurrent-Remissive to Secondary Progressive Multiple Sclerosis
Abstract
1. Introduction
2. miRNAs—Biological Origins
3. Early miRNA Biomarkers Explored in MS
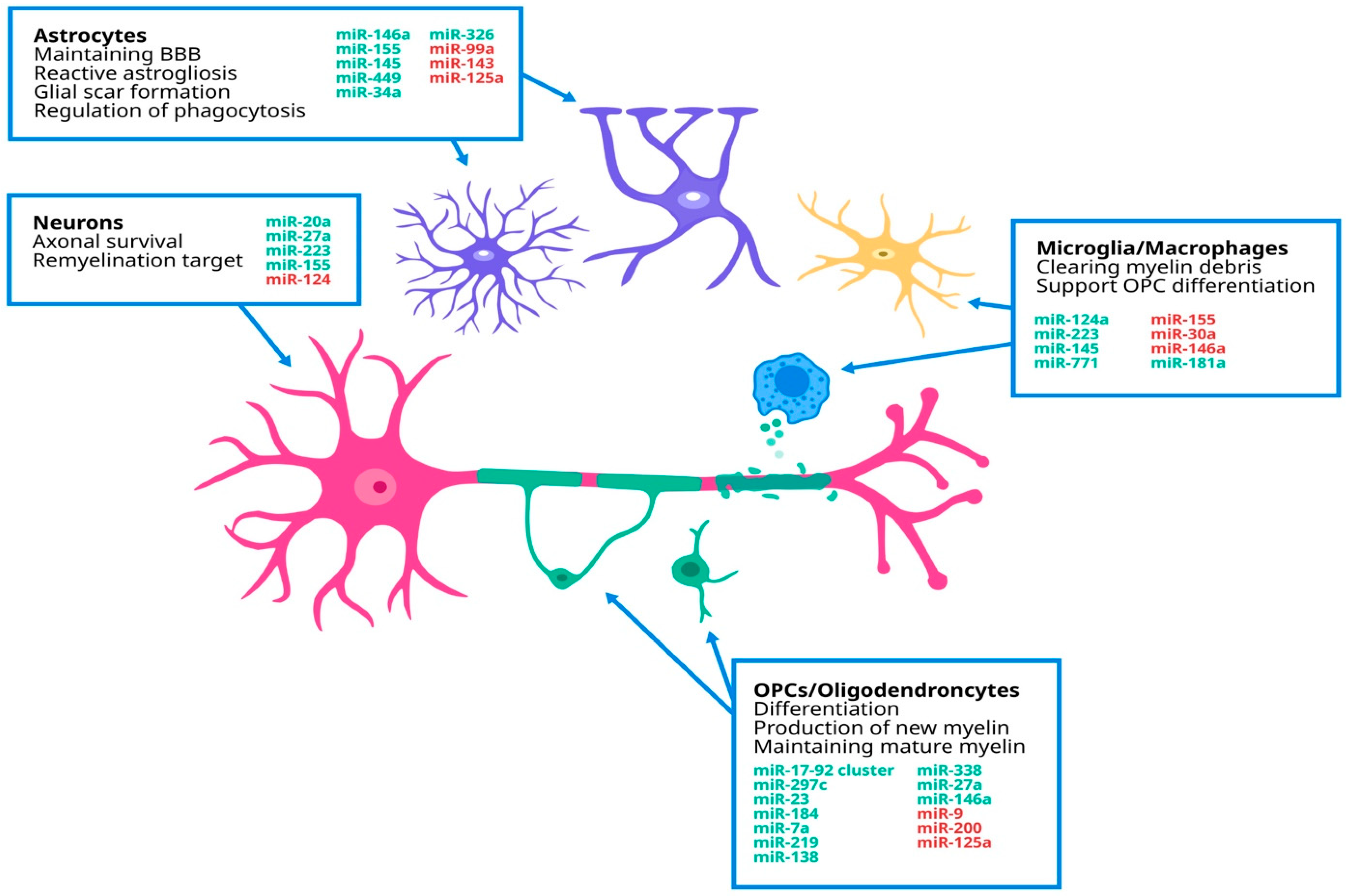
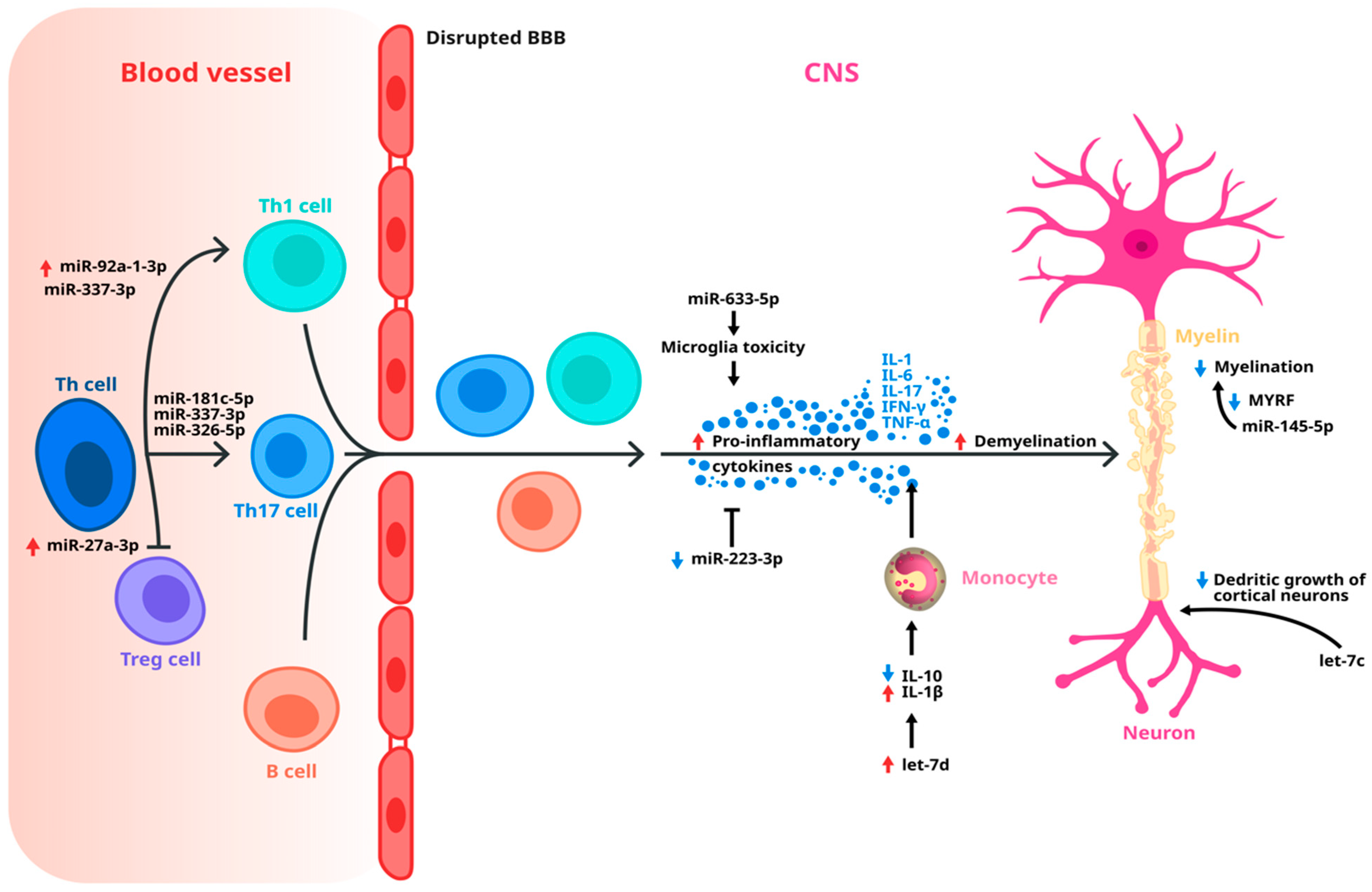
4. Dysregulated miRNAs Implicated in MS Pathology
| Cell Type | miRNA | Role |
|---|---|---|
| CD4+T cells | miR-34a | -⇓ related to the late stage of neuron mitosis [51] |
| miR-146a | -⇓ regulates T cell adhesion [50] | |
| Let-7e | - inhibits the proliferation of Th2 cells and makes polarization of CD4+ T cells towards Th1 cells and Th17 cells [48] | |
| Th 1 and Th 17 cells | miR-155 | -⇑ promotes the down-regulation of CD47 expression, and triggers macrophage-mediated myelin phagocytosis [56] |
| miR-30a | - negatively regulates the expression level of IL-17 [49] | |
| miR-132 | - promotes the differentiation of Th17 cells [52] | |
| CD 8+ B cells | miR-17-92 | - affects phosphatidylinositol-3 kinase in B-cell receptor [57] |
| miR-106b | ||
| Neuron | miR-155 | -neuroprotective [56] |
| miR-20a | ||
| miR-124 | -⇑ regulation demyelination of axons [55] | |
| miR-223 | -⇑ regulation neuroprotective [54] | |
| Astrocyte | miR-146a | -reduces astrocyte activation [59] |
| miR-155 | -expression in astrocytes releases microglia of inhibitory control of phagocytosis [56] | |
| miR-145 | -inhibits astrogliosis [59] | |
| miR-99a | -⇓ regulation inhibits astrocyte proliferation [59] | |
| miR-143 | -⇓ regulation inhibits astrocyte proliferation [59] | |
| miR-449 | -attenuates glial scar formation [59] | |
| miR-34a | -expression in astrocytes releases microglia of inhibitory control of phagocytosis [60] | |
| miR-326 | ||
| Microglia | miR-124 | -promotes inactive state [55] |
| miR-223 | -phagocytosis of myelin debris [54] | |
| miR-30a | -⇑ regulation promotes release of factors that induce OPC apoptosis [64] | |
| miR-145 | -associated with anti-inflammatory microglia activation [63] | |
| miR-771 | ||
| miR-146a | -promotes anti-inflammatory activation [62] | |
| Oligodendrocyte | miR-219 | -maintain mature myelin sheath [58] |
| miR-138 | -promotes OPC differentiation [66] | |
| miR-338 | ||
| miR-125a | ||
| miR-27a | -proliferation and differentiation of OPC [66] | |
| miR-146a | -promotes OPC differentiation [70] | |
| miR-17-92-cluster | ||
| miR-297c | ||
| miR-9 | -⇑ regulation impairs differentiation [67] | |
| miR-200 | ||
| miR-23 | -supports myelin maintenance [68] | |
| miR-7a | -promotes generation of OPCs [69] |
5. Potential miRNAs That Could Differentiate RRMS from SPMS
| Study | Reported Study Population | Biological Material | Selected Reported Results |
|---|---|---|---|
| Ebrahimkhani et al., 2017 [75] | 14 RRMS, 7 SPMS, 4 PPMS, 11 health controls (HCs) | Exosomes | -Nine miRNAs were identified with a different expression between RRMS and progressive MS patients: miR-15b-5p, miR-23a-3p, miR-223-3p, miR-374a-5p, miR-30b-5p, miR-433-3p, miR-485-3p, miR-342-3p, miR-432-5p). -A combination of miR-223-3p, miR-485-3p and miR-30b-5p showed a 95% accuracy rate in distinguishing progressive forms of MS from RRMS. |
| Niwald et al., 2017 [76] | 13 RRMS-relapse, 23 RRMS- no relapse in 2 years | Exosomes | -Decreased expression of miR-155 and miR-301a (in 94% and 51% of samples, respectively) and an increased expression of miR-326 (in 72% samples) in RRMS patients. -Levels of miR-301a and miR155 were higher in RRMS patients in post-acute vs. stable phase of remission. |
| Selmaj et al., 2017 [77] | 19 RRMS, 10 HCs | Exosomes | -miR-122-5p, miR-196b-5p, miR-301a-3p, and miR-532-5p decreased during relapse. |
| Kacperska M., 2015 [78] | 37 RRMS, 20 HC | Exosomes | -Expression levels of miR-let-7a in patients in remission were lower than in the control group. - miR-648a in patients in MS remission was lower than in the control group. |
| Kramer et al., 2019 [79] | 218 MS patients, 211 patients with other neurological diseases (OND) | Cerebral spinal fluid | -Up-regulation of miR-181 in patients with MS vs. patients with OND; no difference in patients with RRMS and SPMS, but difference between SPMS and PPMS. -Up-regulation of miR-633 in patients with MS vs. OND; higher levels in SPMS than in PPMS. |
| Haghikia et al., 2012 [80] | 53 MS patients (17 RRMS, 30 SPMS, 6 PPMS), 39 OND | Cerebral spinal fluid | -miR-922 (p = 0.0001) was down-regulated, whereas miR-181c (p = 0.0007) and miR-633 (p = 0.0014) were up-regulated as compared with OND. |
| Ibrahim et al., 2020 [81] | 39 RRMS, 35 SPMS, 10 HC | Serum | -miR-300 and miR-450b-5p expressions were significantly lower in SPMS patients, regardless of the EDSS score, compared with RRMS patients with EDSS 5.0. |
| Sharaf-Eldin et al., 2017 [82] | 19 RRMS, 18 SPMS, 10 NMSOD, 23 HC | Serum | -miR-145 and miR-223 up-regulated in MS patients compared to healthy subjects. -miR-326 showed insignificant up-regulation, but is significantly lower in SPMS, compared with RRMS with EDSS < 5.0. |
| Gandhi et al., 2013 [83] | 10 RRMS, 9 SPMS, 9 HC | Serum | -miR-92a-1, miR-135a, miR-454, miR-500, and miR-574-3p showed a significant association with RRMS. -let-7d, miR-145, and let-7c showed an association with SPMS. |
| Geiger et al., 2023 [84] | 29 RRMS, 14 SPMS, 7 PPMS | Serum | -miR-143-3p levels were significantly lower in the SPMS. -higher miR-92a-3p and miR-486-5p levels were associated with greater total white matter lesion volumes within the cervical spine. |
| Vistbakka J., 2016 [85] | 31 PPMS, 31 SPMS, 21 HC | Serum | -miR-191-5p showed the strongest up-regulation in progressive MS. -up-regulation of miR-128-3p and miR-24-3p in PPMS. -up-regulation of miR-375 in SPMS. |
| Regev et al., 2016 [86] | Discovery phase: 7 RRMS, 9 SPMS, 10 PPMS, 20 HC Validation phase: 29 RRMS, 19 SPMS, 10 PPMS, 30 HC | Serum | -miR-27a-3p and miR-376b-3p were the only miRNAs showing significantly different expressions between RRMS and SPMS. After multiple comparisons only miR-27a-3p remained significant. |
| Regev et al., 2018 [87] | Discovery phase: 7 RRMS, 9 SPMS, 10 PPMS, 20 HC Validation phase: 29 RRMS, 19 SPMS, 10 PPMS, 30 HC Reproductibility phase: 24 RRMS, 18 SPMS, 30 HC Transportability phase: 91 RRMS, 33 SPMS, 58 HC | Serum | -Significant down-regulation of miR-337-3p in patients with SSPMS compared with RRMS |
| Agostini et al., 2023 [88] | 33 RRMS, 36 RRMS patients converted in SPMS patients, 30HCs -10-year observation period | Serum | -miR34a-5p, miR-103a-3p, and miR-376a-3p are significantly more expressed in RRMS that will convert to SPMS within 10 years compared to RRMS. |
| Al-Temaimi et al., 2024 [89] | 76 MS, 75 HCs | Serum | -miR-24-3p was down-regulated in all MS patients compared to healthy controls. -miR-484 was significantly up-regulated in RRMS patients compared to HCs. -mir-146-5p and miR-484 were significantly down-regulated in SPMS compared to RRMS. |
| Gonzalez-Martinez et al., 2023 [90] | 144 MS patients: 104 benign, 40 not benign with a 10- year follow-up | Serum | -Patients who retained benign MS had lower values of miR-25-3p (p = 0.047) and higher miR-320b (p = 0.025) values. -Development of SPMS was associated with higher miR-320b (p = 0.002) levels. -Brain parenchymal fraction at year 10 was negatively correlated with miR-25-3p (p = 0.0004) and positively correlated with miR-320b (p = 0.006). |
| Mohammadinasr et al., 2023 [91] | 30 RRMS untreated patients, 30HCs | Cerebral spinal fluid and serum exosomes | -Let-7 g-5p, miR-18a-5p, miR-145-5p, miR-374a-5p, miR-150-5p, and miR-342-3p were significantly up-regulated in both CSF and serum-derived exosomes of RRMS patients compared to corresponding HCs. -miR-132-5p and miR-320a-5p were significantly down-regulated in both CSF and serum-derived exosomes of RRMS patients compared to HCs. |
6. Future Perspectives and Limitations of Exosomal miRNAs as a Biomarker for MS
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pietrasik, S.; Dziedzic, A.; Miller, E.; Starosta, M.; Saluk-Bijak, J. Circulating miRNAs as Potential Biomarkers Distin-guishing Relapsing–Remitting from Secondary Progressive Multiple Sclerosis. A Review. Int. J. Mol. Sc. 2021, 22, 11887. [Google Scholar] [CrossRef] [PubMed]
- Portaccio, E.; Magyari, M.; Havrdova, E.K.; Ruet, A.; Brochet, B.; Scalfari, A.; Di Filippo, M.; Tur, C.; Montalban, X.; Amato, M.P. Multiple sclerosis: Emerging epidemiological trends and redefining the clinical course. Lancet Reg. Health Eur. 2024, 44, 100977. [Google Scholar] [CrossRef]
- Belbasis, L.; Bellou, V.; Evangelou, E.; Ioannidis, J.P.; Tzoulaki, I. Environmental risk factors and multiple sclerosis: An um-brella review of systematic reviews and meta-analyses. Lancet Neurol. 2015, 14, 263–273. [Google Scholar] [CrossRef]
- Lassmann, H. Multiple Sclerosis Pathology. Cold Spring Harb. Perspect. Med. 2018, 8, a028936. [Google Scholar] [CrossRef] [PubMed]
- Trapp, B.D.; Vignos, M.; Dudman, J.; Chang, A.; Fisher, E.; Staugaitis, S.M.; Battapady, H.; Mork, S.; Ontaneda, D.; Jones, E.S.; et al. Cortical neuronal densities and cerebral white matter demyelination in multiple sclerosis: A retrospective study. Lancet Neurol. 2018, 17, 870–884. [Google Scholar] [CrossRef] [PubMed]
- Jakimovski, D.; Bittner, S.; Zivadinov, R.; Morrow, S.A.; Benedict, R.H.; Zipp, F.; Weinstock-Guttman, B. Multiple sclerosis. Lancet 2023, 403, 183–202. [Google Scholar] [CrossRef]
- Davies, F.; Wood, F.; Brain, K.E.; Edwards, M.; Jones, R.; Wallbank, R.; Robertson, N.P.; Edwards, A. The Transition to Secondary Progressive Multiple Sclerosis: An Exploratory Qualitative Study of Health Professionals’ Experiences. Int. J. MS Care 2016, 18, 257–264. [Google Scholar] [CrossRef]
- Kuhlmann, T.; Moccia, M.; Coetzee, T.; Cohen, J.A.; Correale, J.; Graves, J.; Marrie, R.A.; Montalban, X.; Yong, V.W.; Thompson, A.J.; et al. Multiple sclerosis progression: Time for a new mechanism-driven framework. Lancet Neurol. 2023, 22, 78–88. [Google Scholar] [CrossRef]
- Polman, C.H.; Reingold, S.C.; Banwell, B.; Clanet, M.; Cohen, J.A.; Filippi, M.; Fujihara, K.; Havrdova, E.; Hutchinson, M.; Kappos, L.; et al. Diagnostic criteria for multiple sclerosis revisions to the McDonald criteria. Ann. Neurol. 2011, 69, 292–302. [Google Scholar] [CrossRef]
- Balasa, R.; Barcutean, L.; Mosora, O.; Manu, D. Reviewing the Significance of Blood–Brain Barrier Disruption in Multiple Sclerosis Pathology and Treatment. Int. J. Mol. Sci. 2021, 22, 8370. [Google Scholar] [CrossRef]
- Kappos, L.; Butzkueven, H.; Wiendl, H.; Spelman, T.; Pellegrini, F.; Chen, Y.; Dong, Q.; Koendgen, H.; Belachew, S.; Trojano, M.; et al. Greater sensitivity to multiple sclerosis disability worsening and progression events using a roving versus a fixed reference value in a prospective cohort study. Mult. Scler. 2018, 24, 963–973. [Google Scholar] [CrossRef] [PubMed]
- Kappos, L.; Wolinsky, J.S.; Giovannoni, G.; Arnold, D.L.; Wang, Q.; Bernasconi, C.; Model, F.; Koendgen, H.; Manfrini, M.; Belachew, S.; et al. Contribution of relapse-independent progression vs relapse-associated worsening to overall confirmed disability accumulation in typical relapsing multiple sclerosis in a pooled analysis of 2 randomized clinical trials. JAMA Neurol. 2020, 77, 1132–1140. [Google Scholar] [CrossRef] [PubMed]
- Londono, A.C.; Mora, C.A. Evidence of disease control: A realistic concept beyond NEDA in the treatment of multiple sclerosis. F1000Research 2017, 6, 566. [Google Scholar] [CrossRef] [PubMed]
- University of California, San Francisco MS-EPIC Team; Cree, B.A.C.; Hollenbach, J.A.; Bove, R.; Kirkish, G.; Sacco, S.; Caverzasi, E.; Bischof, A.; Gundel, T.; Zhu, A.H.; et al. Silent progression in disease activity-free relapsing multiple scle-rosis. Ann. Neurol. 2019, 85, 653–666. [Google Scholar]
- Treabă, C.A.; Bălaşa, R.; Podeanu, D.M.; Simu, I.P.; Buruian, M.M. Cerebral lesions of multiple sclerosis: Is gadolinium always irreplaceable in assessing lesion activity? Diagn. Interv. Radiol. 2014, 20, 178–184. [Google Scholar] [CrossRef]
- Hollen, C.W.; Paz Soldán, M.M.; Rinker, J.R., 2nd; Spain, R.I. The Future of Progressive Multiple Sclerosis Therapies. Fed. Pract. 2020, 37 (Suppl. 1), S43–S49. [Google Scholar] [PubMed]
- Adamczyk-Sowa, M.; Adamczyk, B.; Kułakowska, A.; Rejdak, K.; Nowacki, P. Secondary Progressive Multiple Sclerosis—From Neuropathology to Definition and Effective Treatment. Neurol. Neurochir. Pol. 2020, 54, 384–398. [Google Scholar] [CrossRef]
- Gross, H.J.; Watson, C. Characteristics, Burden of Illness, and Physical Functioning of Patients with Relapsing-Remitting and Secondary Progressive Multiple Sclerosis: A Cross-Sectional US Survey. Neuropsychiatr. Dis. Treat. 2017, 13, 1349–1357. [Google Scholar] [CrossRef]
- Thompson, A.J.; Banwell, B.L.; Barkhof, F.; Carroll, W.M.; Coetzee, T.; Comi, G.; Correale, J.; Fazekas, F.; Filippi, M.; Freedman, M.S.; et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018, 17, 162–173. [Google Scholar] [CrossRef]
- Maier, S.; Barcutean, L.; Andone, S.; Manu, D.; Sarmasan, E.; Bajko, Z.; Balasa, R. Recent Progress in the Identification of Early Transition Biomarkers from Relapsing-Remitting to Progressive Multiple Sclerosis. Int. J. Mol. Sci. 2023, 24, 4375. [Google Scholar] [CrossRef]
- Inojosa, H.; Proschmann, U.; Akgün, K.; Ziemssen, T. A focus on secondary progressive multiple sclerosis (SPMS): Challenges in diagnosis and definition. J. Neurol. 2021, 268, 1210–1221. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Comabella, M.; Gandhi, R. Biomarkers in multiple sclerosis. Cold Spring Harb. Perspect. Med. 2019, 9, a029058. [Google Scholar] [CrossRef]
- Bărcuţean, L.I.; Romaniuc, A.; Maier, S.; Bajko, Z.; Moţăţăianu, A.; Adina, H.; Simu, I.; Andone, S.; Bălaşa, R. Clinical and Serological Biomarkers of Treatment’s Response in Multiple Sclerosis Patients Treated Continuously with Interferonβ-1b for More than a Decade. CNS Neurol. Disord. Drug Targets 2018, 17, 780–792. [Google Scholar] [CrossRef] [PubMed]
- Piket, E.; Zheleznyakova, G.Y.; Kular, L.; Jagodic, M. Small non-coding RNAs as important players, biomarkers and therapeutic targets in multiple sclerosis: A comprehensive overview. J. Autoimmun. 2019, 101, 17–25. [Google Scholar] [CrossRef]
- Gao, Y.; Han, D.; Feng, J. MicroRNA in multiple sclerosis. Clin. Chim. Acta 2021, 516, 92–99. [Google Scholar] [CrossRef]
- Dolati, S.; Marofi, F.; Babaloo, Z.; Aghebati-Maleki, L.; Roshangar, L.; Ahmadi, M.; Rikhtegar, R.; Yousefi, M. Dysregulated Network of miRNAs Involved in the Pathogenesis of Multiple Sclerosis. Biomed. Pharmacother. 2018, 104, 280–290. [Google Scholar] [CrossRef]
- Chen, C.; Zhou, Y.; Wang, J.; Yan, Y.; Peng, L.; Qiu, W. Dysregulated MicroRNA Involvement in Multiple Sclerosis by Induction of T Helper 17 Cell Differentiation. Front. Immunol. 2018, 9, 1256. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Lai, X.; Wang, X.; Ying, J.; Zhang, L.; Zhou, B.; Liu, X.; Zhang, J.; Wei, G.; Hua, F. Long Non-coding RNAs and Circular RNAs: Insights into Microglia and Astrocyte Mediated Neurological Diseases. Front. Mol. Neurosci. 2021, 14, 745066. [Google Scholar] [CrossRef]
- Du, C.; Liu, C.; Kang, J.; Zhao, G.; Ye, Z.; Huang, S.; Li, Z.; Wu, Z.; Pei, G. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat. Immunol. 2009, 10, 1252–1259. [Google Scholar] [CrossRef]
- Abolghasemi, M.; Ali Ashrafi, S.; Asadi, M.; Shanehbandi, D.; Sadigh Etehad, S.; Poursaei, E.; Nejadghaderi, S.A.; Shaafi, S. MicroRNAs expression in peripheral blood mononuclear cells of patients with multiple sclerosis propose. Mol. Biol. Rep. 2023, 50, 167–172. [Google Scholar] [CrossRef]
- Backes, C.; Meese, E.; Keller, A. Specific miRNA Disease Biomarkers in Blood, Serum and Plasma: Challenges and Prospects. Mol. Diagn. Ther. 2016, 20, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Tandon, M.; Alevizos, I.; Illei, G.G. The Majority of MicroRNAs Detectable in Serum and Saliva Is Concentrated in Exosomes. PLoS ONE 2012, 7, e30679. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Wang, J.; Zhang, X.; Rang, X.; Xu, C.; Fu, J. Identification and functional analysis of specific MS risk miRNAs and their target genes. Mult. Scle. Relat. Disord. 2020, 41, 102044. [Google Scholar] [CrossRef]
- Manna, I.; Iaccino, E.; Dattilo, V.; Barone, S.; Vecchio, E.; Mimmi, S.; Filippelli, E.; Demonte, G.; Polidoro, S.; Granata, A.; et al. Exosome-Associated MiRNA Profile as a Prognostic Tool for Therapy Response Monitoring in Multiple Sclerosis Patients. FASEB 2018, 32, 4241–4246. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. MiRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed]
- Selmaj, I.; Mycko, M.P.; Raine, C.S.; Selmaj, K.W. The role of exosomes in CNS inflammation and their involvement in multiple sclerosis. J. Neuroimmunol. 2017, 306, 1–10. [Google Scholar] [CrossRef]
- Griffiths-Jones, S. The microRNA Registry. Nucleic Acids Res. 2004, 32, D109–D111. [Google Scholar] [CrossRef]
- Arruda, L.; Lorenzi, J.; Sousa, A.; Zanette, D.L.; Palma, P.V.; Panepucci, R.A.; Brum, D.S.; Barreira, A.A.; Covas, D.T.; Simões, B.P.; et al. Autologous hematopoietic SCT normalizes miR-16, -155 and -142-3p expression in multiple sclerosis patients. Bone Marrow Transpl. 2015, 50, 380–389. [Google Scholar] [CrossRef]
- Waschbisch, A.; Atiya, M.; Linker, R.A.; Potapov, S.; Schwab, S.; Derfuss, T. Glatiramer acetate treatment normalizes deregu-lated microRNA expression in relapsing remitting multiple sclerosis. PLoS ONE 2011, 6, e24604. [Google Scholar] [CrossRef]
- Louafi, F.; Martinez-Nunez, R.T.; Sanchez-Elsner, T. MicroRNA-155 targets SMAD2 and modulates the response of macro-phages to transforming growth factor-{beta}. J. Biol. Chem. 2010, 285, 41328–41336. [Google Scholar] [CrossRef]
- Shademan, B.; Zakeri, M.; Abbasi, S.; Biray Avci, C.; Karamad, V.; Sogutlu, F.; Laghousi, D.; Nouri, M.; Hassanpour, M.; Nourazarian, A. Relationship between miRNA-21, miRNA-155, and miRNA-182 expression and inflammatory factors in cerebrospinal fluid from patients with multiple sclerosis. Clin. Neurol. Neurosurg. 2023, 232, 107873. [Google Scholar] [CrossRef]
- Rezaee, D.; Saadatpour, F.; Akbari, N.; Zoghi, A.; Najafi, S.; Beyranvand, P.; Zamani-Rarani, F.; Rashidi, M.A.; Bagheri-Mohammadi, S.; Bakhtiari, M. The role of microRNAs in the pathophysiology of human central nervous system: A focus on neurodegenerative diseases. Ageing Res. Rev. 2023, 92, 102090. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, A.D.; Fonken, L.K.; Watkins, L.R.; Nelson, R.J.; Popovich, P.G. MicroRNAs: Roles in Regulating Neuroinflammation. Neuroscientist 2018, 24, 221–245. [Google Scholar] [CrossRef]
- Schonrock, N.; Ke, Y.D.; Humphreys, D.; Staufenbiel, M.; Ittner, L.M.; Preiss, T.; Götz, J. Neuronal microRNA deregulation in response to Alzheimer’s disease amy-loid-beta. PLoS ONE 2010, 5, e11070. [Google Scholar] [CrossRef] [PubMed]
- Ridolfi, E.; Fenoglio, C.; Cantoni, C.; Calvi, A.; De Riz, M.; Pietroboni, A.; Villa, C.; Serpente, M.; Bonsi, R.; Vercellino, M.; et al. Expression and Genetic Analysis of MicroRNAs Involved in Multiple Sclerosis. Int. J. Mol. Sci. 2013, 14, 4375–4384. [Google Scholar] [CrossRef]
- O’Connell, M.; Kahn, D.; Gibson, W.S.J.; Round, L.; Scholz, L.; Chaudhuri, A.; Kahn, M.; Rao, D.S.; Baltimore, D. MicroRNA-155 Promotes Autoimmune Inflammation by Enhancing Inflammatory T Cell Development. Immunity 2010, 33, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, R.L.; Hoffmann, F.; Mehling, M.; Kuhle, J.; Kappos, L. Altered expression of miR-17-5p in CD4+ lymphocytes of relapsing-remitting multiple sclerosis patients. Eur. J. Immunol. 2010, 40, 888–898. [Google Scholar] [CrossRef]
- Guan, H.; Fan, D.; Mrelashvili, D.; Hao, H.; Singh, N.P.; Singh, U.P.; Nagarkatti, P.S.; Nagarkatti, M. MicroRNA let-7e is associated with the pathogenesis of experimental autoimmune encephalomyelitis. Eur. J. Immunol. 2013, 43, 104–114. [Google Scholar] [CrossRef]
- Zhao, M.; Sun, D.; Guan, Y.; Wang, Z.; Sang, D.; Liu, M.; Pu, Y.; Fang, X.; Wang, D.; Huang, A.; et al. Disulfiram and Diphenhydramine Hydrochloride Upregulate miR-30a to Suppress IL-17-Associated Autoimmune Inflammation. J. Neurosci. 2016, 36, 9253–9266. [Google Scholar] [CrossRef]
- Wu, D.; Cerutti, C.; Lopez-Ramirez, M.A.; Pryce, G.; King-Robson, J.; Simpson, J.E.; van der Pol, S.M.; Hirst, M.C.; de Vries, H.E.; Sharrack, B.; et al. Brain endothelial miR-146a negatively modulates T-cell adhesion through repressing multiple targets to inhibit NF-κB activation. J. Cereb. Blood Flow Metab. 2015, 35, 412–423. [Google Scholar] [CrossRef]
- Chen, F.; Hu, S.J. Effect of microRNA-34a in cell cycle, differentiation, and apoptosis: A review. J. Biochem. Mol. Toxicol. 2012, 26, 79–86. [Google Scholar] [CrossRef]
- Feng, R.; Cui, Z.; Liu, Z.; Zhang, Y. Upregulated microRNA-132 in T helper 17 cells activates hepatic stellate cells to promote hepatocellular carcinoma cell migration in vitro. Scand. J. Immunol. 2021, 93, e13007. [Google Scholar] [CrossRef] [PubMed]
- Nakahama, T.; Hanieh, H.; Nguyen, N.T.; Chinen, I.; Ripley, B.; Millrine, D.; Lee, S.; Nyati, K.K.; Dubey, P.K.; Chowdhury, K.; et al. Aryl hydrocarbon receptor-mediated induction of the microRNA-132/212 cluster promotes interleukin-17-producing T-helper cell differentiation. Proc. Natl. Acad. Sci. USA 2013, 110, 11964–11969. [Google Scholar] [CrossRef] [PubMed]
- Morquette, B.; Juźwik, C.A.; Drake, S.S.; Charabati, M.; Zhang, Y.; Lécuyer, M.A.; Galloway, D.A.; Dumas, A.; de Faria Junior, O.; Paradis-Isler, N.; et al. MicroRNA-223 protects neurons from degeneration in experimental autoim-mune encephalomyelitis. Brain 2019, 142, 2979–2995. [Google Scholar] [CrossRef] [PubMed]
- Son, G.; Na, Y.; Kim, Y.; Son, J.H.; Clemenson, G.D.; Schafer, S.T.; Yoo, J.Y.; Parylak, S.L.; Paquola, A.; Do, H.; et al. miR-124 coordinates metabolic regulators acting at early stages of human neurogenesis. Commun. Biol. 2024, 7, 1393. [Google Scholar] [CrossRef]
- Zhang, J.; Cheng, Y.; Cui, W.; Li, M.; Li, B.; Guo, L. MicroRNA-155 modulates Th1 and Th17 cell differentiation and is associated with multiple sclerosis and experimental autoimmune encephalomyelitis. J. Neuroimmunol. 2014, 266, 56–63. [Google Scholar] [CrossRef]
- Mandelbaum, A.D.; Kredo-Russo, S.; Aronowitz, D.; Myers, N.; Yanowski, E.; Klochendler, A.; Swisa, A.; Dor, Y.; Hornstein, E. miR-17-92 and miR-106b-25 clusters regulate beta cell mitotic checkpoint and insulin secretion in mice. Diabetologia 2019, 62, 1653–1666. [Google Scholar] [CrossRef]
- Dugas, J.C.; Cuellar, T.L.; Scholze, A.; Ason, B.; Ibrahim, A.; Emery, B.; Zamanian, J.L.; Foo, L.C.; McManus, M.T.; Barres, B.A. Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron 2010, 65, 597–611. [Google Scholar] [CrossRef]
- Rao, V.T.S.; Fuh, S.C.; Karamchandani, J.R.; Woulfe, J.M.J.; Munoz, D.G.; Ellezam, B.; Blain, M.; Ho, M.K.; Bedell, B.J.; Antel, J.P.; et al. Astrocytes in the Pathogenesis of Multiple Sclerosis: An In Situ MicroRNA Study. J. Neuropathol. Exp. Neurol. 2019, 78, 1130–1146. [Google Scholar] [CrossRef]
- Sofroniew, M.V.; Vinters, H.V. Astrocytes: Biology and pathology. Acta Neuropathol. 2010, 119, 7–35. [Google Scholar] [CrossRef]
- Junker, A.; Krumbholz, M.; Eisele, S.; Mohan, H.; Augstein, F.; Bittner, R.; Lassmann, H.; Wekerle, H.; Hohlfeld, R.; Meinl, E. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain 2009, 132 Pt 12, 3342–3352. [Google Scholar] [CrossRef] [PubMed]
- Martin, N.A.; Molnar, V.; Szilagyi, G.T.; Elkjaer, M.L.; Nawrocki, A.; Okarmus, J.; Wlodarczyk, A.; Thygesen, E.K.; Palkovits, M.; Gallyas, F., Jr.; et al. Experimental Demyelination and Axonal Loss Are Reduced in MicroRNA-146a Deficient Mice. Front. Immunol. 2018, 9, 490. [Google Scholar] [CrossRef] [PubMed]
- Prinz, M.; Priller, J. Microglia and brain macrophages in the molecular age: From origin to neuropsychiatric disease. Nat. Rev. Neurosci. 2014, 15, 300–312. [Google Scholar] [CrossRef]
- Freilich, R.W.; Woodbury, M.E.; Ikezu, T. Integrated expression profiles of mRNA and miRNA in polarized primary murine microglia. PLoS ONE 2013, 8, e79416. [Google Scholar] [CrossRef]
- Fang, X.; Sun, D.; Wang, Z.; Yu, Z.; Liu, W.; Pu, Y.; Wang, D.; Huang, A.; Liu, M.; Xiang, Z.; et al. MiR-30a Positively Regulates the Inflammatory Response of Microglia in Experimental Autoimmune Encephalomyelitis. Neurosci. Bull. 2017, 33, 603–615. [Google Scholar] [CrossRef]
- Patel, J.R.; Klein, R.S. Mediators of oligodendrocyte differentiation during remyelination. FEBS Lett. 2011, 585, 3730–3737. [Google Scholar] [CrossRef]
- Buller, B.; Chopp, M.; Ueno, Y.; Zhang, L.; Zhang, R.L.; Morris, D.; Zhang, Y.; Zhang, Z.G. Regulation of serum response factor by miRNA-200 and miRNA-9 modulates oli-godendrocyte progenitor cell differentiation. Glia 2012, 60, 1906–1914. [Google Scholar] [CrossRef] [PubMed]
- Lau, P.; Verrier, J.D.; Nielsen, J.A.; Johnson, K.R.; Notterpek, L.; Hudson, L.D. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J. Neurosci. 2008, 28, 11720–11730. [Google Scholar] [CrossRef]
- Lin, S.T.; Fu, Y.H. miR-23 regulation of lamin B1 is crucial for oligodendrocyte development and myelination. Dis. Model. Mech. 2009, 2, 178–188. [Google Scholar] [CrossRef]
- Zhao, X.; Wu, J.; Zheng, M.; Gao, F.; Ju, G. Specification and maintenance of oligodendrocyte precursor cells from neural progenitor cells: Involvement of microRNA-7a. Mol. Biol. Cell. 2012, 23, 2867–2878. [Google Scholar] [CrossRef]
- Liguori, M.; Nuzziello, N.; Licciulli, F.; Consiglio, A.; Simone, M.; Viterbo, R.G.; Creanza, T.M.; Ancona, N.; Tortorella, C.; Margari, L.; et al. Combined microRNA and mRNA expression analysis in pediatric multiple sclerosis: An integrated approach to uncover novel pathogenic mechanisms of the disease. Hum. Mol. Genet. 2018, 27, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Budde, H.; Schmitt, S.; Fitzner, D.; Opitz, L.; Salinas-Riester, G.; Simons, M. Control of oligodendroglial cell number by the miR-17-92 cluster. Development 2010, 137, 2127–2132. [Google Scholar] [CrossRef] [PubMed]
- Tang, N. Exosomes in multiple sclerosis and Alzheimer’s disease—Adversary and ally. Biomed. J. 2024, 47, 100665. [Google Scholar] [CrossRef]
- Duffy, C.P.; McCoy, C.E. The Role of MicroRNAs in Repair Processes in Multiple Sclerosis. Cells 2020, 9, 1711. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimkhani, S.; Vafaee, F.; Young, P.E.; Hur, S.S.J.; Hawke, S.; Devenney, E.; Beadnall, H.; Barnett, M.H.; Suter, C.M.; Buckland, M.E. Exosomal microRNA signatures in multiple sclerosis reflect disease status. Sci. Rep. 2017, 7, 14293. [Google Scholar] [CrossRef]
- Niwald, M.; Migdalska-Sęk, M.; Brzeziańska-Lasota, E.; Miller, E. Evaluation of Selected MicroRNAs Expression in Remission Phase of Multiple Sclerosis and Their Potential Link to Cognition, Depression, and Disability. J. Mol. Neurosci. 2017, 63, 275–282. [Google Scholar] [CrossRef]
- Selmaj, I.; Cichalewska, M.; Namiecinska, M.; Galazka, G.; Horzelski, W.; Selmaj, K.W.; Mycko, M.P. Global exosome transcriptome profiling reveals biomarkers for multiple sclerosis. Ann Neurol. 2017, 81, 703–717. [Google Scholar] [CrossRef]
- Kacperska, M.J.; Jastrzebski, K.; Tomasik, B.; Walenczak, J.; Konarska-Krol, M.; Glabinski, A. Selected extracellular microRNA as potential biomarkers of multiple sclerosis activity—Preliminary study. J. Mol. Neurosci. 2015, 56, 154–163. [Google Scholar] [CrossRef]
- Kramer, S.; Haghikia, A.; Bang, C.; Scherf, K.; Pfanne, A.; Duscha, A.; Kaisler, J.; Gisevius, B.; Gold, R.; Thum, T.; et al. Elevated levels of miR-181c and miR-633 in the CSF of patients with MS: A validation study. Neurol. Neuroimmunol. Neuroinflamm. 2019, 6, e623. [Google Scholar] [CrossRef]
- Haghikia, A.; Haghikia, A.; Hellwig, K.; Baraniskin, A.; Holzmann, A.; Décard, B.F.; Thum, T.; Gold, R. Regulated microRNAs in the CSF of patients with multiple sclerosis: A case-control study. Neurology 2012, 79, 2166–2170. [Google Scholar] [CrossRef]
- Ibrahim, S.H.; El-Mehdawy, K.M.; Seleem, M.; El-Sawalhi, M.M.; Shaheen, A.A. Serum ROCK2, miR-300 and miR-450b-5p levels in two different clinical phenotypes of multiple sclerosis: Relation to patient disability and disease progression. J. Neu-roimmunol. 2020, 347, 577356. [Google Scholar] [CrossRef] [PubMed]
- Sharaf-Eldin, W.E.; Kishk, N.A.; Gad, Y.Z.; Hassan, H.; Ali, M.A.M.; Zaki, M.; Mohamed, M.R.; Essawi, M.L. Extracellular miR-145, miR-223 and miR-326 expression signature allow for differential diagnosis of immune-mediated neuroinflammatory diseases. J. Neurol. Sci. 2017, 383, 188–198. [Google Scholar] [CrossRef]
- Gandhi, R.; Healy, B.; Gholipour, T.; Egorova, S.; Musallam, A.; Hussain, M.S.; Nejad, P.; Patel, B.; Hei, H.; Khoury, S.; et al. Circulating microRNAs as biomarkers for disease staging in multiple sclerosis. Ann. Neurol. 2013, 73, 729–740. [Google Scholar] [CrossRef]
- Geiger, L.; Orsi, G.; Cseh, T.; Gombos, K.; Illés, Z.; Czéh, B. Circulating microRNAs correlate with structural and functional MRI parameters in patients with multiple sclerosis. Front. Mol. Neurosci. 2023, 16, 1173212. [Google Scholar] [CrossRef] [PubMed]
- Vistbakka, J.; Elovaara, I.; Lehtimäki, T.; Hagman, S. Circulating microRNAs as biomarkers in progressive multiple sclerosis. Mult. Scler. 2017, 23, 403–412. [Google Scholar] [CrossRef]
- Regev, K.; Paul, A.; Healy, B.; von Glenn, F.; Diaz-Cruz, C.; Gholipour, T.; Mazzola, M.A.; Raheja, R.; Nejad, P.; Glanz, B.I.; et al. Comprehensive evaluation of serum microRNAs as biomarkers in multiple sclerosis. Neurol. Neuroimmunol. Neuroinflamm. 2016, 3, e296. [Google Scholar] [CrossRef]
- Regev, K.; Healy, B.C.; Paul, A.; Diaz-Cruz, C.; Mazzola, M.A.; Raheja, R.; Glanz, B.I.; Kivisäkk, P.; Chitnis, T.; Jagodic, M.; et al. Identification of MS-specific serum miRNAs in an international multicenter study. Neurol. Neuroimmunol. Neuroinflamm. 2018, 5, e491. [Google Scholar] [CrossRef] [PubMed]
- Agostini, S.; Mancuso, R.; Citterio, L.A.; Caputo, D.; Oreni, L.; Nuzzi, R.; Pasanisi, M.B.; Rovaris, M.; Clerici, M. Serum miR-34a-5p, miR-103a-3p, and miR-376a-3p as possible biomarkers of conversion from relapsing-remitting to secondary progressive multiple sclerosis. Neurobiol. Dis. 2024, 200, 106648. [Google Scholar] [CrossRef]
- Al-Temaimi, R.; Alroughani, R. miR-24-3p and miR-484 are potential biomarkers for neurodegeneration in multiple sclerosis. Heliyon 2024, 10, e32685. [Google Scholar] [CrossRef]
- Gonzalez-Martinez, A.; Bose, G.; Lokhande, H.; Saxena, S.; Healy, B.C.; Polgar-Turcsanyi, M.; Weiner, H.L.; Chitnis, T. Early miR-320b and miR-25-3p miRNA levels correlate with multiple sclerosis severity at 10 years: A cohort study. J. Neuroinflam. 2023, 20, 136. [Google Scholar] [CrossRef]
- Mohammadinasr, M.; Montazersaheb, S.; Molavi, O.; Kahroba, H.; Talebi, M.; Ayromlou, H.; Hejazi, M.S. Multiplex Analysis of Cerebrospinal Fluid and Serum Exosomes MicroRNAs of Untreated Relapsing Remitting Multiple Sclerosis (RRMS) and Proposing Noninvasive Diagnostic Bi-omarkers. Neuromolecular Med. 2023, 25, 402–414. [Google Scholar] [CrossRef] [PubMed]
- Jagot, F.; Davoust, N. Is It worth Considering Circulating microRNAs in Multiple Sclerosis? Front. Immunol. 2016, 7, 129. [Google Scholar] [CrossRef] [PubMed]
- Baulina, N.; Kulakova, O.; Kiselev, I.; Osmak, G.; Popova, E.; Boyko, A.; Favorova, O. Immune-related miRNA expression patterns in peripheral blood mononuclear cells differ in multiple sclerosis relapse and remission. J. Neuroimmunol. 2018, 317, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.J.; Zhao, B.; Zhao, J.; Li, S. Potential Roles of Exosomal MicroRNAs as Diagnostic Biomarkers and Therapeutic Ap-plication in Alzheimer’s Disease. Neural Plast. 2017, 2017, 7027380. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, L. Circulating Exosomal miRNA as Diagnostic Biomarkers of Neurodegenerative Diseases. Front. Mol. Neurosci. 2020, 13, 53. [Google Scholar] [CrossRef]
- Koch, M.W.; Metz, L.M.; Kovalchuk, O. Epigenetics and miRNAs in the diagnosis and treatment of multiple sclerosis. Trends Mol. Med. 2013, 19, 23–30. [Google Scholar] [CrossRef]
- Shan, K.; Pang, R.; Zhao, C.; Liu, X.; Gao, W.; Zhang, J.; Zhao, D.; Wang, Y.; Qiu, W. IL-17-triggered downregulation of miR-497 results in high HIF-1α expression and consequent IL-1β and IL-6 production by astrocytes in EAE mice. Cell Mol. Immunol. 2017, 14, 909–923. [Google Scholar] [CrossRef]
- Søndergaard, H.B.; Hesse, D.; Krakauer, M.; Sørensen, P.S.; Sellebjerg, F. Differential microRNA expression in blood in mul-tiple sclerosis. Mult. Scler. 2013, 19, 1849–1857. [Google Scholar] [CrossRef]
- Martinelli-Boneschi, F.; Fenoglio, C.; Brambilla, P.; Sorosina, M.; Giacalone, G.; Esposito, F.; Serpente, M.; Cantoni, C.; Ridolfi, E.; Rodegher, M.; et al. MicroRNA and mRNA expression profile screening in multiple sclerosis patients to unravel novel pathogenic steps and identify potential biomarkers. Neurosci. Lett. 2012, 508, 4–8. [Google Scholar] [CrossRef]
- Sanders, K.A.; Benton, M.C.; Lea, R.A.; Maltby, V.E.; Agland, S.; Griffin, N.; Scott, R.J.; Tajouri, L.; Lechner-Scott, J. Next-generation sequencing reveals broad down-regulation of microRNAs in secondary progressive multiple sclerosis CD4+ T cells. Clin. Epigenetics 2016, 8, 87. [Google Scholar] [CrossRef]
- Fenoglio, C.; Ridolfi, E.; Galimberti, D.; Scarpini, E. MicroRNAs as active players in the pathogenesis of multiple sclerosis. Int. J. Mol. Sci. 2012, 13, 13227–13239. [Google Scholar] [CrossRef]
- Huang, Q.; Xiao, B.; Ma, X.; Qu, M.; Li, Y.; Nagarkatti, P.; Nagarkatti, M.; Zhou, J. MicroRNAs associated with the pathogenesis of multiple sclerosis. J. Neuroimmunol. 2016, 295–296, 148–161. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Pan, W.; Qian, L. Identification of the miRNA-mRNA regulatory network in multiple sclerosis. Neurol. Res. 2017, 39, 142–151. [Google Scholar] [CrossRef]
- Mansoor, S.R.; Ghasemi-Kasman, M.; Yavarpour-Bali, H. The role of microRNAs in multiple sclerosis. Int. Rev. Immunol. 2022, 41, 57–71. [Google Scholar] [CrossRef] [PubMed]
- Martinez, B.; Peplow, P.V. MicroRNAs in blood and cerebrospinal fluid as diagnostic biomarkers of multiple sclerosis and to monitor disease progression. Neural Regen. Res. 2020, 15, 606–619. [Google Scholar]
- Nuzziello, N.; Vilardo, L.; Pelucchi, P.; Consiglio, A.; Liuni, S.; Trojano, M.; Liguori, M. Investigating the Role of MicroRNA and Transcription Factor Co-regulatory Networks in Multiple Sclerosis Pathogenesis. Int. J. Mol. Sci. 2018, 19, 3652. [Google Scholar] [CrossRef] [PubMed]
- Moutinho, C.; Esteller, M. MicroRNAs and Epigenetics. Adv. Cancer Res. 2017, 135, 189–220. [Google Scholar]
- Disanto, G.; Barro, C.; Benkert, P.; Naegelin, Y.; Schädelin, S.; Giardiello, A.; Zecca, C.; Blennow, K.; Zetterberg, H.; Leppert, D. Serum Neurofilament Light: A Biomarker of Neuronal Damage in Multiple Sclerosis. Ann. Neurol. 2017, 81, 857–870. [Google Scholar] [CrossRef]
- Sá, M.J.; Basílio, C.; Capela, C.; Cerqueira, J.J.; Mendes, I.; Morganho, A.; Correia de Sá, J.; Salgado, V.; Martins Silva, A.; Vale, J.; et al. Consensus for the Early Identification of Secondary Progressive Multiple Sclerosis in Portugal: A Delphi Panel. Acta Med. Port. 2023. Online ahead of print. [Google Scholar] [CrossRef]
- Romanello, A.; Krohn, S.; von Schwanenflug, N.; Chien, C.; Bellmann-Strobl, J.; Ruprecht, K.; Paul, F.; Finke, C. Functional connectivity dynamics reflect disability and mul-ti-domain clinical impairment in patients with relapsing-remitting multiple sclerosis. Neuroimage Clin. 2022, 36, 103203. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mosora, O.; Maier, S.; Manu, D.; Bărcuțean, L.; Roman, M.; Dumitreasă, M.; Bălașa, R. Exosomal microRNAs as Early Transition Biomarkers from Recurrent-Remissive to Secondary Progressive Multiple Sclerosis. Int. J. Mol. Sci. 2025, 26, 3889. https://doi.org/10.3390/ijms26083889
Mosora O, Maier S, Manu D, Bărcuțean L, Roman M, Dumitreasă M, Bălașa R. Exosomal microRNAs as Early Transition Biomarkers from Recurrent-Remissive to Secondary Progressive Multiple Sclerosis. International Journal of Molecular Sciences. 2025; 26(8):3889. https://doi.org/10.3390/ijms26083889
Chicago/Turabian StyleMosora, Oana, Smaranda Maier, Doina Manu, Laura Bărcuțean, Medeea Roman, Mihai Dumitreasă, and Rodica Bălașa. 2025. "Exosomal microRNAs as Early Transition Biomarkers from Recurrent-Remissive to Secondary Progressive Multiple Sclerosis" International Journal of Molecular Sciences 26, no. 8: 3889. https://doi.org/10.3390/ijms26083889
APA StyleMosora, O., Maier, S., Manu, D., Bărcuțean, L., Roman, M., Dumitreasă, M., & Bălașa, R. (2025). Exosomal microRNAs as Early Transition Biomarkers from Recurrent-Remissive to Secondary Progressive Multiple Sclerosis. International Journal of Molecular Sciences, 26(8), 3889. https://doi.org/10.3390/ijms26083889






