Expression Pattern of the AB1-Gal4 Driver in Drosophila Third-Instar Larvae
Abstract
1. Introduction
2. Results
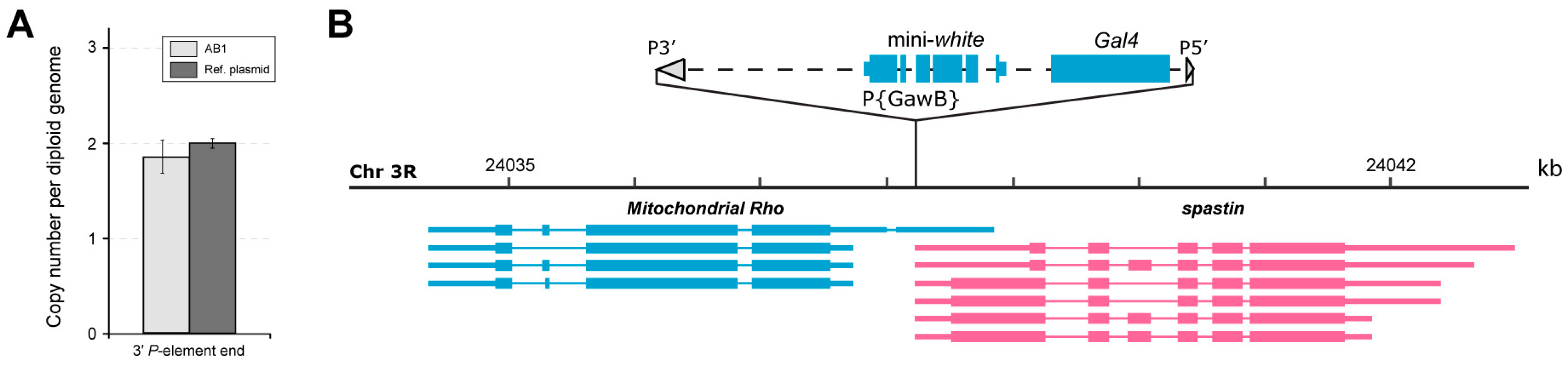
2.1. Molecular Analysis of the AB1-Gal4 Driver Line
2.2. Analysis of the Influence of the P{GawB} Transposon Insertion on the Expression of the Miro and Spas Genes in the Third-Instar Larval Salivary Glands and CNS
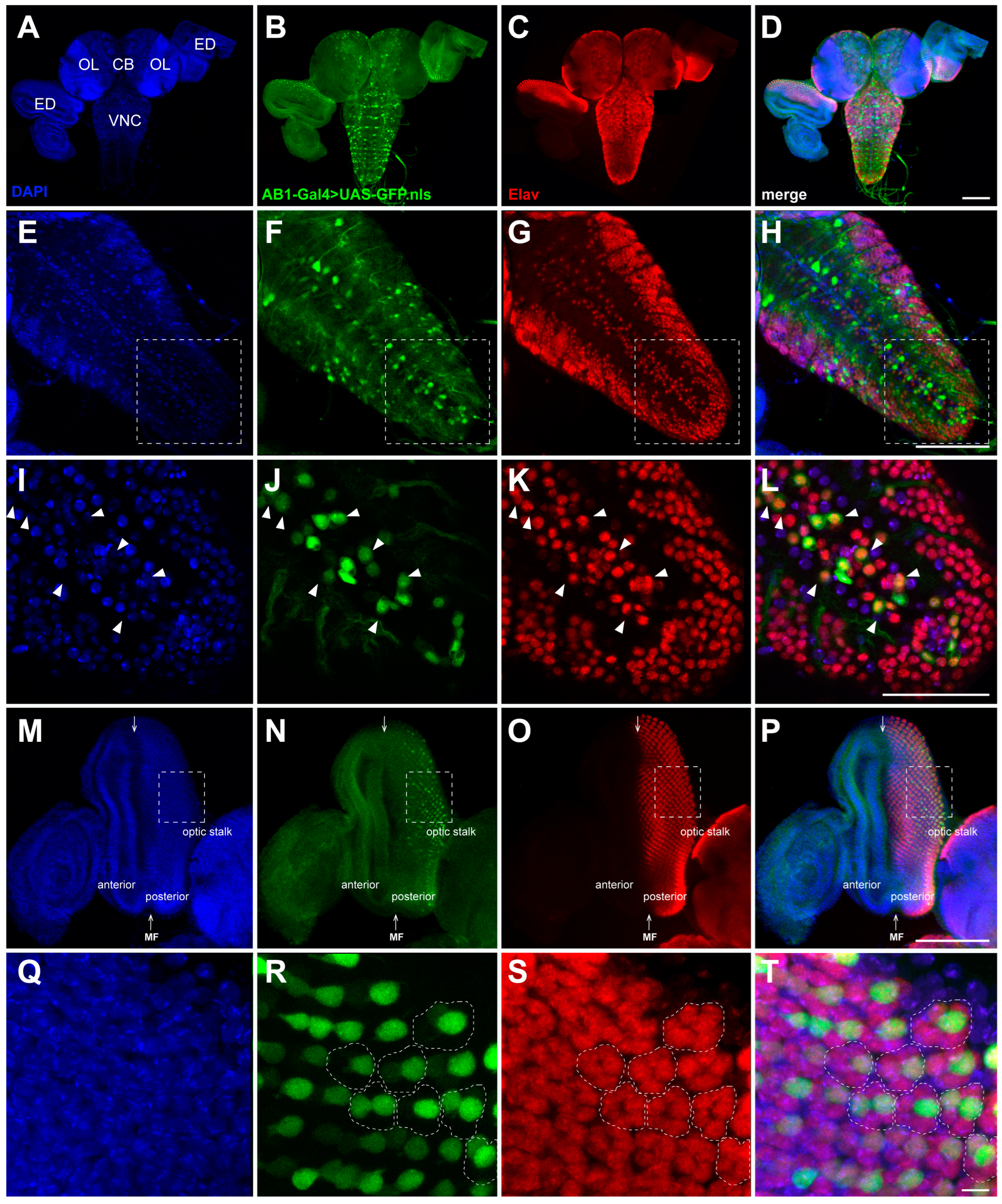
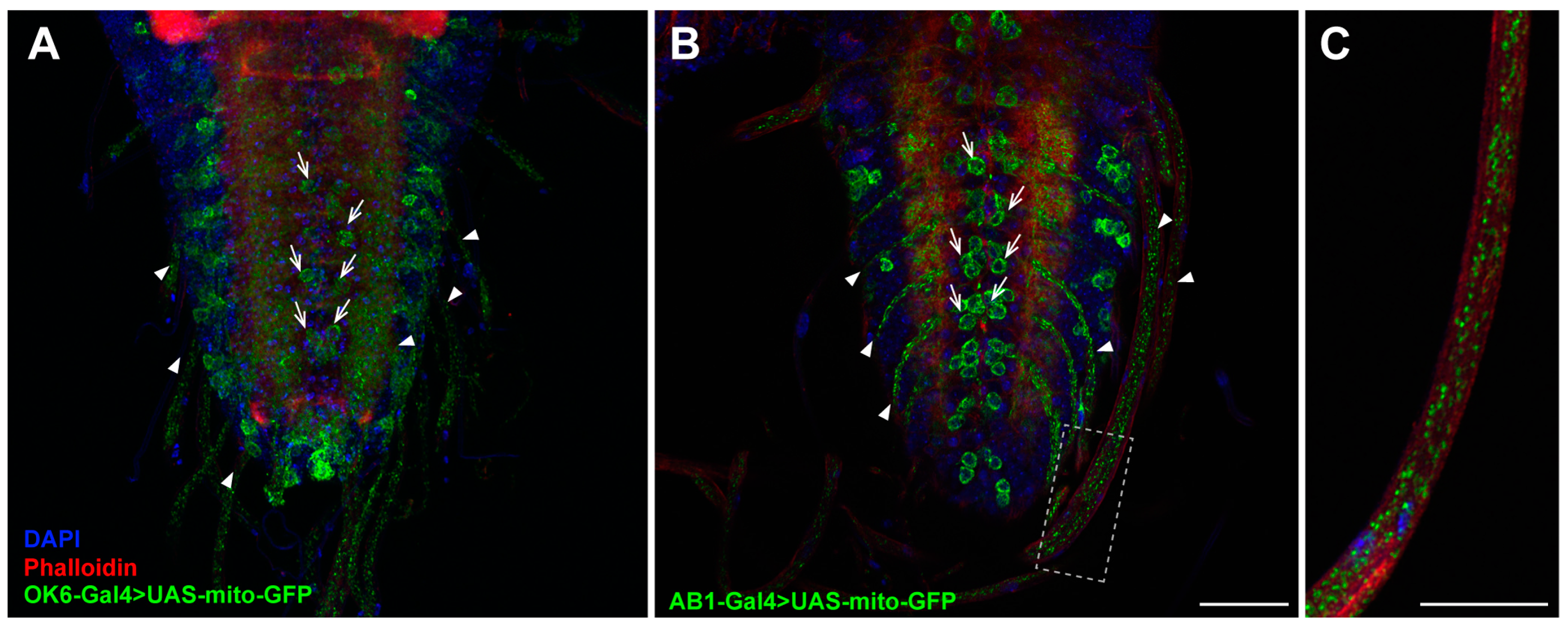
2.3. Expression Pattern of the AB1-Gal4 Driver in the CNS of Third-Instar Larvae
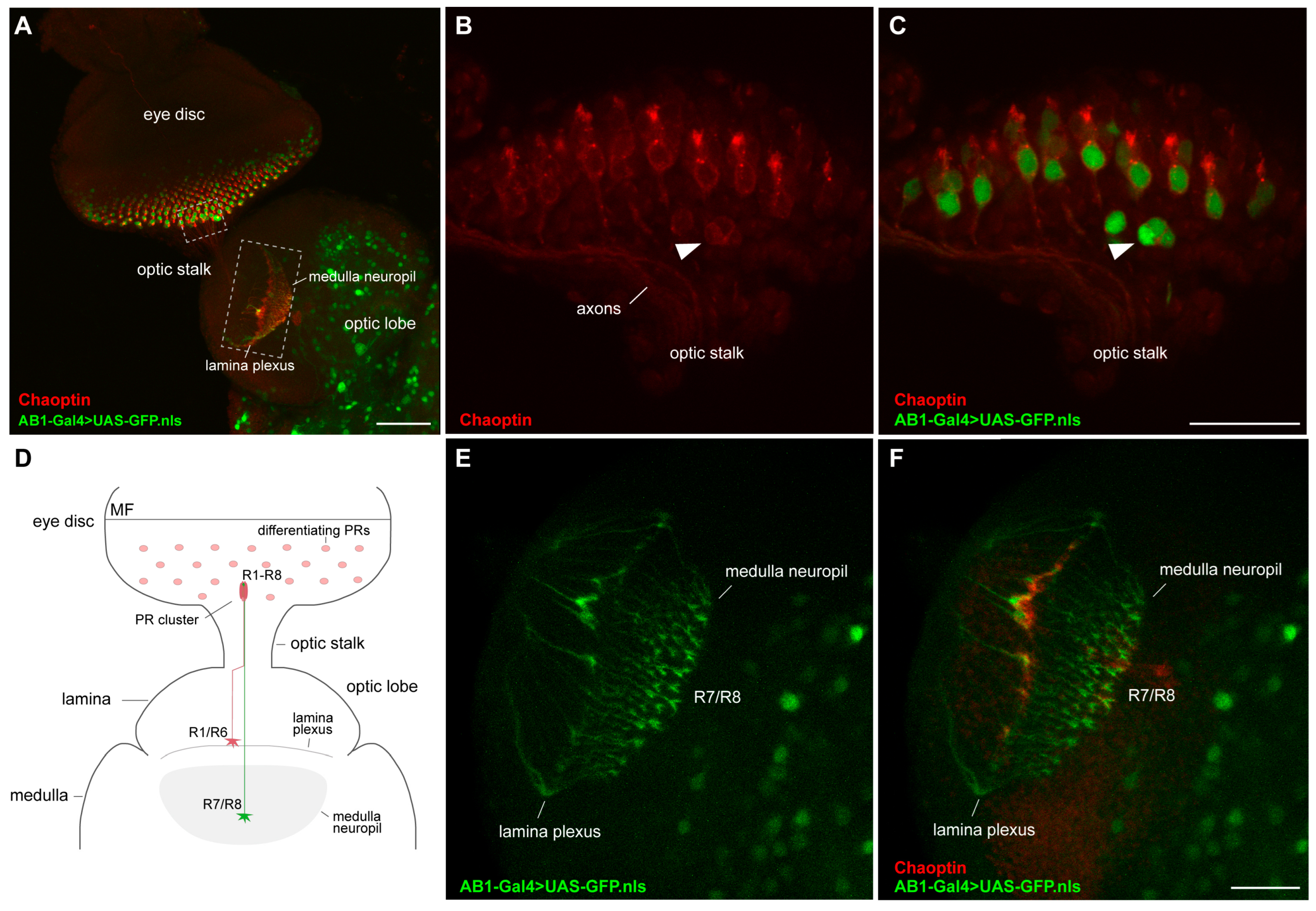
2.4. Expression Pattern of the AB1-Gal4 Driver in Eye Discs of Third-Instar Larvae
3. Discussion
4. Materials and Methods
4.1. Fly Stocks
4.2. Determination of P-Element Transgene Copy Number and Mapping Its Insertion Site
4.3. Molecular Analysis of the P{GawB} Transposon
4.4. RNA Extraction and RT-qPCR Analysis
4.5. Western Blotting
4.6. Immunostaining and Microscopy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fischer, J.A.; Giniger, E.; Maniatis, T.; Ptashne, M. GAL4 Activates Transcription in Drosophila. Nature 1988, 332, 853–856. [Google Scholar] [CrossRef] [PubMed]
- Kakidani, H.; Ptashne, M. GAL4 Activates Gene Expression in Mammalian Cells. Cell 1988, 52, 161–167. [Google Scholar] [CrossRef]
- Webster, N.; Jin, J.R.; Green, S.; Hollis, M.; Chambon, P. The Yeast UASG Is a Transcriptional Enhancer in Human HeLa Cells in the Presence of the GAL4 Trans-Activator. Cell 1988, 52, 169–178. [Google Scholar] [CrossRef]
- Brand, A.H.; Perrimon, N. Targeted Gene Expression as a Means of Altering Cell Fates and Generating Dominant Phenotypes. Development 1993, 118, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Perkins, L.A.; Holderbaum, L.; Tao, R.; Hu, Y.; Sopko, R.; McCall, K.; Yang-Zhou, D.; Flockhart, I.; Binari, R.; Shim, H.-S.; et al. The Transgenic RNAi Project at Harvard Medical School: Resources and Validation. Genetics 2015, 201, 843–852. [Google Scholar] [CrossRef]
- Dietzl, G.; Chen, D.; Schnorrer, F.; Su, K.-C.; Barinova, Y.; Fellner, M.; Gasser, B.; Kinsey, K.; Oppel, S.; Scheiblauer, S.; et al. A Genome-Wide Transgenic RNAi Library for Conditional Gene Inactivation in Drosophila. Nature 2007, 448, 151–156. [Google Scholar] [CrossRef]
- Ni, J.-Q.; Liu, L.-P.; Binari, R.; Hardy, R.; Shim, H.-S.; Cavallaro, A.; Booker, M.; Pfeiffer, B.D.; Markstein, M.; Wang, H.; et al. A Drosophila Resource of Transgenic RNAi Lines for Neurogenetics. Genetics 2009, 182, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.-T.; Zirin, J.; Kanca, O.; Lin, W.-W.; Schulze, K.L.; Li-Kroeger, D.; Tao, R.; Devereaux, C.; Hu, Y.; Chung, V.; et al. A Gene-Specific T2A-GAL4 Library for Drosophila. eLife 2018, 7, e35574. [Google Scholar] [CrossRef]
- Fisher, Y.E.; Yang, H.H.; Isaacman-Beck, J.; Xie, M.; Gohl, D.M.; Clandinin, T.R. FlpStop, a Tool for Conditional Gene Control in Drosophila. eLife 2017, 6, e22279. [Google Scholar] [CrossRef] [PubMed]
- Port, F.; Chen, H.-M.; Lee, T.; Bullock, S.L. Optimized CRISPR/Cas Tools for Efficient Germline and Somatic Genome Engineering in Drosophila. Proc. Natl. Acad. Sci. USA 2014, 111, E2967–E2976. [Google Scholar] [CrossRef]
- Lin, S.; Ewen-Campen, B.; Ni, X.; Housden, B.E.; Perrimon, N. In Vivo Transcriptional Activation Using CRISPR/Cas9 in Drosophila. Genetics 2015, 201, 433–442. [Google Scholar] [CrossRef]
- Port, F.; Bullock, S.L. Augmenting CRISPR Applications in Drosophila with tRNA-Flanked sgRNAs. Nat. Methods 2016, 13, 852–854. [Google Scholar] [CrossRef]
- Port, F.; Starostecka, M.; Boutros, M. Multiplexed Conditional Genome Editing with Cas12a in Drosophila. Proc. Natl. Acad. Sci. USA 2020, 117, 22890–22899. [Google Scholar] [CrossRef]
- Xu, R.; Deng, K.; Zhu, Y.; Wu, Y.; Ren, J.; Wan, M.; Zhao, S.; Wu, X.; Han, M.; Zhuang, Y.; et al. A Large-Scale Functional Approach to Uncover Human Genes and Pathways in Drosophila. Cell Res. 2008, 18, 1114–1127. [Google Scholar] [CrossRef]
- Bischof, J.; Björklund, M.; Furger, E.; Schertel, C.; Taipale, J.; Basler, K. A Versatile Platform for Creating a Comprehensive UAS-ORFeome Library in Drosophila. Development 2013, 140, 2434–2442. [Google Scholar] [CrossRef]
- Brand, A.H.; Manoukian, A.S.; Perrimon, N. Ectopic Expression in Drosophila. Methods Cell Biol. 1994, 44, 635–654. [Google Scholar] [CrossRef]
- Pfeiffer, B.D.; Ngo, T.-T.B.; Hibbard, K.L.; Murphy, C.; Jenett, A.; Truman, J.W.; Rubin, G.M. Refinement of Tools for Targeted Gene Expression in Drosophila. Genetics 2010, 186, 735–755. [Google Scholar] [CrossRef]
- Gerlitz, O.; Nellen, D.; Ottiger, M.; Basler, K. A Screen for Genes Expressed in Drosophila Imaginal Discs. Int. J. Dev. Biol. 2002, 46, 173–176. [Google Scholar]
- Hrdlicka, L.; Gibson, M.; Kiger, A.; Micchelli, C.; Schober, M.; Schöck, F.; Perrimon, N. Analysis of Twenty-Four Gal4 Lines in Drosophila Melanogaster. Genesis 2002, 34, 51–57. [Google Scholar] [CrossRef]
- Pfeiffer, B.D.; Jenett, A.; Hammonds, A.S.; Ngo, T.-T.B.; Misra, S.; Murphy, C.; Scully, A.; Carlson, J.W.; Wan, K.H.; Laverty, T.R.; et al. Tools for Neuroanatomy and Neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA 2008, 105, 9715–9720. [Google Scholar] [CrossRef] [PubMed]
- Munro, S.; Freeman, M. The Notch Signalling Regulator Fringe Acts in the Golgi Apparatus and Requires the Glycosyltransferase Signature Motif DXD. Curr. Biol. 2000, 10, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Atienza-Manuel, A.; Castillo-Mancho, V.; De Renzis, S.; Culi, J.; Ruiz-Gómez, M. Endocytosis Mediated by an Atypical CUBAM Complex Modulates Slit Diaphragm Dynamics in Nephrocytes. Development 2021, 148, dev199894. [Google Scholar] [CrossRef] [PubMed]
- Hevia, C.F.; López-Varea, A.; Esteban, N.; de Celis, J.F. A Search for Genes Mediating the Growth-Promoting Function of TGFβ in the Drosophila Melanogaster Wing Disc. Genetics 2017, 206, 231–249. [Google Scholar] [CrossRef][Green Version]
- Sun, J.; Wei, H.-M.; Xu, J.; Chang, J.-F.; Yang, Z.; Ren, X.; Lv, W.-W.; Liu, L.-P.; Pan, L.-X.; Wang, X.; et al. Histone H1-Mediated Epigenetic Regulation Controls Germline Stem Cell Self-Renewal by Modulating H4K16 Acetylation. Nat. Commun. 2015, 6, 8856. [Google Scholar] [CrossRef] [PubMed]
- Costantino, B.F.B.; Bricker, D.K.; Alexandre, K.; Shen, K.; Merriam, J.R.; Antoniewski, C.; Callender, J.L.; Henrich, V.C.; Presente, A.; Andres, A.J. A Novel Ecdysone Receptor Mediates Steroid-Regulated Developmental Events during the Mid-Third Instar of Drosophila. PLoS Genet. 2008, 4, e1000102. [Google Scholar] [CrossRef]
- Ozaki, K.; Nagatani, H.; Ozaki, M.; Tokunaga, F. Maturation of Major Drosophila Rhodopsin, ninaE, Requires Chromophore 3-Hydroxyretinal. Neuron 1993, 10, 1113–1119. [Google Scholar] [CrossRef]
- Gu, G.; Yang, J.; Mitchell, K.A.; O’Tousa, J.E. Drosophila ninaB and ninaD Act Outside of Retina to Produce Rhodopsin Chromophore. J. Biol. Chem. 2004, 279, 18608–18613. [Google Scholar] [CrossRef]
- Yang, J.; O’Tousa, J.E. Cellular Sites of Drosophila NinaB and NinaD Activity in Vitamin A Metabolism. Mol. Cell Neurosci. 2007, 35, 49–56. [Google Scholar] [CrossRef]
- Ogienko, A.A.; Yarinich, L.A.; Fedorova, E.V.; Lebedev, M.O.; Andreyeva, E.N.; Pindyurin, A.V.; Baricheva, E.M. New Slbo-Gal4 Driver Lines for the Analysis of Border Cell Migration during Drosophila Oogenesis. Chromosoma 2018, 127, 475–487. [Google Scholar] [CrossRef]
- Ogienko, A.A.; Andreyeva, E.N.; Omelina, E.S.; Oshchepkova, A.L.; Pindyurin, A.V. Molecular and Cytological Analysis of Widely-Used Gal4 Driver Lines for Drosophila Neurobiology. BMC Genet. 2020, 21, 96. [Google Scholar] [CrossRef]
- Ochman, H.; Gerber, A.S.; Hartl, D.L. Genetic Applications of an Inverse Polymerase Chain Reaction. Genetics 1988, 120, 621–623. [Google Scholar] [CrossRef]
- Hoskins, R.A.; Carlson, J.W.; Wan, K.H.; Park, S.; Mendez, I.; Galle, S.E.; Booth, B.W.; Pfeiffer, B.D.; George, R.A.; Svirskas, R.; et al. The Release 6 Reference Sequence of the Drosophila Melanogaster Genome. Genome Res. 2015, 25, 445–458. [Google Scholar] [CrossRef] [PubMed]
- Fransson, A.; Ruusala, A.; Aspenström, P. Atypical Rho GTPases Have Roles in Mitochondrial Homeostasis and Apoptosis. J. Biol. Chem. 2003, 278, 6495–6502. [Google Scholar] [CrossRef]
- Guo, X.; Macleod, G.T.; Wellington, A.; Hu, F.; Panchumarthi, S.; Schoenfield, M.; Marin, L.; Charlton, M.P.; Atwood, H.L.; Zinsmaier, K.E. The GTPase dMiro Is Required for Axonal Transport of Mitochondria to Drosophila Synapses. Neuron 2005, 47, 379–393. [Google Scholar] [CrossRef]
- Kammermeier, L.; Spring, J.; Stierwald, M.; Burgunder, J.-M.; Reichert, H. Identification of the Drosophila Melanogaster Homolog of the Human Spastin Gene. Dev. Genes. Evol. 2003, 213, 412–415. [Google Scholar] [CrossRef] [PubMed]
- Sherwood, N.T.; Sun, Q.; Xue, M.; Zhang, B.; Zinn, K. Drosophila Spastin Regulates Synaptic Microtubule Networks and Is Required for Normal Motor Function. PLoS Biol. 2004, 2, e429. [Google Scholar] [CrossRef] [PubMed]
- Babic, M.; Russo, G.J.; Wellington, A.J.; Sangston, R.M.; Gonzalez, M.; Zinsmaier, K.E. Miro’s N-Terminal GTPase Domain Is Required for Transport of Mitochondria into Axons and Dendrites. J. Neurosci. 2015, 35, 5754–5771. [Google Scholar] [CrossRef]
- Lee, K.-S.; Lu, B. The Myriad Roles of Miro in the Nervous System: Axonal Transport of Mitochondria and Beyond. Front. Cell Neurosci. 2014, 8, 330. [Google Scholar] [CrossRef]
- Mishra, P.; Chan, D.C. Mitochondrial Dynamics and Inheritance during Cell Division, Development and Disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 634–646. [Google Scholar] [CrossRef]
- Trotta, N.; Orso, G.; Rossetto, M.G.; Daga, A.; Broadie, K. The Hereditary Spastic Paraplegia Gene, Spastin, Regulates Microtubule Stability to Modulate Synaptic Structure and Function. Curr. Biol. 2004, 14, 1135–1147. [Google Scholar] [CrossRef]
- Errico, A.; Ballabio, A.; Rugarli, E.I. Spastin, the Protein Mutated in Autosomal Dominant Hereditary Spastic Paraplegia, Is Involved in Microtubule Dynamics. Hum. Mol. Genet. 2002, 11, 153–163. [Google Scholar] [CrossRef]
- Chintapalli, V.R.; Wang, J.; Dow, J.A.T. Using FlyAtlas to Identify Better Drosophila Melanogaster Models of Human Disease. Nat. Genet. 2007, 39, 715–720. [Google Scholar] [CrossRef]
- Brown, J.B.; Boley, N.; Eisman, R.; May, G.E.; Stoiber, M.H.; Duff, M.O.; Booth, B.W.; Wen, J.; Park, S.; Suzuki, A.M.; et al. Diversity and Dynamics of the Drosophila Transcriptome. Nature 2014, 512, 393–399. [Google Scholar] [CrossRef]
- Öztürk-Çolak, A.; Marygold, S.J.; Antonazzo, G.; Attrill, H.; Goutte-Gattat, D.; Jenkins, V.K.; Matthews, B.B.; Millburn, G.; Dos Santos, G.; Tabone, C.J.; et al. FlyBase: Updates to the Drosophila Genes and Genomes Database. Genetics 2024, 227, iyad211. [Google Scholar] [CrossRef]
- Robinow, S.; White, K. Characterization and Spatial Distribution of the ELAV Protein during Drosophila Melanogaster Development. J. Neurobiol. 1991, 22, 443–461. [Google Scholar] [CrossRef]
- Hunter, I.; Coulson, B.; Zarin, A.A.; Baines, R.A. The Drosophila Larval Locomotor Circuit Provides a Model to Understand Neural Circuit Development and Function. Front. Neural Circuits 2021, 15, 684969. [Google Scholar] [CrossRef]
- Aberle, H.; Haghighi, A.P.; Fetter, R.D.; McCabe, B.D.; Magalhães, T.R.; Goodman, C.S. Wishful Thinking Encodes a BMP Type II Receptor That Regulates Synaptic Growth in Drosophila. Neuron 2002, 33, 545–558. [Google Scholar] [CrossRef]
- Sanyal, S.; Narayanan, R.; Consoulas, C.; Ramaswami, M. Evidence for Cell Autonomous AP1 Function in Regulation of Drosophila Motor-Neuron Plasticity. BMC Neurosci. 2003, 4, 20. [Google Scholar] [CrossRef]
- Cagan, R. Principles of Drosophila Eye Differentiation. Curr. Top. Dev. Biol. 2009, 89, 115–135. [Google Scholar] [CrossRef]
- Tomlinson, A. Cellular Interactions in the Developing Drosophila Eye. Development 1988, 104, 183–193. [Google Scholar] [CrossRef]
- Tomlinson, A.; Ready, D.F. Neuronal Differentiation in Drosophila Ommatidium. Dev. Biol. 1987, 120, 366–376. [Google Scholar] [CrossRef]
- Freeman, M. Reiterative Use of the EGF Receptor Triggers Differentiation of All Cell Types in the Drosophila Eye. Cell 1996, 87, 651–660. [Google Scholar] [CrossRef]
- Tomlinson, A.; Struhl, G. Delta/Notch and Boss/Sevenless Signals Act Combinatorially to Specify the Drosophila R7 Photoreceptor. Mol. Cell 2001, 7, 487–495. [Google Scholar] [CrossRef]
- Flores, G.V.; Duan, H.; Yan, H.; Nagaraj, R.; Fu, W.; Zou, Y.; Noll, M.; Banerjee, U. Combinatorial Signaling in the Specification of Unique Cell Fates. Cell 2000, 103, 75–85. [Google Scholar] [CrossRef]
- Roignant, J.-Y.; Treisman, J.E. Pattern Formation in the Drosophila Eye Disc. Int. J. Dev. Biol. 2009, 53, 795–804. [Google Scholar] [CrossRef]
- Wernet, M.F.; Mazzoni, E.O.; Çelik, A.; Duncan, D.M.; Duncan, I.; Desplan, C. Stochastic Spineless Expression Creates the Retinal Mosaic for Colour Vision. Nature 2006, 440, 174–180. [Google Scholar] [CrossRef]
- Katz, B.; Minke, B. Drosophila Photoreceptors and Signaling Mechanisms. Front. Cell Neurosci. 2009, 3, 2. [Google Scholar] [CrossRef]
- Kanie, Y.; Yamamoto-Hino, M.; Karino, Y.; Yokozawa, H.; Nishihara, S.; Ueda, R.; Goto, S.; Kanie, O. Insight into the Regulation of Glycan Synthesis in Drosophila Chaoptin Based on Mass Spectrometry. PLoS ONE 2009, 4, e5434. [Google Scholar] [CrossRef]
- Richard, M.; Doubková, K.; Nitta, Y.; Kawai, H.; Sugie, A.; Tavosanis, G. A Quantitative Model of Sporadic Axonal Degeneration in the Drosophila Visual System. J. Neurosci. 2022, 42, 4937–4952. [Google Scholar] [CrossRef]
- Rass, M.; Gizler, L.; Bayersdorfer, F.; Irlbeck, C.; Schramm, M.; Schneuwly, S. The Drosophila Functional Smad Suppressing Element Fuss, a Homologue of the Human Skor Genes, Retains pro-Oncogenic Properties of the Ski/Sno Family. PLoS ONE 2022, 17, e0262360. [Google Scholar] [CrossRef]
- Zhang, T.; Du, W. Groucho Restricts Rhomboid Expression and Couples EGFR Activation with R8 Selection during Drosophila Photoreceptor Differentiation. Dev. Biol. 2015, 407, 246–255. [Google Scholar] [CrossRef]
- Griffith, L.C. Identifying Behavioral Circuits in Drosophila Melanogaster: Moving Targets in a Flying Insect. Curr. Opin. Neurobiol. 2012, 22, 609–614. [Google Scholar] [CrossRef]
- Venken, K.J.T.; Simpson, J.H.; Bellen, H.J. Genetic Manipulation of Genes and Cells in the Nervous System of the Fruit Fly. Neuron 2011, 72, 202–230. [Google Scholar] [CrossRef]
- Koushika, S.P.; Lisbin, M.J.; White, K. ELAV, a Drosophila Neuron-Specific Protein, Mediates the Generation of an Alternatively Spliced Neural Protein Isoform. Curr. Biol. 1996, 6, 1634–1641. [Google Scholar] [CrossRef]
- Krantz, D.E.; Zipursky, S.L. Drosophila Chaoptin, a Member of the Leucine-Rich Repeat Family, Is a Photoreceptor Cell-Specific Adhesion Molecule. EMBO J. 1990, 9, 1969–1977. [Google Scholar] [CrossRef]
- Goldstein, A.; Falk, M.J. Single Large-Scale Mitochondrial DNA Deletion Syndromes. In GeneReviews®; Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Amemiya, A., Eds.; University of Washington, Seattle: Seattle, WA, USA, 1993. [Google Scholar]
- Crosby, A.H.; Proukakis, C. Is the Transportation Highway the Right Road for Hereditary Spastic Paraplegia? Am. J. Hum. Genet. 2002, 71, 1009–1016. [Google Scholar] [CrossRef]
- Yan, M.H.; Wang, X.; Zhu, X. Mitochondrial Defects and Oxidative Stress in Alzheimer Disease and Parkinson Disease. Free Radic. Biol. Med. 2013, 62, 90–101. [Google Scholar] [CrossRef]
- Birsa, N.; Norkett, R.; Wauer, T.; Mevissen, T.E.T.; Wu, H.-C.; Foltynie, T.; Bhatia, K.; Hirst, W.D.; Komander, D.; Plun-Favreau, H.; et al. Lysine 27 Ubiquitination of the Mitochondrial Transport Protein Miro Is Dependent on Serine 65 of the Parkin Ubiquitin Ligase. J. Biol. Chem. 2014, 289, 14569–14582. [Google Scholar] [CrossRef]
- Liu, S.; Sawada, T.; Lee, S.; Yu, W.; Silverio, G.; Alapatt, P.; Millan, I.; Shen, A.; Saxton, W.; Kanao, T.; et al. Parkinson’s Disease-Associated Kinase PINK1 Regulates Miro Protein Level and Axonal Transport of Mitochondria. PLoS Genet. 2012, 8, e1002537. [Google Scholar] [CrossRef]
- Tang, B.L. MIRO GTPases in Mitochondrial Transport, Homeostasis and Pathology. Cells 2015, 5, 1. [Google Scholar] [CrossRef]
- Pavlowsky, A.; Comyn, T.; Minatchy, J.; Geny, D.; Bun, P.; Danglot, L.; Preat, T.; Plaçais, P.-Y. Spaced Training Activates Miro/Milton-Dependent Mitochondrial Dynamics in Neuronal Axons to Sustain Long-Term Memory. Curr. Biol. 2024, 34, 1904–1917.e6. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, W.; Siedlak, S.L.; Liu, Y.; Liu, J.; Jiang, K.; Perry, G.; Zhu, X.; Wang, X. Miro1 Deficiency in Amyotrophic Lateral Sclerosis. Front. Aging Neurosci. 2015, 7, 100. [Google Scholar] [CrossRef]
- Mórotz, G.M.; De Vos, K.J.; Vagnoni, A.; Ackerley, S.; Shaw, C.E.; Miller, C.C.J. Amyotrophic Lateral Sclerosis-Associated Mutant VAPBP56S Perturbs Calcium Homeostasis to Disrupt Axonal Transport of Mitochondria. Hum. Mol. Genet. 2012, 21, 1979–1988. [Google Scholar] [CrossRef]
- Hazan, J.; Fonknechten, N.; Mavel, D.; Paternotte, C.; Samson, D.; Artiguenave, F.; Davoine, C.S.; Cruaud, C.; Dürr, A.; Wincker, P.; et al. Spastin, a New AAA Protein, Is Altered in the Most Frequent Form of Autosomal Dominant Spastic Paraplegia. Nat. Genet. 1999, 23, 296–303. [Google Scholar] [CrossRef]
- Orso, G.; Martinuzzi, A.; Rossetto, M.G.; Sartori, E.; Feany, M.; Daga, A. Disease-Related Phenotypes in a Drosophila Model of Hereditary Spastic Paraplegia Are Ameliorated by Treatment with Vinblastine. J. Clin. Investig. 2005, 115, 3026–3034. [Google Scholar] [CrossRef]
- Omelina, E.S.; Letiagina, A.E.; Boldyreva, L.V.; Ogienko, A.A.; Galimova, Y.A.; Yarinich, L.A.; Pindyurin, A.V.; Andreyeva, E.N. Slight Variations in the Sequence Downstream of the Polyadenylation Signal Significantly Increase Transgene Expression in HEK293T and CHO Cells. Int. J. Mol. Sci. 2022, 23, 15485. [Google Scholar] [CrossRef]
- Andreyeva, E.N.; Ogienko, A.A.; Yushkova, A.A.; Popova, J.V.; Pavlova, G.A.; Kozhevnikova, E.N.; Ivankin, A.V.; Gatti, M.; Pindyurin, A.V. Non3 Is an Essential Drosophila Gene Required for Proper Nucleolus Assembly. Vestn. VOGiS 2019, 23, 190–198. [Google Scholar] [CrossRef]
- Guild, G.M.; Connelly, P.S.; Shaw, M.K.; Tilney, L.G. Actin Filament Cables in Drosophila Nurse Cells Are Composed of Modules That Slide Passively Past One Another during Dumping. J. Cell Biol. 1997, 138, 783–797. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogienko, A.A.; Andreyeva, E.N.; Yarinich, L.A.; Pindyurin, A.V.; Battulina, N.V.; Omelina, E.S. Expression Pattern of the AB1-Gal4 Driver in Drosophila Third-Instar Larvae. Int. J. Mol. Sci. 2025, 26, 3923. https://doi.org/10.3390/ijms26093923
Ogienko AA, Andreyeva EN, Yarinich LA, Pindyurin AV, Battulina NV, Omelina ES. Expression Pattern of the AB1-Gal4 Driver in Drosophila Third-Instar Larvae. International Journal of Molecular Sciences. 2025; 26(9):3923. https://doi.org/10.3390/ijms26093923
Chicago/Turabian StyleOgienko, Anna A., Evgeniya N. Andreyeva, Lyubov A. Yarinich, Alexey V. Pindyurin, Nadezhda V. Battulina, and Evgeniya S. Omelina. 2025. "Expression Pattern of the AB1-Gal4 Driver in Drosophila Third-Instar Larvae" International Journal of Molecular Sciences 26, no. 9: 3923. https://doi.org/10.3390/ijms26093923
APA StyleOgienko, A. A., Andreyeva, E. N., Yarinich, L. A., Pindyurin, A. V., Battulina, N. V., & Omelina, E. S. (2025). Expression Pattern of the AB1-Gal4 Driver in Drosophila Third-Instar Larvae. International Journal of Molecular Sciences, 26(9), 3923. https://doi.org/10.3390/ijms26093923







