A Comprehensive Analysis of the Alternative Splicing Co-Factor U2AF65B Gene Family Reveals Its Role in Stress Responses and Root Development
Abstract
:1. Introduction
2. Results
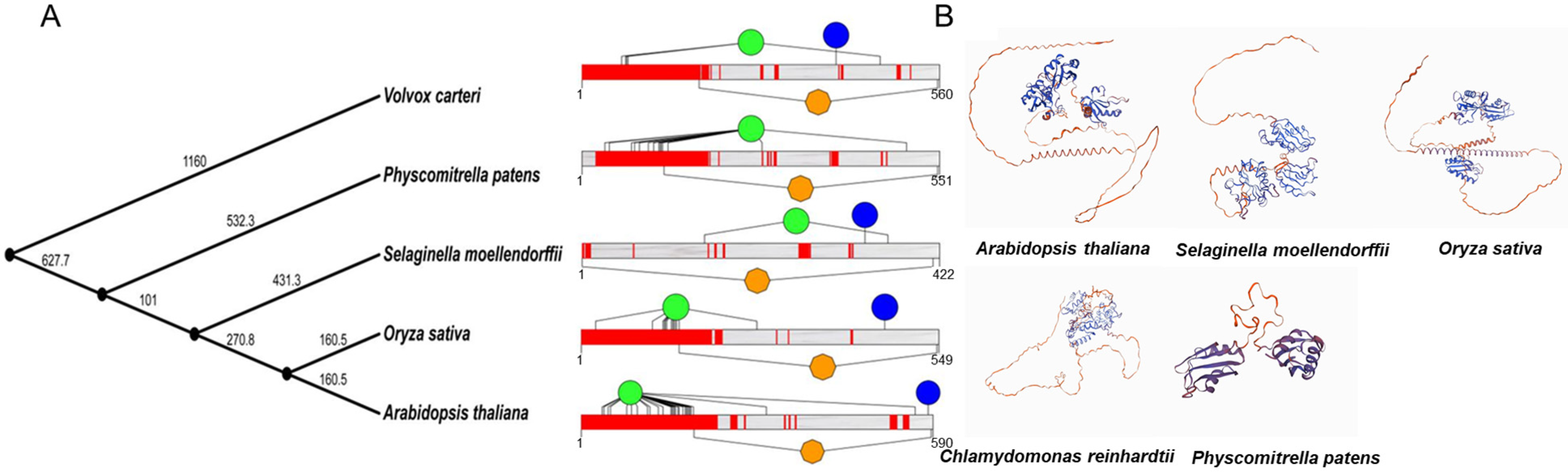
2.1. Genome-Wide Identification and Phylogenetic Analysis of the Plant U2AF65B Gene Family
2.2. Comparative Analysis of Gene Structure and Conserved Motifs in the U2AF65B Gene Family
2.3. Homology Modeling and Amino Acid Conservation Estimation
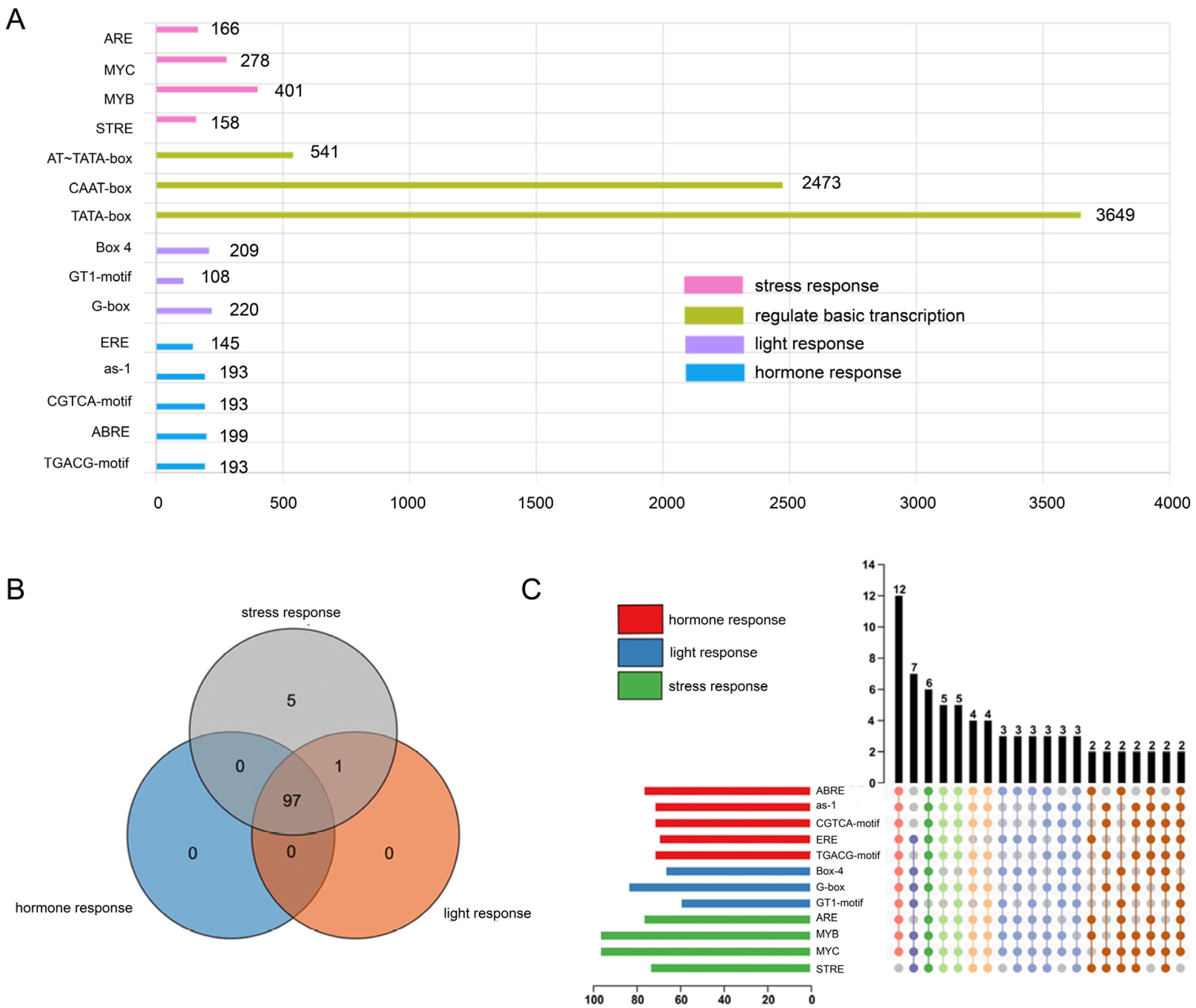
2.4. Analysis of Cis-Acting Elements in U2AF65B Promoter Sequences
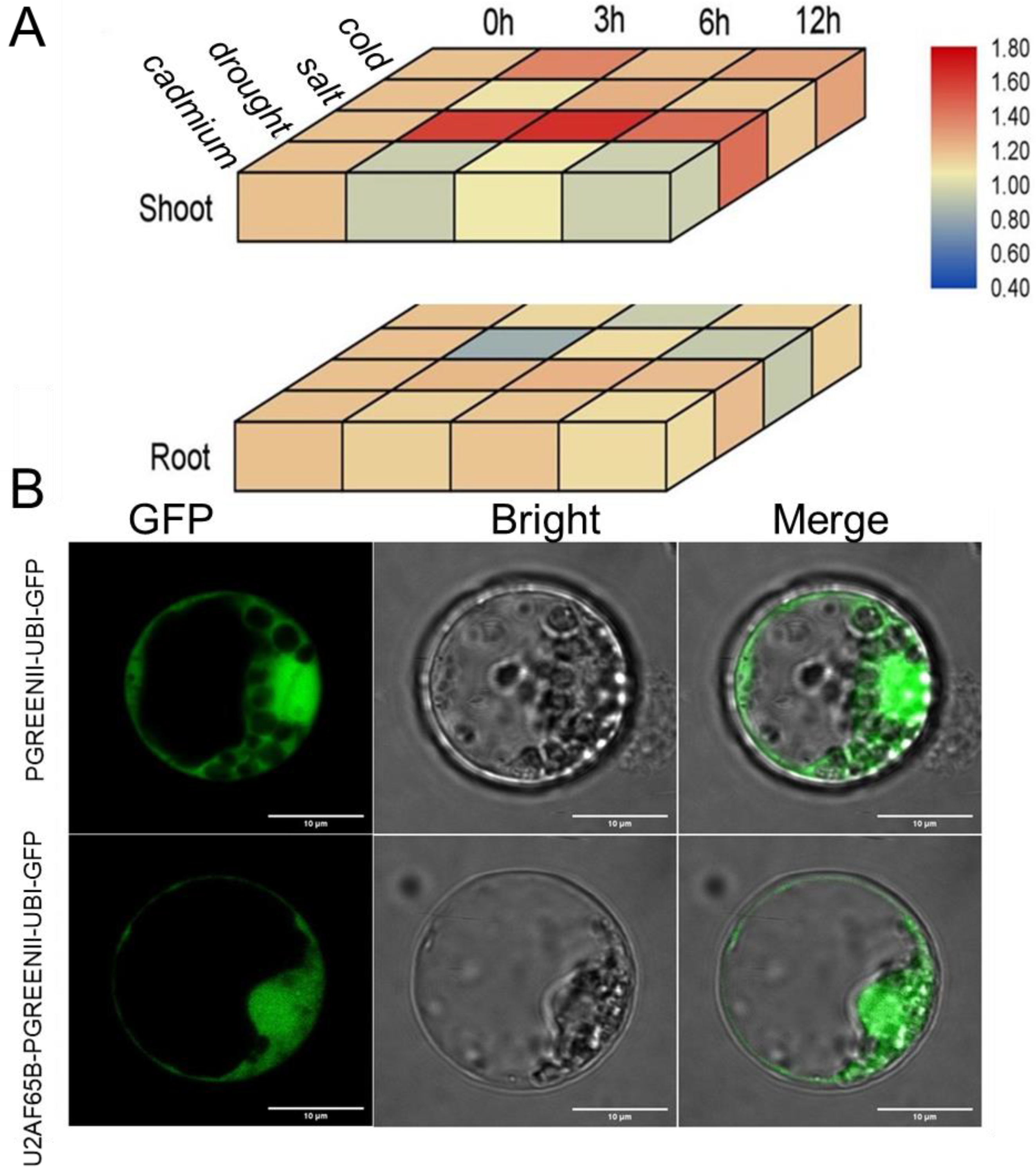
2.5. Expression and Subcellular Localization of Plant U2AF65B Genes
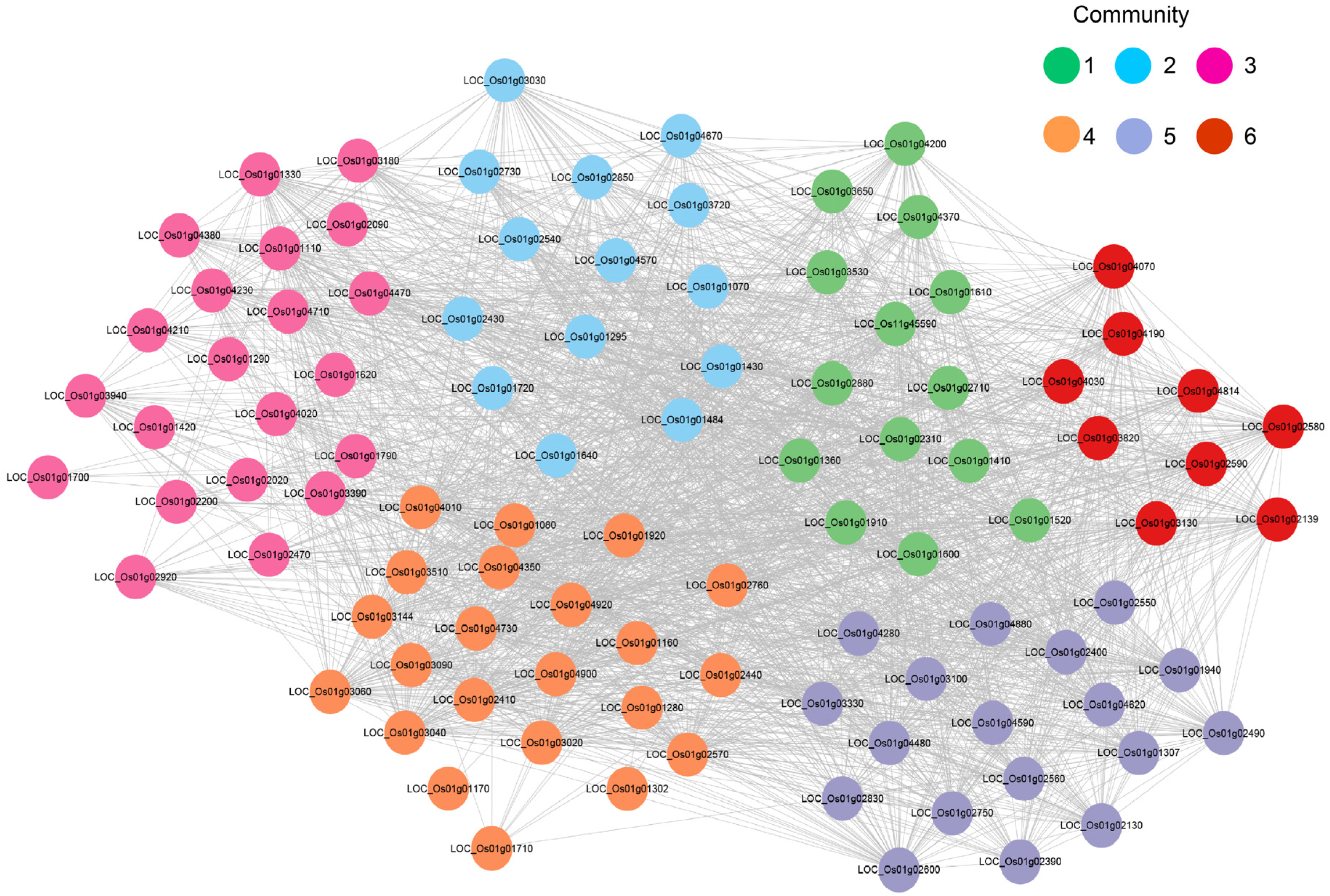
2.6. Analysis of the Co-Expression Network of the OsU2AF65B Gene Under Drought Stress
2.7. Analysis of AS Profile and Splicing Isoforms
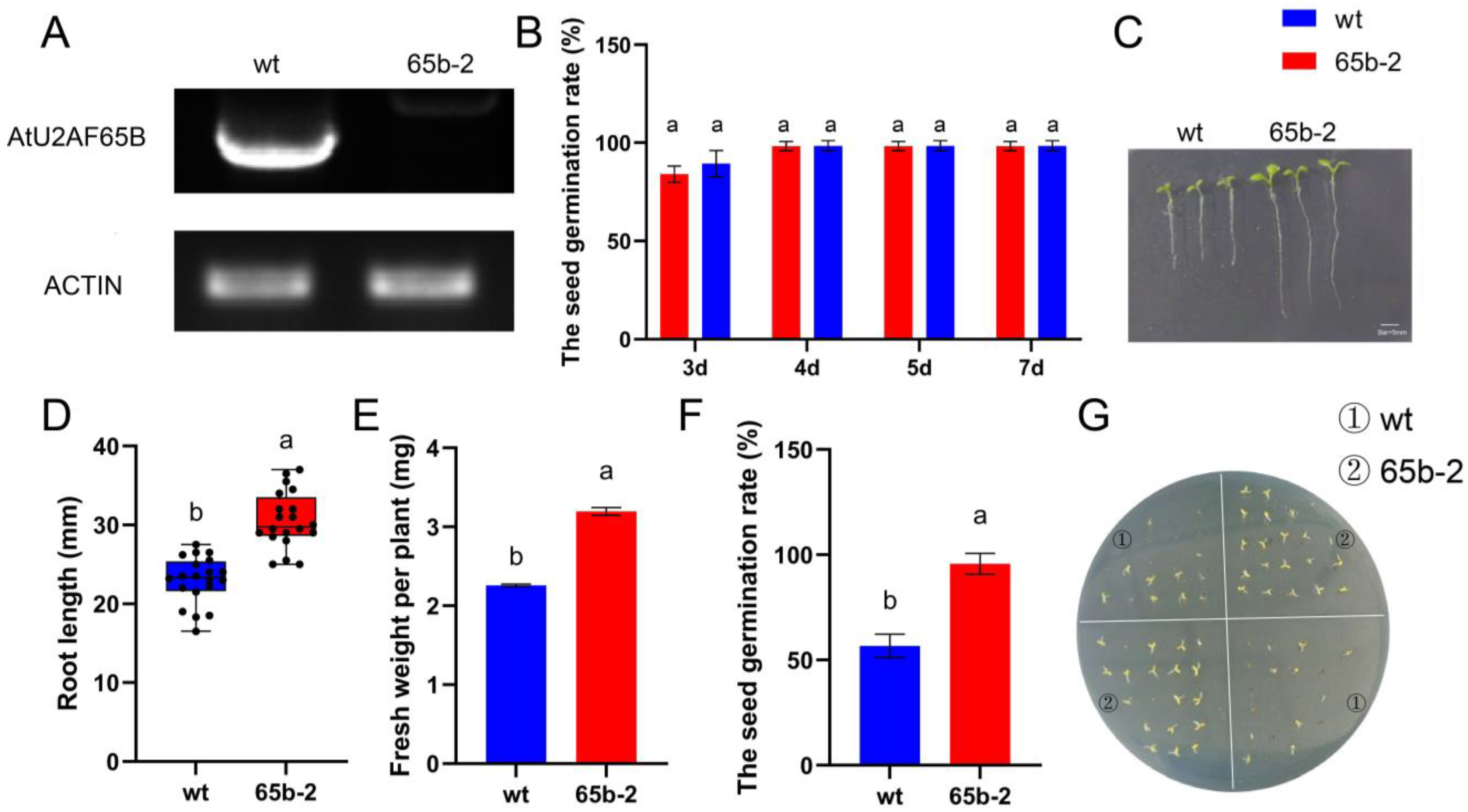
2.8. Loss of AtU2AF65B Function Enhances Root Elongation and Attenuates ABA-Dependent Germination Suppression
3. Discussion
3.1. The U2AF65B Genes Exhibited a Conserved Structure and Function
3.2. Regulatory Roles of U2AF65B Genes in Plant Stress Responses
4. Materials and Methods
4.1. Sequence Identification of Plant U2AF65B Genes
4.2. Phylogenetic Analysis of U2AF65B Gene Families
4.3. Gene Structure, Protein Domain, and Motif Analysis
4.4. Homology Modeling and Amino Acid Conservation Estimation
4.5. Cis-Acting Element Prediction
4.6. Plant Materials, Stress Treatments, RNA Extraction, and RT-qPCR Analysis
4.7. Subcellular Localization Analysis of U2AF65B Genes
4.8. Transcriptomic Data Analysis and Co-Expression Network Construction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AS | Alternative splicing |
| ABA | Abscisic acid |
| pre-mRNA | Precursor messenger RNA |
| RRM | RNA recognition motif |
| ERE | Estrogen response element |
| MeJA | Methyl jasmonate |
| ABRE | ABA-responsive elements |
| ARE | Antioxidant Response Element |
| DEG | Differentially expressed genes |
References
- Goremykin, V.V.; Hirsch-Ernst, K.I.; Wölfl, S.; Hellwig, F.H. The Chloroplast Genome of Nymphaea alba: Whole-Genome Analyses and the Problem of Identifying the Most Basal Angiosperm. Mol. Biol. Evol. 2004, 21, 1445–1454. [Google Scholar] [CrossRef]
- Ferrari, C.; Shivhare, D.; Hansen, B.O.; Pasha, A.; Esteban, E.; Provart, N.J.; Kragler, F.; Fernie, A.; Tohge, T.; Mutwil, M. Expression Atlas of Selaginella moellendorffii Provides Insights into the Evolution of Vasculature, Secondary Metabolism, and Roots. Plant Cell 2020, 32, 853–870. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Xian, W.; Fu, Y.; Marin, B.; Keller, J.; Wu, T.; Sun, W.; Li, X.; Xu, Y.; Zhang, Y.; et al. Genomes of Subaerial Zygnematophyceae Provide Insights into Land Plant Evolution. Cell 2019, 179, 1057–1067. [Google Scholar] [CrossRef]
- Sharp, P.A. The discovery of split genes and RNA splicing. Trends Biochem. Sci. 2005, 30, 279–281. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.A. Split genes and RNA splicing. Cell 1994, 77, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Rogalska, M.E.; Mancini, E.; Bonnal, S.; Gohr, A.; Dunyak, B.M.; Arecco, N.; Smith, P.G.; Vaillancourt, F.H.; Valcárcel, J. Transcriptome-wide splicing network reveals specialized regulatory functions of the core spliceosome. Science 2024, 386, 551–560. [Google Scholar] [CrossRef]
- Will, C.L.; Lührmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011, 3, a003707. [Google Scholar] [CrossRef]
- Liu, Y.; Do, S.; Huynh, H.; Li, J.-X.; Liu, Y.-G.; Du, Z.-Y.; Chen, M.-X. Importance of pre-mRNA splicing and its study tools in plants. Adv. Biotechnol. 2024, 2, 4. [Google Scholar] [CrossRef]
- Wahl, M.C.; Will, C.L.; Lührmann, R. The spliceosome: Design principles of a dynamic RNP machine. Cell 2009, 136, 701–718. [Google Scholar] [CrossRef]
- Kaida, D.; Berg, M.G.; Younis, I.; Kasim, M.; Singh, L.N.; Wan, L.; Dreyfuss, G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 2010, 468, 664–668. [Google Scholar] [CrossRef]
- König, H.; Matter, N.; Bader, R.; Thiele, W.; Müller, F. Splicing segregation: The minor spliceosome acts outside the nucleus and controls cell proliferation. Cell 2007, 131, 718–729. [Google Scholar] [CrossRef]
- Tidow, H.; Andreeva, A.; Rutherford, T.J.; Fersht, A.R. Solution structure of the U11-48K CHHC zinc-finger domain that specifically binds the 5′ splice site of U12-type introns. Structure 2009, 17, 294–302. [Google Scholar] [CrossRef]
- Côté, J.; Beaudoin, J.; Tacke, R.; Chabot, B. The U1 small nuclear ribonucleoprotein/5′ splice site interaction affects U2AF65 binding to the downstream 3′ splice site. J. Biol. Chem. 1995, 270, 4031–4036. [Google Scholar] [CrossRef] [PubMed]
- Corrionero, A.; Raker, V.A.; Izquierdo, J.M.; Valcárcel, J. Strict 3′ splice site sequence requirements for U2 snRNP recruitment after U2AF binding underlie a genetic defect leading to autoimmune disease. Rna 2011, 17, 401–411. [Google Scholar] [CrossRef]
- Zhang, M.; Zamore, P.D.; Carmo-Fonseca, M.; Lamond, A.I.; Green, M.R. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc. Natl. Acad. Sci. USA 1992, 89, 8769–8773. [Google Scholar] [CrossRef] [PubMed]
- Fairbrother, W.G.; Chasin, L.A. Human genomic sequences that inhibit splicing. Mol. Cell Biol. 2000, 20, 6816–6825. [Google Scholar] [CrossRef]
- Kellenberger, E.; Stier, G.; Sattler, M. Induced folding of the U2AF35 RRM upon binding to U2AF65. FEBS Lett. 2002, 528, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Zamore, P.D.; Patton, J.G.; Green, M.R. Cloning and domain structure of the mammalian splicing factor U2AF. Nature 1992, 355, 609–614. [Google Scholar] [CrossRef]
- Thickman, K.R.; Sickmier, E.A.; Kielkopf, C.L. Alternative conformations at the RNA-binding surface of the N-terminal U2AF(65) RNA recognition motif. J. Mol. Biol. 2007, 366, 703–710. [Google Scholar] [CrossRef]
- Kielkopf, C.L.; Lücke, S.; Green, M.R. U2AF homology motifs: Protein recognition in the RRM world. Genes Dev. 2004, 18, 1513–1526. [Google Scholar] [CrossRef]
- Loerch, S.; Kielkopf, C.L. Unmasking the U2AF homology motif family: A bona fide protein-protein interaction motif in disguise. Rna 2016, 22, 1795–1807. [Google Scholar] [CrossRef] [PubMed]
- Jagtap, P.K.; Garg, D.; Kapp, T.G.; Will, C.L.; Demmer, O.; Lührmann, R.; Kessler, H.; Sattler, M. Rational Design of Cyclic Peptide Inhibitors of U2AF Homology Motif (UHM) Domains To Modulate Pre-mRNA Splicing. J. Med. Chem. 2016, 59, 10190–10197. [Google Scholar] [CrossRef]
- Chen, C.; Zhao, X.; Kierzek, R.; Yu, Y.T. A flexible RNA backbone within the polypyrimidine tract is required for U2AF65 binding and pre-mRNA splicing in vivo. Mol. Cell Biol. 2010, 30, 4108–4119. [Google Scholar] [CrossRef]
- Yoshida, H.; Park, S.Y.; Oda, T.; Akiyoshi, T.; Sato, M.; Shirouzu, M.; Tsuda, K.; Kuwasako, K.; Unzai, S.; Muto, Y.; et al. A novel 3′ splice site recognition by the two zinc fingers in the U2AF small subunit. Genes Dev. 2015, 29, 1649–1660. [Google Scholar] [CrossRef]
- Saha, K.; Fernandez, M.M.; Biswas, T.; Lumba, C.L.M.; Ghosh, G. In vitro assembly of an early spliceosome defining both splice sites. Biorxiv 2018. [Google Scholar]
- Zhang, Y.; Madl, T.; Bagdiul, I.; Kern, T.; Kang, H.-S.; Zou, P.; Mäusbacher, N.; Sieber, S.A.; Krämer, A.; Sattler, M. Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3′-splice site recognition. Nucleic Acids Res. 2012, 41, 1343–1354. [Google Scholar] [CrossRef] [PubMed]
- Sridharan, V.; Heimiller, J.; Singh, R. Genomic mRNA profiling reveals compensatory mechanisms for the requirement of the essential splicing factor U2AF. Mol. Cell Biol. 2011, 31, 652–661. [Google Scholar] [CrossRef]
- Kent, O.A.; Ritchie, D.B.; Macmillan, A.M. Characterization of a U2AF-independent commitment complex (E’) in the mammalian spliceosome assembly pathway. Mol. Cell Biol. 2005, 25, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Moon, H.; Loh, T.J.; Jang, H.N.; Liu, Y.; Zhou, J.; Ohn, T.; Zheng, X.; Shen, H. Splicing inhibition of U2AF65 leads to alternative exon skipping. Proc. Natl. Acad. Sci. USA 2015, 112, 9926–9931. [Google Scholar] [CrossRef]
- Fleckner, J.; Zhang, M.; Valcárcel, J.; Green, M.R. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 1997, 11, 1864–1872. [Google Scholar] [CrossRef]
- Lee, H.T.; Park, H.Y.; Lee, K.C.; Lee, J.H.; Kim, J.K. Two Arabidopsis Splicing Factors, U2AF65a and U2AF65b, Differentially Control Flowering Time by Modulating the Expression or Alternative Splicing of a Subset of FLC Upstream Regulators. Plants 2023, 12, 1655. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Gao, C.; Wang, Y.; He, Y.; Du, J.; Chen, M.; Zhao, H.; Fang, H.; Wang, B.; Cao, Y. Phylogenetic Analysis of the Plant U2 snRNP Auxiliary Factor Large Subunit A Gene Family in Response to Developmental Cues and Environmental Stimuli. Front. Plant Sci. 2021, 12, 739671. [Google Scholar] [CrossRef] [PubMed]
- Xiong, F.; Ren, J.J.; Yu, Q.; Wang, Y.Y.; Lu, C.C.; Kong, L.J.; Otegui, M.S.; Wang, X.L. AtU2AF65b functions in abscisic acid mediated flowering via regulating the precursor messenger RNA splicing of ABI5 and FLC in Arabidopsis. New Phytol. 2019, 223, 277–292. [Google Scholar] [CrossRef] [PubMed]
- Domon, C.; Lorković, Z.J.; Valcárcel, J.; Filipowicz, W. Multiple forms of the U2 small nuclear ribonucleoprotein auxiliary factor U2AF subunits expressed in higher plants. J. Biol. Chem. 1998, 273, 34603–34610. [Google Scholar] [CrossRef]
- Park, H.Y.; Lee, K.C.; Jang, Y.H.; Kim, S.K.; Thu, M.P.; Lee, J.H.; Kim, J.K. The Arabidopsis splicing factors, AtU2AF65, AtU2AF35, and AtSF1 shuttle between nuclei and cytoplasms. Plant Cell Rep. 2017, 36, 1113–1123. [Google Scholar] [CrossRef]
- Park, H.Y.; Lee, H.T.; Lee, J.H.; Kim, J.K. Arabidopsis U2AF65 Regulates Flowering Time and the Growth of Pollen Tubes. Front. Plant Sci. 2019, 10, 569. [Google Scholar] [CrossRef]
- Yang, X.; Jia, Z.; Pu, Q.; Tian, Y.; Zhu, F.; Liu, Y. ABA Mediates Plant Development and Abiotic Stress via Alternative Splicing. Int. J. Mol. Sci. 2022, 23, 3796. [Google Scholar] [CrossRef]
- Verhage, L.; Severing, E.I.; Bucher, J.; Lammers, M.; Busscher-Lange, J.; Bonnema, G.; Rodenburg, N.; Proveniers, M.C.; Angenent, G.C.; Immink, R.G. Splicing-related genes are alternatively spliced upon changes in ambient temperatures in plants. PLoS ONE 2017, 12, e0172950. [Google Scholar] [CrossRef]
- Guan, Q.; Wu, J.; Zhang, Y.; Jiang, C.; Liu, R.; Chai, C.; Zhu, J. A DEAD box RNA helicase is critical for pre-mRNA splicing, cold-responsive gene regulation, and cold tolerance in Arabidopsis. Plant Cell 2013, 25, 342–356. [Google Scholar] [CrossRef]
- Feng, J.; Li, J.; Gao, Z.; Lu, Y.; Yu, J.; Zheng, Q.; Yan, S.; Zhang, W.; He, H.; Ma, L.; et al. SKIP Confers Osmotic Tolerance during Salt Stress by Controlling Alternative Gene Splicing in Arabidopsis. Mol. Plant 2015, 8, 1038–1052. [Google Scholar] [CrossRef]
- Zhu, F.Y.; Chen, X.; Song, Y.C.; Lam, L.P.Y.; Tobimatsu, Y.; Gao, B.; Chen, M.X.; Cao, F.L. SWATH-MS-based proteogenomic analysis reveals the involvement of alternative splicing in poplar upon lead stress. Genome Res. 2023, 33, 371–385. [Google Scholar] [CrossRef] [PubMed]
- Alhabsi, A.; Butt, H.; Kirschner, G.K.; Blilou, I.; Mahfouz, M.M. SCR106 splicing factor modulates abiotic stress responses by maintaining RNA splicing in rice. J. Exp. Bot. 2024, 75, 802–818. [Google Scholar] [CrossRef]
- Tiwari, S.B.; Belachew, A.; Ma, S.F.; Young, M.; Ade, J.; Shen, Y.; Marion, C.M.; Holtan, H.E.; Bailey, A.; Stone, J.K.; et al. The EDLL motif: A potent plant transcriptional activation domain from AP2/ERF transcription factors. Plant J. 2012, 70, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jia, J.; Zhai, J. Plant Intron Splicing Efficiency Database (PISE): Exploring splicing of ~1,650,000 introns in Arabidopsis, maize, rice and soybean from ~57,000 public RNA-seq libraries. Sci. China Life Sci. 2023, 66, 602–611. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Sun, W.; Duan, Y.; Xu, Y.; Wang, H.; Hao, J.; Han, Y.; Liu, C. Genome-Wide Identification and Expression Analysis of HSP70 Gene Family Under High-Temperature Stress in Lettuce (Lactuca sativa L.). Int. J. Mol. Sci. 2024, 26, 102. [Google Scholar] [CrossRef]
- Koo, S.C.; Yoon, H.W.; Kim, C.Y.; Moon, B.C.; Cheong, Y.H.; Han, H.J.; Lee, S.M.; Kang, K.Y.; Kim, M.C.; Lee, S.Y.; et al. Alternative splicing of the OsBWMK1 gene generates three transcript variants showing differential subcellular localizations. Biochem. Biophys. Res. Commun. 2007, 360, 188–193. [Google Scholar] [CrossRef]
- Miao, H.; Wu, F.; Li, Y.; Qin, C.; Zhao, Y.; Xie, M.; Dai, H.; Yao, H.; Cai, H.; Wang, Q.; et al. MALAT1 modulates alternative splicing by cooperating with the splicing factors PTBP1 and PSF. Sci. Adv. 2022, 8, eabq7289. [Google Scholar] [CrossRef]
- Zheng, H.C.; Jiang, H.M. Shuttling of cellular proteins between the plasma membrane and nucleus (Review). Mol. Med. Rep. 2022, 25, 14. [Google Scholar] [CrossRef]
- Wang, Z.; Ren, Z.; Cheng, C.; Wang, T.; Ji, H.; Zhao, Y.; Deng, Z.; Zhi, L.; Lu, J.; Wu, X.; et al. Counteraction of ABA-Mediated Inhibition of Seed Germination and Seedling Establishment by ABA Signaling Terminator in Arabidopsis. Mol. Plant 2020, 13, 1284–1297. [Google Scholar] [CrossRef]
- Tognacca, R.S.; Servi, L.; Hernando, C.E.; Saura-Sanchez, M.; Yanovsky, M.J.; Petrillo, E.; Botto, J.F. Alternative Splicing Regulation During Light-Induced Germination of Arabidopsis thaliana Seeds. Front. Plant Sci. 2019, 10, 1076. [Google Scholar] [CrossRef]
- Lv, B.; Wei, K.; Hu, K.; Tian, T.; Zhang, F.; Yu, Z.; Zhang, D.; Su, Y.; Sang, Y.; Zhang, X.; et al. MPK14-mediated auxin signaling controls lateral root development via ERF13-regulated very-long-chain fatty acid biosynthesis. Mol. Plant 2021, 14, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Han, G.; Sun, C.; Sui, N. Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signal Behav. 2019, 14, 1613131. [Google Scholar] [CrossRef]
- Freitas, F.Z.; Virgilio, S.; Cupertino, F.B.; Kowbel, D.J.; Fioramonte, M.; Gozzo, F.C.; Glass, N.L.; Bertolini, M.C. The SEB-1 Transcription Factor Binds to the STRE Motif in Neurospora crassa and Regulates a Variety of Cellular Processes Including the Stress Response and Reserve Carbohydrate Metabolism. G3 2016, 6, 1327–1343. [Google Scholar] [CrossRef] [PubMed]
- Niggeweg, R.; Thurow, C.; Kegler, C.; Gatz, C. Tobacco transcription factor TGA2.2 is the main component of as-1-binding factor ASF-1 and is involved in salicylic acid- and auxin-inducible expression of as-1-containing target promoters. J. Biol. Chem. 2000, 275, 19897–19905. [Google Scholar] [CrossRef]
- Govind, A.P.; Thampan, R.V. Proteins interacting with the mammalian estrogen receptor: Proposal for an integrated model for estrogen receptor mediated regulation of transcription. J. Cell Biochem. 2001, 80, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Wang, S.; Dao, Y.; Wang, J.; Wang, K. Arabidopsis thaliana trehalose-6-phosphate phosphatase gene TPPI enhances drought tolerance by regulating stomatal apertures. J. Exp. Bot. 2020, 71, 4285–4297. [Google Scholar] [CrossRef]
- Day, I.S.; Golovkin, M.; Palusa, S.G.; Link, A.; Ali, G.S.; Thomas, J.; Richardson, D.N.; Reddy, A.S. Interactions of SR45, an SR-like protein, with spliceosomal proteins and an intronic sequence: Insights into regulated splicing. Plant J. 2012, 71, 936–947. [Google Scholar] [CrossRef]
- Huang, R.; Jin, Z.; Zhang, D.; Lee, L.; Zhou, J.; Xiao, L.; Li, P.; Zhang, M.; Tian, C.; Zhang, W.; et al. Rare variations within the serine/arginine-rich splicing factor PtoRSZ21 modulate stomatal size to determine drought tolerance in Populus. New Phytol. 2024, 243, 1776–1794. [Google Scholar] [CrossRef]
- Xiang, W.; Jin, Y.; Wang, Y.; Han, S.; He, L.; Fan, Y.; Zhou, J.; Shi, H.; Yang, W. The splicing factor U2AF65B regulates cytosine methylation through interacting with DEFECTIVE IN MERISTEM SILENCING 3 in Arabidopsis. New Phytol. 2025, 3, 21. [Google Scholar] [CrossRef]
- Seregin, I.V.; Kozhevnikova, A.D. Roles of root and shoot tissues in transport and accumulation of cadmium, lead, nickel, and strontium. Russ. J. Plant Physiol. 2008, 55, 1–22. [Google Scholar] [CrossRef]
- Lei, G.J.; Fujii-Kashino, M.; Wu, D.Z.; Hisano, H.; Saisho, D.; Deng, F.; Yamaji, N.; Sato, K.; Zhao, F.; Ma, J.F. Breeding for low cadmium barley by introgression of a Sukkula-like transposable element. Nat. Food 2020, 1, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.L.; Zhang, M.M.; Li, C.S.; Li, B.Y.; Zhuo, S.L.; Yang, Y.S.; Chen, Y.D.; Zhong, A.N.; Liu, H.Y.; Lai, W.F.; et al. Response mechanism of water status and photosynthetic characteristics of Cotoneaster multiflorus under drought stress and rehydrated conditions. Front. Plant Sci. 2024, 15, 1457955. [Google Scholar] [CrossRef]
- Aanniz, T.; El Baaboua, A.; Aboulaghras, S.; Bouyahya, A.; Benali, T.; Balahbib, A.; El Omari, N.; Butnariu, M.; Muzammil, K.; Yadav, K.K.; et al. Impact of water stress to plant epigenetic mechanisms in stress and adaptation. Physiol. Plant 2025, 177, e70058. [Google Scholar] [CrossRef]
- Camacho, C.G.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J. BLAST plus: Architecture and applications. BMC Bioinf. 2009, 10, 1–9. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids. Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinf. 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Morariu, V.I.; Srinivasan, B.; Raykar, V.; Davis, L. Automatic online tuning for fast Gaussian summation. Adv. Neural Inf. Process. Syst. 2008, 21, 1113–1120. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids. Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids. Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids. Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Kondo, Y.; Oubridge, C.; van Roon, A.M.; Nagai, K. Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. Elife 2015, 4, e04986. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Zhang, Q.; Chen, Y.; Zhao, H.; Lang, Y.; Yu, C.; Yang, J. Identification of differential expression genes in leaves of rice (Oryza sativa L.) in response to heat stress by cDNA-AFLP analysis. Biomed. Res. Int. 2013, 2013, 576189. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, X.; Wang, Y.; Tang, B.; Zhou, J.; Gu, Y.; Shen, Q.; Zhou, Y.; Wang, B.; Fang, H.; Cao, Y. A Comprehensive Analysis of the Alternative Splicing Co-Factor U2AF65B Gene Family Reveals Its Role in Stress Responses and Root Development. Int. J. Mol. Sci. 2025, 26, 3901. https://doi.org/10.3390/ijms26083901
Meng X, Wang Y, Tang B, Zhou J, Gu Y, Shen Q, Zhou Y, Wang B, Fang H, Cao Y. A Comprehensive Analysis of the Alternative Splicing Co-Factor U2AF65B Gene Family Reveals Its Role in Stress Responses and Root Development. International Journal of Molecular Sciences. 2025; 26(8):3901. https://doi.org/10.3390/ijms26083901
Chicago/Turabian StyleMeng, Xiangfeng, Yongzhou Wang, Bei Tang, Jie Zhou, Yangfan Gu, Qingqiu Shen, Yaqun Zhou, Baohua Wang, Hui Fang, and Yunying Cao. 2025. "A Comprehensive Analysis of the Alternative Splicing Co-Factor U2AF65B Gene Family Reveals Its Role in Stress Responses and Root Development" International Journal of Molecular Sciences 26, no. 8: 3901. https://doi.org/10.3390/ijms26083901
APA StyleMeng, X., Wang, Y., Tang, B., Zhou, J., Gu, Y., Shen, Q., Zhou, Y., Wang, B., Fang, H., & Cao, Y. (2025). A Comprehensive Analysis of the Alternative Splicing Co-Factor U2AF65B Gene Family Reveals Its Role in Stress Responses and Root Development. International Journal of Molecular Sciences, 26(8), 3901. https://doi.org/10.3390/ijms26083901




