Drug Resistance Analysis of Pancreatic Cancer Based on Universally Differentially Expressed Genes
Abstract
1. Introduction
2. Results
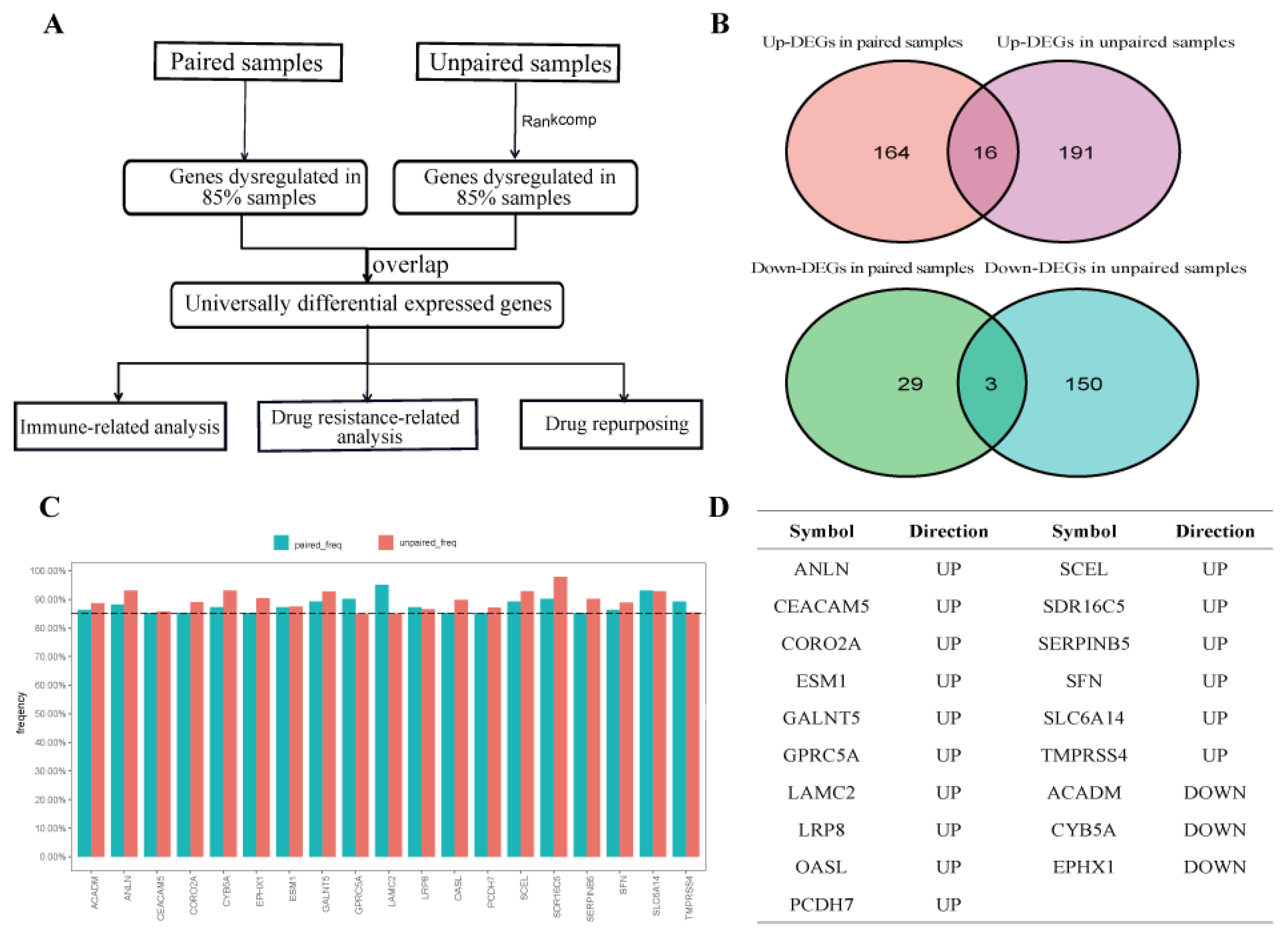
2.1. Identification of UDEGs in PDAC
2.2. Functions and Roles of UDEGs in PDAC
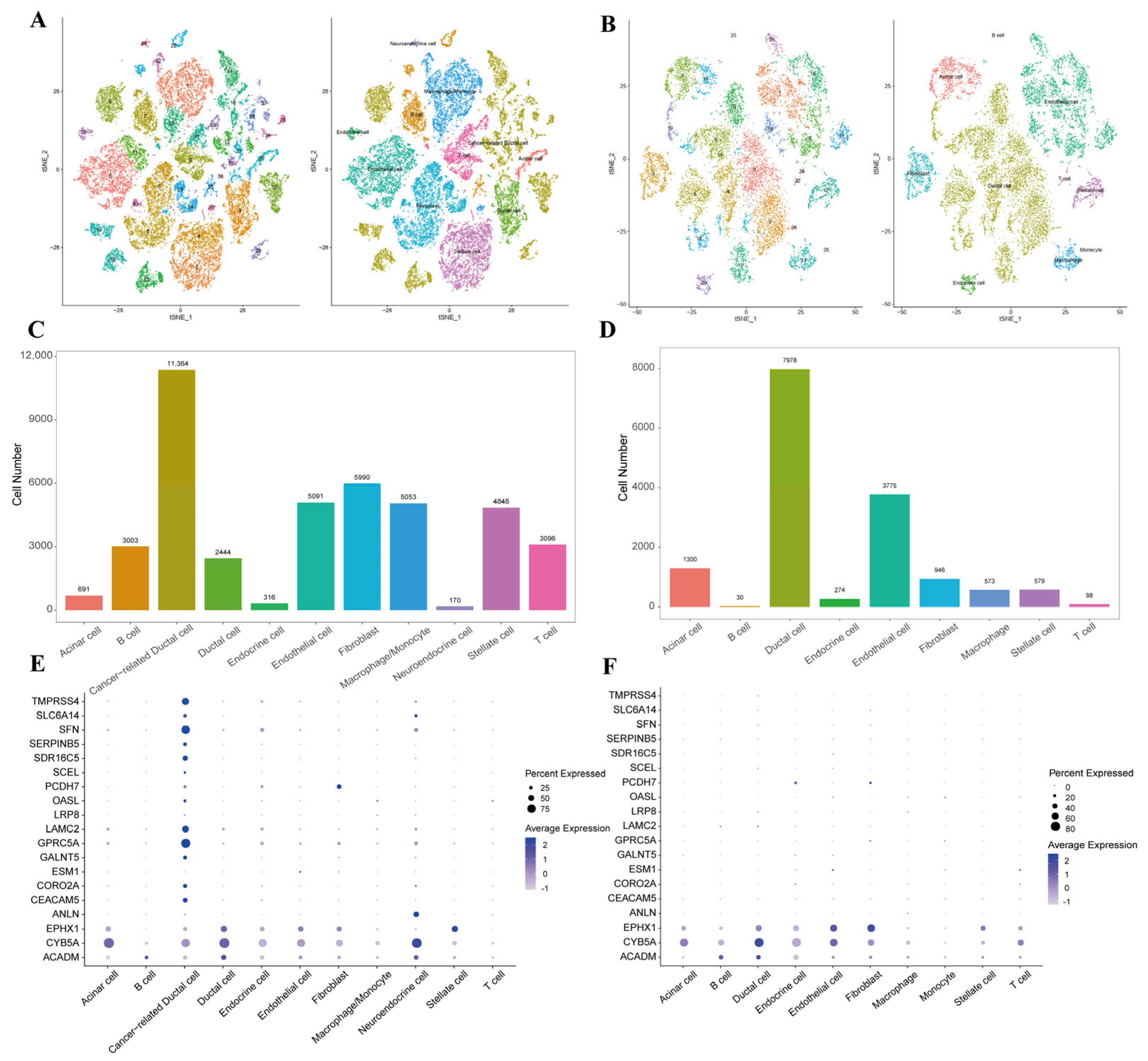
2.3. Characteristics of UDEGs at Single-Cell Level
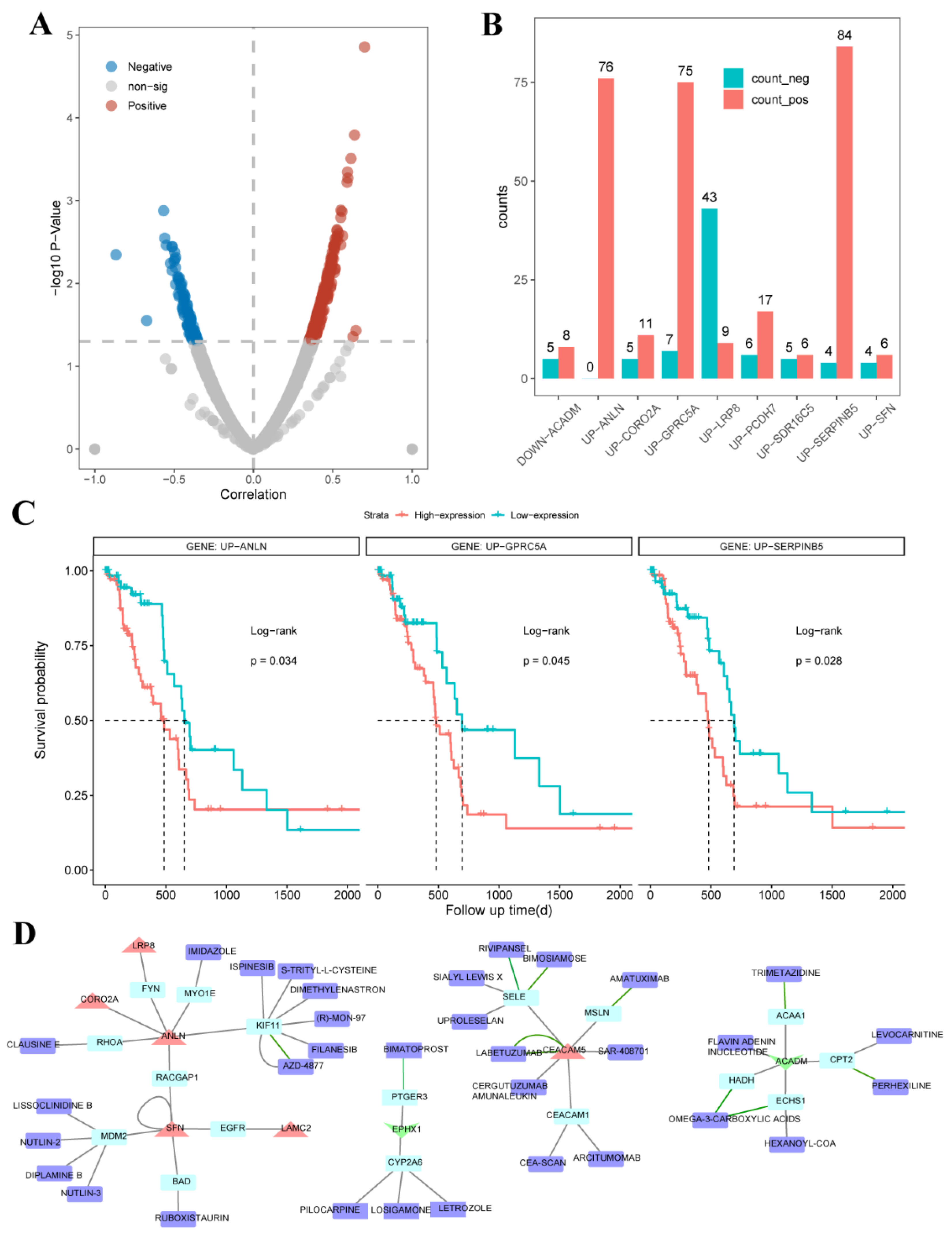
2.4. UDEGs Involved in Resistance to Anticancer Drugs
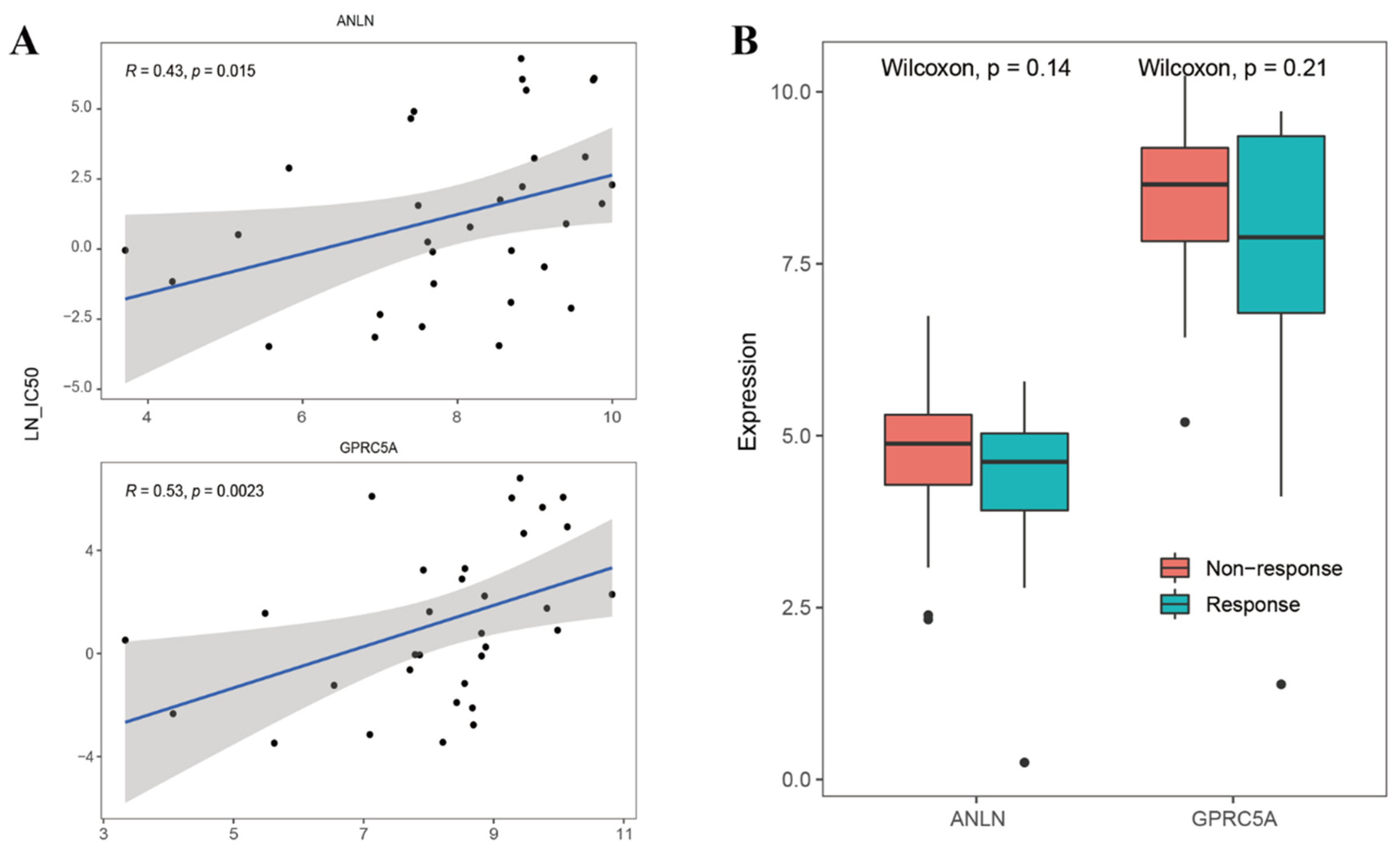
2.5. Roles of UDEGs in Response to Immunotherapy for PDAC Patients
3. Discussion
4. Materials and Methods
4.1. The Sources and Preprocessing of Expression Profiles
4.2. Identification of Universally Dysregulated Genes in PDAC
4.3. Protein–Protein Interaction Network
4.4. Prognostic Efficiency and Functions of UDEGs
4.5. Processing of scRNA-Seq Data
4.6. Cluster Identification and Cell Type Assignment
4.7. Drug Resistance
4.8. Drug–Gene Interaction Network
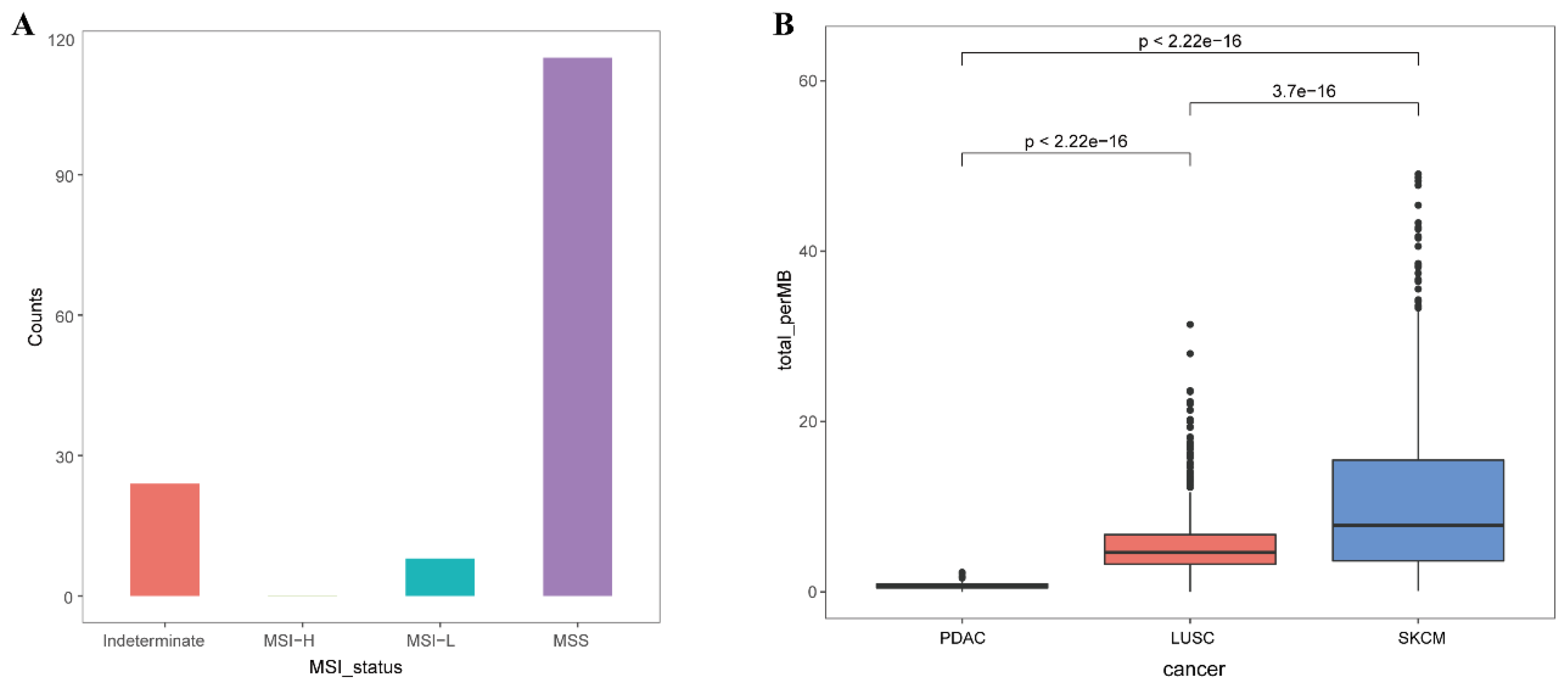
4.9. TMB and MSI Information
4.10. Immune Cell Proportion of TCGA PDAC Patients
4.11. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| UDEGs | universally differentially expressed genes |
| PDAC | pancreatic ductal adenocarcinoma |
| ICB | immune checkpoint blockade |
| TME | tumor microenvironment |
| DEG | differentially expressed gene |
| PPI | protein–protein interaction |
| DGI | drug–gene interaction |
| TMB | tumor mutation burden |
| MSI | microsatellite instability |
| GEO | Gene Expression Omnibus |
| TCGA | The Cancer Genome Atlas |
| GTEX | The Genotype-Tissue Expression |
| GDSC | Genomics of Drug Sensitivity in Cancer |
Appendix A
| Gene | Cancer Type | Role |
|---|---|---|
| CEACAM5 | colorectal cancer | Driver |
| ESM1 | esophageal cancer | Oncogene |
| SFN | prostate cancer, kidney cancer, pancreatic cancer, lung adenocarcinoma, malignant glioma, osteosarcoma | Oncogene |
| SFN | breast cancer, tongue squamous cell carcinoma | Tumor_Suppressor |
| LAMC2 | pancreatic cancer, penis squamous cell carcinoma, esophagus squamous cell carcinoma, ovarian cancer | Oncogene |
| PCDH7 | colon cancer | Oncogene |
| SERPINB5 | breast cancer, stomach cancer, breast carcinoma | Oncogene |
| SERPINB5 | breast cancer, prostate cancer, squamous cell carcinoma, melanoma, breast carcinoma, colorectal cancer, urinary bladder cancer, transitional cell carcinoma, stomach cancer, laryngeal carcinoma, lung squamous cell carcinoma, invasive lobular carcinoma, lung adenocarcinoma | Tumor_Suppressor |
| ANLN | pancreatic cancer | Oncogene |
| CORO2A | breast cancer | Oncogene |
| GPRC5A | lung non-small cell carcinoma, lung cancer, head and neck squamous cell carcinoma, stomach cancer, breast cancer | Tumor_Suppressor |
| GPRC5A | pancreatic ductal adenocarcinoma, pancreatic cancer, prostate cancer | Oncogene |
| ACADM | breast cancer | Oncogene |
| Drug | Gene | Threshold | Drug | Gene | Threshold |
|---|---|---|---|---|---|
| CAMPTOTHECIN | GPRC5A | Positive | OSU-03012 | SERPINB5 | Positive |
| VINBLASTINE | GPRC5A | Positive | EMBELIN | ANLN | Positive |
| VINBLASTINE | SERPINB5 | Positive | EMBELIN | GPRC5A | Positive |
| CISPLATIN | ANLN | Positive | FH535 | GPRC5A | Positive |
| CISPLATIN | GPRC5A | Positive | FH535 | LRP8 | Negative |
| CISPLATIN | SERPINB5 | Positive | FH535 | SDR16C5 | Positive |
| CYTARABINE | ANLN | Positive | FH535 | SERPINB5 | Positive |
| CYTARABINE | GPRC5A | Positive | IPA-3 | ANLN | Positive |
| CYTARABINE | SERPINB5 | Positive | IPA-3 | SERPINB5 | Positive |
| GEFITINIB | GPRC5A | Positive | GSK650394 | ANLN | Positive |
| NILOTINIB | GPRC5A | Positive | GSK650394 | SERPINB5 | Positive |
| OLAPARIB | ANLN | Positive | BAY-61-3606 | SERPINB5 | Positive |
| OLAPARIB | GPRC5A | Positive | THAPSIGARGIN | CORO2A | Positive |
| AZD7762 | LRP8 | Negative | BEXAROTENE | GPRC5A | Positive |
| AZD7762 | SERPINB5 | Negative | BEXAROTENE | SERPINB5 | Positive |
| AZD7762 | SFN | Negative | BEXAROTENE | SFN | Positive |
| AFATINIB | ACADM | Positive | BLEOMYCIN | ANLN | Positive |
| STAUROSPORINE | GPRC5A | Positive | BLEOMYCIN | SERPINB5 | Positive |
| NUTLIN-3A (-) | GPRC5A | Positive | LFM-A13 | GPRC5A | Positive |
| PD173074 | ANLN | Positive | GW-2580 | ANLN | Positive |
| PD173074 | GPRC5A | Positive | PHENFORMIN | ANLN | Positive |
| PD173074 | LRP8 | Negative | PHENFORMIN | SERPINB5 | Positive |
| PALBOCICLIB | GPRC5A | Positive | PAZOPANIB | CORO2A | Positive |
| PALBOCICLIB | SERPINB5 | Positive | PAZOPANIB | LRP8 | Negative |
| PD0325901 | PCDH7 | Positive | DACINOSTAT | ANLN | Positive |
| DASATINIB | PCDH7 | Positive | EPOTHILONE B | ANLN | Positive |
| PACLITAXEL | ACADM | Positive | EPOTHILONE B | SERPINB5 | Positive |
| PACLITAXEL | GPRC5A | Positive | TIPIFARNIB | ANLN | Positive |
| PACLITAXEL | PCDH7 | Positive | AVAGACESTAT | ANLN | Positive |
| RAPAMYCIN | GPRC5A | Positive | AS601245 | SERPINB5 | Positive |
| RAPAMYCIN | LRP8 | Negative | ISPINESIB MESYLATE | SERPINB5 | Positive |
| RAPAMYCIN | PCDH7 | Positive | TL-2-105 | ANLN | Positive |
| SORAFENIB | ANLN | Positive | TL-2-105 | SERPINB5 | Positive |
| SORAFENIB | GPRC5A | Positive | TAK-715 | ANLN | Positive |
| SORAFENIB | SERPINB5 | Positive | BX-912 | ANLN | Positive |
| IRINOTECAN | GPRC5A | Positive | BX-912 | SERPINB5 | Positive |
| GSK1904529A | GPRC5A | Positive | AS605240 | SERPINB5 | Positive |
| ERLOTINIB | CORO2A | Negative | GENENTECH CPD 10 | SERPINB5 | Positive |
| ERLOTINIB | SDR16C5 | Negative | GSK1070916 | ANLN | Positive |
| MK-1775 | GPRC5A | Positive | GSK1070916 | SERPINB5 | Positive |
| MK-1775 | LRP8 | Negative | ENZASTAURIN | PCDH7 | Negative |
| DINACICLIB | LRP8 | Negative | GSK429286A | ANLN | Positive |
| GEMCITABINE | ANLN | Positive | GSK429286A | GPRC5A | Positive |
| GEMCITABINE | GPRC5A | Positive | GSK429286A | SERPINB5 | Positive |
| TAMOXIFEN | LRP8 | Negative | GSK429286A | SFN | Positive |
| EPZ004777 | LRP8 | Negative | QL-XII-47 | SERPINB5 | Positive |
| YK-4-279 | GPRC5A | Positive | IC-87114 | SERPINB5 | Positive |
| YK-4-279 | SERPINB5 | Positive | UNC0638 | SERPINB5 | Positive |
| DAPORINAD | CORO2A | Positive | XMD14-99 | ANLN | Positive |
| DAPORINAD | SERPINB5 | Positive | XMD14-99 | SERPINB5 | Positive |
| TALAZOPARIB | ANLN | Positive | CP724714 | ANLN | Positive |
| TALAZOPARIB | GPRC5A | Positive | CP724714 | SERPINB5 | Positive |
| XAV939 | ACADM | Positive | JW-7-24-1 | SERPINB5 | Positive |
| DABRAFENIB | ANLN | Positive | NPK76-II-72-1 | ANLN | Positive |
| DABRAFENIB | GPRC5A | Positive | NG-25 | ANLN | Positive |
| DABRAFENIB | LRP8 | Negative | TL-1-85 | ANLN | Positive |
| TEMOZOLOMIDE | GPRC5A | Positive | ACY-1215 | ANLN | Positive |
| TEMOZOLOMIDE | LRP8 | Negative | ACY-1215 | LRP8 | Positive |
| IAP_5620 | GPRC5A | Positive | ACY-1215 | SERPINB5 | Positive |
| AZD1208 | GPRC5A | Positive | ACY-1215 | SFN | Positive |
| EPIRUBICIN | ANLN | Positive | TUBASTATIN A | ANLN | Positive |
| EPIRUBICIN | GPRC5A | Positive | TUBASTATIN A | GPRC5A | Positive |
| CYCLOPHOSPHAMIDE | GPRC5A | Positive | TUBASTATIN A | SERPINB5 | Positive |
| PEVONEDISTAT | ANLN | Positive | ZIBOTENTAN | ANLN | Positive |
| PEVONEDISTAT | SERPINB5 | Positive | ZIBOTENTAN | GPRC5A | Positive |
| SAPITINIB | SDR16C5 | Negative | ZIBOTENTAN | SERPINB5 | Positive |
| SAPITINIB | SERPINB5 | Negative | NSC-207895 | ANLN | Positive |
| SAPITINIB | SFN | Negative | VNLG/124 | SERPINB5 | Positive |
| LCL161 | SERPINB5 | Negative | AR-42 | ANLN | Positive |
| LAPATINIB | GPRC5A | Positive | AR-42 | SERPINB5 | Positive |
| LAPATINIB | LRP8 | Negative | CUDC-101 | ANLN | Positive |
| LUMINESPIB | CORO2A | Positive | CUDC-101 | SERPINB5 | Positive |
| ALPELISIB | ACADM | Positive | BELINOSTAT | ANLN | Positive |
| TASELISIB | ACADM | Positive | BELINOSTAT | SERPINB5 | Positive |
| EPZ5676 | GPRC5A | Positive | CAY10603 | ANLN | Positive |
| IWP-2 | LRP8 | Negative | CAY10603 | SERPINB5 | Positive |
| LGK974 | CORO2A | Negative | JNJ38877605 | LRP8 | Positive |
| WZ4003 | LRP8 | Negative | SU11274 | SERPINB5 | Positive |
| CZC24832 | GPRC5A | Positive | KIN001-236 | ANLN | Positive |
| CZC24832 | LRP8 | Negative | KIN001-260 | ANLN | Positive |
| AZD5582 | LRP8 | Negative | NVP-BHG712 | ANLN | Positive |
| GSK2606414 | CORO2A | Negative | OSI-930 | ANLN | Positive |
| PFI3 | PCDH7 | Positive | OSI-930 | SERPINB5 | Positive |
| PCI-34051 | LRP8 | Negative | CX-5461 | ANLN | Positive |
| PCI-34051 | PCDH7 | Positive | CX-5461 | SERPINB5 | Positive |
| WNT-C59 | SERPINB5 | Negative | PHA-793887 | SERPINB5 | Positive |
| I-BET-762 | ANLN | Positive | PI-103 | CORO2A | Positive |
| RVX-208 | PCDH7 | Positive | FEDRATINIB | ANLN | Positive |
| GSK343 | ACADM | Positive | Y-39983 | ANLN | Positive |
| ML323 | ANLN | Positive | Y-39983 | GPRC5A | Positive |
| ML323 | GPRC5A | Positive | Y-39983 | SERPINB5 | Positive |
| PRT062607 | LRP8 | Negative | YM201636 | ANLN | Positive |
| AGI-6780 | SFN | Negative | WYE-125132 | PCDH7 | Negative |
| CDK9_5576 | LRP8 | Negative | GSK690693 | SERPINB5 | Positive |
| CDK9_5038 | LRP8 | Negative | SNX-2112 | ANLN | Positive |
| EG5_9814 | GPRC5A | Positive | SNX-2112 | GPRC5A | Positive |
| EG5_9814 | LRP8 | Negative | XMD13-2 | ANLN | Positive |
| EG5_9814 | SERPINB5 | Positive | XMD13-2 | SERPINB5 | Positive |
| ERK_2440 | LRP8 | Negative | QL-X-138 | SERPINB5 | Positive |
| ERK_2440 | SDR16C5 | Negative | XMD15-27 | ANLN | Positive |
| AZD5991 | GPRC5A | Positive | T0901317 | GPRC5A | Positive |
| AZD5991 | LRP8 | Negative | SELISISTAT | SERPINB5 | Positive |
| PAK_5339 | LRP8 | Negative | KIN001-270 | SERPINB5 | Positive |
| TAF1_5496 | ACADM | Negative | THZ-2-102-1 | ANLN | Positive |
| TAF1_5496 | ANLN | Positive | AT7867 | SERPINB5 | Positive |
| TAF1_5496 | GPRC5A | Positive | AT7867 | SFN | Positive |
| SELUMETINIB | GPRC5A | Negative | CI-1033 | PCDH7 | Negative |
| TENIPOSIDE | ANLN | Positive | TWS119 | GPRC5A | Negative |
| TENIPOSIDE | GPRC5A | Positive | TWS119 | LRP8 | Positive |
| TENIPOSIDE | LRP8 | Negative | TORIN 2 | PCDH7 | Negative |
| MITOXANTRONE | GPRC5A | Positive | PILARALISIB | SERPINB5 | Positive |
| MITOXANTRONE | LRP8 | Negative | GSK1059615 | ANLN | Positive |
| DACTINOMYCIN | PCDH7 | Positive | BRIVANIB, BMS-540215 | SERPINB5 | Positive |
| NELARABINE | ACADM | Positive | BIBF-1120 | CORO2A | Negative |
| NELARABINE | GPRC5A | Positive | AST-1306 | CORO2A | Negative |
| VINCRISTINE | GPRC5A | Positive | AST-1306 | SDR16C5 | Negative |
| VINCRISTINE | LRP8 | Negative | APITOLISIB | LRP8 | Positive |
| PODOPHYLLOTOXIN BROMIDE | GPRC5A | Positive | APITOLISIB | PCDH7 | Negative |
| PODOPHYLLOTOXIN BROMIDE | LRP8 | Negative | LIMK1 INHIBITOR BMS4 | ACADM | Negative |
| DIHYDROROTENONE | ANLN | Positive | KB NB 142-70 | CORO2A | Positive |
| ELEPHANTIN | LRP8 | Negative | SPHINGOSINE KINASE 1 INHIBITOR II | SERPINB5 | Positive |
| SABUTOCLAX | CORO2A | Positive | EEF2K INHIBITOR, A-484954 | SERPINB5 | Positive |
| MG-132 | GPRC5A | Positive | ARA-G | GPRC5A | Positive |
| MG-132 | PCDH7 | Positive | C-75 | SERPINB5 | Positive |
| BDP-00009066 | GPRC5A | Positive | CAP-232, TT-232, TLN-232 | GPRC5A | Positive |
| BDP-00009066 | LRP8 | Negative | TRICHOSTATIN A | ANLN | Positive |
| BUPARLISIB | GPRC5A | Positive | TRICHOSTATIN A | GPRC5A | Positive |
| ULIXERTINIB | ANLN | Positive | PANOBINOSTAT | ANLN | Positive |
| ULIXERTINIB | LRP8 | Negative | IMD-0354 | SERPINB5 | Positive |
| AFURESERTIB | GPRC5A | Positive | ETP-45835 | SERPINB5 | Positive |
| AFURESERTIB | LRP8 | Negative | CD532 | ANLN | Positive |
| AFURESERTIB | PCDH7 | Positive | CD532 | SERPINB5 | Positive |
| AGI-5198 | ANLN | Positive | ARRY-520 | SERPINB5 | Positive |
| AGI-5198 | GPRC5A | Positive | SB505124 | GPRC5A | Positive |
| AZD3759 | SDR16C5 | Negative | A-83-01 | LRP8 | Positive |
| AZD5363 | GPRC5A | Positive | FTY-720 | GPRC5A | Positive |
| AZD5363 | LRP8 | Negative | KOBE2602 | SERPINB5 | Positive |
| IPATASERTIB | LRP8 | Negative | TRETINOIN | GPRC5A | Positive |
| IPATASERTIB | PCDH7 | Positive | CI-1040 | SDR16C5 | Positive |
| GDC0810 | GPRC5A | Positive | TEMSIROLIMUS | ACADM | Negative |
| GSK2578215A | GPRC5A | Positive | TEMSIROLIMUS | SDR16C5 | Positive |
| GSK2578215A | LRP8 | Negative | TEMSIROLIMUS | SERPINB5 | Positive |
| GSK2578215A | PCDH7 | Positive | BOSUTINIB | ACADM | Negative |
| I-BRD9 | LRP8 | Negative | AXITINIB | GPRC5A | Positive |
| I-BRD9 | PCDH7 | Positive | AXITINIB | SERPINB5 | Positive |
| TELOMERASE INHIBITOR IX | PCDH7 | Positive | GW441756 | SERPINB5 | Positive |
| MIRA-1 | GPRC5A | Positive | VX-702 | ANLN | Positive |
| MIRA-1 | LRP8 | Negative | ELESCLOMOL | ACADM | Negative |
| SAVOLITINIB | ANLN | Positive | ELESCLOMOL | SDR16C5 | Positive |
| UMI-77 | PCDH7 | Positive | VISMODEGIB | ANLN | Positive |
| SEPANTRONIUM BROMIDE | ANLN | Positive | VISMODEGIB | SERPINB5 | Positive |
| SEPANTRONIUM BROMIDE | CORO2A | Positive | BX795 | SDR16C5 | Positive |
| MIM1 | GPRC5A | Positive | BX795 | SERPINB5 | Positive |
| BPD-00008900 | GPRC5A | Positive | SL0101 | ANLN | Positive |
| BPD-00008900 | PCDH7 | Positive | SL0101 | SERPINB5 | Positive |
| BPD-00008900 | SERPINB5 | Positive | JNK INHIBITOR VIII | LRP8 | Negative |
| FORETINIB | ANLN | Positive | SB590885 | ANLN | Positive |
| BIBR-1532 | ANLN | Positive | CETUXIMAB | GPRC5A | Negative |
| PYRIDOSTATIN | GPRC5A | Positive | HG-5-88-01 | LRP8 | Positive |
| PYRIDOSTATIN | PCDH7 | Positive | TW 37 | SERPINB5 | Positive |
| AMG-319 | GPRC5A | Positive | TW 37 | SFN | Positive |
| AMG-319 | LRP8 | Negative | XMD8-92 | LRP8 | Positive |
| MK-8776 | GPRC5A | Positive | QL-VIII-58 | PCDH7 | Negative |
| VINORELBINE | ANLN | Positive | CCT-018159 | CORO2A | Positive |
| VINORELBINE | GPRC5A | Positive | CCT-018159 | SERPINB5 | Positive |
| VINORELBINE | SERPINB5 | Positive | CCT-018159 | SFN | Positive |
| VX-11E | ANLN | Positive | QL-XII-61 | GPRC5A | Negative |
| VX-11E | LRP8 | Negative | IOX2 | SERPINB5 | Positive |
| LJI308 | LRP8 | Negative | (5Z)-7-OXOZEAENOL | SDR16C5 | Positive |
| AT13148 | GPRC5A | Positive | UNC1215 | SERPINB5 | Positive |
| AT13148 | LRP8 | Negative | UNC0642 | CORO2A | Positive |
| GSK269962A | ANLN | Positive | UNC0642 | SERPINB5 | Positive |
| GSK269962A | GPRC5A | Positive | SGC0946 | SERPINB5 | Positive |
| GSK269962A | SERPINB5 | Positive | ICL1100013 | CORO2A | Positive |
| DOXORUBICIN | ANLN | Positive | ICL1100013 | SERPINB5 | Positive |
| DOXORUBICIN | SERPINB5 | Positive | AZD4877 | SERPINB5 | Positive |
| ETOPOSIDE | ANLN | Positive | EPHB4_9721 | ACADM | Positive |
| ETOPOSIDE | SERPINB5 | Positive | EPHB4_9721 | GPRC5A | Negative |
| MITOMYCIN-C | ANLN | Positive | EPHB4_9721 | SFN | Negative |
| MITOMYCIN-C | SERPINB5 | Positive | BPTES | LRP8 | Positive |
| CP466722 | SERPINB5 | Positive | IGFR_3801 | LRP8 | Positive |
| PONATINIB | LRP8 | Negative | JAK1_3715 | SERPINB5 | Positive |
| PF-562271 | SERPINB5 | Positive | PARP_0108 | GPRC5A | Positive |
| DMOG | GPRC5A | Negative | PARP_9482 | GPRC5A | Positive |
| OSU-03012 | ANLN | Positive | TANK_1366 | GPRC5A | Negative |
| OSU-03012 | GPRC5A | Positive |
References
- Bliss, L.A.; Witkowski, E.R.; Yang, C.J.; Tseng, J.F. Outcomes in operative management of pancreatic cancer. J. Surg. Oncol. 2014, 110, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Conroy, T.; Hammel, P.; Hebbar, M.; Ben Abdelghani, M.; Wei, A.C.; Raoul, J.L.; Chone, L.; Francois, E.; Artru, P.; Biagi, J.J.; et al. FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer. N. Engl. J. Med. 2018, 379, 2395–2406. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Blair, A.B.; Groot, V.P.; Javed, A.A.; Burkhart, R.A.; Gemenetzis, G.; Hruban, R.H.; Waters, K.M.; Poling, J.; Zheng, L.; et al. Is a Pathological Complete Response Following Neoadjuvant Chemoradiation Associated With Prolonged Survival in Patients With Pancreatic Cancer? Ann. Surg. 2018, 268, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Lu, W.; Qin, W.; Wu, Y. Neoadjuvant therapy for patients with borderline resectable pancreatic cancer: A systematic review and meta-analysis of response and resection percentages. Pancreatol. Off. J. Int. Assoc. Pancreatol. 2016, 16, 28–37. [Google Scholar] [CrossRef]
- Chuong, M.D.; Springett, G.M.; Freilich, J.M.; Park, C.K.; Weber, J.M.; Mellon, E.A.; Hodul, P.J.; Malafa, M.P.; Meredith, K.L.; Hoffe, S.E.; et al. Stereotactic body radiation therapy for locally advanced and borderline resectable pancreatic cancer is effective and well tolerated. Int. J. Radiat. Oncol. Biol. Phys. 2013, 86, 516–522. [Google Scholar] [CrossRef]
- Moutardier, V.; Magnin, V.; Turrini, O.; Viret, F.; Hennekinne-Mucci, S.; Goncalves, A.; Pesenti, C.; Guiramand, J.; Lelong, B.; Giovannini, M.; et al. Assessment of pathologic response after preoperative chemoradiotherapy and surgery in pancreatic adenocarcinoma. Int. J. Radiat. Oncol. Biol. Phys. 2004, 60, 437–443. [Google Scholar] [CrossRef]
- Chuong, M.D.; Frakes, J.M.; Figura, N.; Hoffe, S.E.; Shridhar, R.; Mellon, E.A.; Hodul, P.J.; Malafa, M.P.; Springett, G.M.; Centeno, B.A. Histopathologic tumor response after induction chemotherapy and stereotactic body radiation therapy for borderline resectable pancreatic cancer. J. Gastrointest. Oncol. 2016, 7, 221–227. [Google Scholar] [CrossRef]
- Morrison, A.H.; Byrne, K.T.; Vonderheide, R.H. Immunotherapy and Prevention of Pancreatic Cancer. Trends Cancer 2018, 4, 418–428. [Google Scholar] [CrossRef]
- Royal, R.E.; Levy, C.; Turner, K.; Mathur, A.; Hughes, M.; Kammula, U.S.; Sherry, R.M.; Topalian, S.L.; Yang, J.C.; Lowy, I.; et al. Phase 2 trial of single agent Ipilimumab (anti-CTLA-4) for locally advanced or metastatic pancreatic adenocarcinoma. J. Immunother. 2010, 33, 828–833. [Google Scholar] [CrossRef]
- Kim, J.M.; Chen, D.S. Immune escape to PD-L1/PD-1 blockade: Seven steps to success (or failure). Ann. Oncol. 2016, 27, 1492–1504. [Google Scholar] [CrossRef]
- Wang, H.; Sun, Q.; Zhao, W.; Qi, L.; Gu, Y.; Li, P.; Zhang, M.; Li, Y.; Liu, S.L.; Guo, Z. Individual-level analysis of differential expression of genes and pathways for personalized medicine. Bioinformatics 2015, 31, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Shema, E.; Bernstein, B.E.; Buenrostro, J.D. Single-cell and single-molecule epigenomics to uncover genome regulation at unprecedented resolution. Nat. Genet. 2019, 51, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Lawson, D.A.; Kessenbrock, K.; Davis, R.T.; Pervolarakis, N.; Werb, Z. Tumour heterogeneity and metastasis at single-cell resolution. Nat. Cell Biol. 2018, 20, 1349–1360. [Google Scholar] [CrossRef]
- Sato, N.; Maitra, A.; Fukushima, N.; van Heek, N.T.; Matsubayashi, H.; Iacobuzio-Donahue, C.A.; Rosty, C.; Goggins, M. Frequent hypomethylation of multiple genes overexpressed in pancreatic ductal adenocarcinoma. Cancer Res. 2003, 63, 4158–4166. [Google Scholar]
- Fukushige, S.; Horii, A. Road to early detection of pancreatic cancer: Attempts to utilize epigenetic biomarkers. Cancer Lett. 2014, 342, 231–237. [Google Scholar] [CrossRef]
- Pyke, C.; Romer, J.; Kallunki, P.; Lund, L.R.; Ralfkiaer, E.; Dano, K.; Tryggvason, K. The gamma 2 chain of kalinin/laminin 5 is preferentially expressed in invading malignant cells in human cancers. Am. J. Pathol. 1994, 145, 782–791. [Google Scholar] [PubMed]
- Wang, H.; Cai, J.; Du, S.; Wei, W.; Shen, X. LAMC2 modulates the acidity of microenvironments to promote invasion and migration of pancreatic cancer cells via regulating AKT-dependent NHE1 activity. Exp. Cell Res. 2020, 391, 111984. [Google Scholar] [CrossRef]
- Kosanam, H.; Prassas, I.; Chrystoja, C.C.; Soleas, I.; Chan, A.; Dimitromanolakis, A.; Blasutig, I.M.; Ruckert, F.; Gruetzmann, R.; Pilarsky, C.; et al. Laminin, gamma 2 (LAMC2): A promising new putative pancreatic cancer biomarker identified by proteomic analysis of pancreatic adenocarcinoma tissues. Mol. Cell. Proteom. MCP 2013, 12, 2820–2832. [Google Scholar] [CrossRef]
- Liu, B.; Yang, H.; Pilarsky, C.; Weber, G.F. The Effect of GPRC5a on the Proliferation, Migration Ability, Chemotherapy Resistance, and Phosphorylation of GSK-3beta in Pancreatic Cancer. Int. J. Mol. Sci. 2018, 19, 1870. [Google Scholar] [CrossRef]
- Rozengurt, E.; Sinnett-Smith, J.; Eibl, G. Yes-associated protein (YAP) in pancreatic cancer: At the epicenter of a targetable signaling network associated with patient survival. Signal Transduct. Target. Ther. 2018, 3, 11. [Google Scholar] [CrossRef]
- Zhou, S.; Yan, Y.; Chen, X.; Zeng, S.; Wei, J.; Wang, X.; Gong, Z.; Xu, Z. A two-gene-based prognostic signature for pancreatic cancer. Aging 2020, 12, 18322–18342. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Dai, H.; Gong, Y.; Zhang, C.; Shu, J.; Luo, Y.; Jiang, Y.; Liu, W.; Bie, P. ANLN-induced EZH2 upregulation promotes pancreatic cancer progression by mediating miR-218-5p/LASP1 signaling axis. J. Exp. Clin. Cancer Res. 2019, 38, 347. [Google Scholar] [CrossRef]
- Lin, L.Y.; Yeh, Y.C.; Chu, C.H.; Won, J.G.S.; Shyr, Y.M.; Chao, Y.; Li, C.P.; Wang, S.E.; Chen, M.H. Endocan expression is correlated with poor progression-free survival in patients with pancreatic neuroendocrine tumors. Medicine 2017, 96, e8262. [Google Scholar] [CrossRef]
- Tian, C.; Ohlund, D.; Rickelt, S.; Lidstrom, T.; Huang, Y.; Hao, L.; Zhao, R.T.; Franklin, O.; Bhatia, S.N.; Tuveson, D.A.; et al. Cancer Cell-Derived Matrisome Proteins Promote Metastasis in Pancreatic Ductal Adenocarcinoma. Cancer Res. 2020, 80, 1461–1474. [Google Scholar] [CrossRef]
- Penheiter, A.R.; Erdogan, S.; Murphy, S.J.; Hart, S.N.; Felipe Lima, J.; Rakhshan Rohakhtar, F.; O’Brien, D.R.; Bamlet, W.R.; Wuertz, R.E.; Smyrk, T.C.; et al. Transcriptomic and Immunohistochemical Profiling of SLC6A14 in Pancreatic Ductal Adenocarcinoma. Biomed. Res. Int. 2015, 2015, 593572. [Google Scholar] [CrossRef]
- Coothankandaswamy, V.; Cao, S.; Xu, Y.; Prasad, P.D.; Singh, P.K.; Reynolds, C.P.; Yang, S.; Ogura, J.; Ganapathy, V.; Bhutia, Y.D. Amino acid transporter SLC6A14 is a novel and effective drug target for pancreatic cancer. Br. J. Pharmacol. 2016, 173, 3292–3306. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhu, Y.Y.; Dai, G.P.; Wu, D.J.; Gao, Z.Z.; Zhang, L.; Fan, Y.H. Screening and validating the core biomarkers in patients with pancreatic ductal adenocarcinoma. Math. Biosci. Eng. 2019, 17, 910–927. [Google Scholar] [CrossRef]
- Cortes, E.; Sarper, M.; Robinson, B.; Lachowski, D.; Chronopoulos, A.; Thorpe, S.D.; Lee, D.A.; Del Rio Hernandez, A.E. GPER is a mechanoregulator of pancreatic stellate cells and the tumor microenvironment. EMBO Rep. 2019, 20, e46556. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Yang, J.; Lu, Z.; Liu, B.; Liu, F. A study on the mechanism of rapamycin mediating the sensitivity of pancreatic cancer cells to cisplatin through PI3K/AKT/mTOR signaling pathway. J. BUON 2019, 24, 739–745. [Google Scholar]
- Spencer, A.; Yoon, S.S.; Harrison, S.J.; Morris, S.R.; Smith, D.A.; Brigandi, R.A.; Gauvin, J.; Kumar, R.; Opalinska, J.B.; Chen, C. The novel AKT inhibitor afuresertib shows favorable safety, pharmacokinetics, and clinical activity in multiple myeloma. Blood 2014, 124, 2190–2195. [Google Scholar] [CrossRef]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Raghavan, S.; Winter, P.S.; Navia, A.W.; Williams, H.L.; DenAdel, A.; Lowder, K.E.; Galvez-Reyes, J.; Kalekar, R.L.; Mulugeta, N.; Kapner, K.S.; et al. Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer. Cell 2021, 184, 6119–6137.e26. [Google Scholar] [CrossRef]
- Terry, S.; Queires, L.; Gil-Diez-de-Medina, S.; Chen, M.W.; de la Taille, A.; Allory, Y.; Tran, P.L.; Abbou, C.C.; Buttyan, R.; Vacherot, F. Protocadherin-PC promotes androgen-independent prostate cancer cell growth. Prostate 2006, 66, 1100–1113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Fu, X. The Clinical Significance and Biological Function of PCDH7 in Cervical Cancer. Cancer Manag. Res. 2021, 13, 3841–3847. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Wang, M.; Wang, T. CCAAT enhancer binding protein beta has a crucial role in regulating breast cancer cell growth via activating the TGF-beta-Smad3 signaling pathway. Exp. Ther. Med. 2017, 14, 1554–1560. [Google Scholar] [CrossRef]
- Deng, J.L.; Zhang, H.B.; Zeng, Y.; Xu, Y.H.; Huang, Y.; Wang, G. Effects of CORO2A on Cell Migration and Proliferation and Its Potential Regulatory Network in Breast Cancer. Front. Oncol. 2020, 10, 916. [Google Scholar] [CrossRef]
- Ye, L.; Yang, Y.; Ma, X.Y.; Li, D.; Xu, M.L.; Tan, P.; Long, L.M.; Wang, H.Q.; Liu, T.; Guo, Y.H. Construction of a novel vector expressing Survivin-shRNA and fusion suicide gene yCDglyTK and its application in inhibiting proliferation and migration of colon cancer cells. Exp. Ther. Med. 2017, 14, 4721–4728. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, J.K.; Bangayan, N.J.; Chai, T.; Smith, B.A.; Pariva, T.E.; Yun, S.; Vashisht, A.; Zhang, Q.; Park, J.W.; Corey, E.; et al. Systemic surfaceome profiling identifies target antigens for immune-based therapy in subtypes of advanced prostate cancer. Proc. Natl. Acad. Sci. USA 2018, 115, E4473–E4482. [Google Scholar] [CrossRef]
- Powell, E.; Shao, J.; Picon, H.M.; Bristow, C.; Ge, Z.; Peoples, M.; Robinson, F.; Jeter-Jones, S.L.; Schlosberg, C.; Grzeskowiak, C.L.; et al. A functional genomic screen in vivo identifies CEACAM5 as a clinically relevant driver of breast cancer metastasis. NPJ Breast Cancer 2018, 4, 9. [Google Scholar] [CrossRef]
- Detarya, M.; Sawanyawisuth, K.; Aphivatanasiri, C.; Chuangchaiya, S.; Saranaruk, P.; Sukprasert, L.; Silsirivanit, A.; Araki, N.; Wongkham, S.; Wongkham, C. The O-GalNAcylating enzyme GALNT5 mediates carcinogenesis and progression of cholangiocarcinoma via activation of AKT/ERK signaling. Glycobiology 2020, 30, 312–324. [Google Scholar] [CrossRef]
- Guo, H.; Zhao, L.; Shi, B.; Bao, J.; Zheng, D.; Zhou, B.; Shi, J. GALNT5 uaRNA promotes gastric cancer progression through its interaction with HSP90. Oncogene 2018, 37, 4505–4517. [Google Scholar] [CrossRef] [PubMed]
- Qiu, H.; Shen, X.; Chen, B.; Chen, T.; Feng, G.; Chen, S.; Feng, D.; Xu, Q. miR-30b-5p inhibits cancer progression and enhances cisplatin sensitivity in lung cancer through targeting LRP8. Apoptosis 2021, 26, 261–276. [Google Scholar] [CrossRef]
- Cai, J.; Chen, J.; Wu, T.; Cheng, Z.; Tian, Y.; Pu, C.; Shi, W.; Suo, X.; Wu, X.; Zhang, K. Genome-scale CRISPR activation screening identifies a role of LRP8 in Sorafenib resistance in Hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2020, 526, 1170–1176. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Qu, W.H.; Ma, H.P.; Wang, L.L.; Zhang, Y.B.; Ma, Y. LRP8, modulated by miR-1262, promotes tumour progression and forecasts the prognosis of patients in breast cancer. Arch. Physiol. Biochem. 2020, 128, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.C.; Lo, M.C.; Moody, R.; Jiang, H.; Harouaka, R.; Stevers, N.; Tinsley, S.; Gasparyan, M.; Wicha, M.; Sun, D. Targeting LRP8 inhibits breast cancer stem cells in triple-negative breast cancer. Cancer Lett. 2018, 438, 165–173. [Google Scholar] [CrossRef]
- Kim, S.; Kang, H.Y.; Nam, E.H.; Choi, M.S.; Zhao, X.F.; Hong, C.S.; Lee, J.W.; Lee, J.H.; Park, Y.K. TMPRSS4 induces invasion and epithelial-mesenchymal transition through upregulation of integrin alpha5 and its signaling pathways. Carcinogenesis 2010, 31, 597–606. [Google Scholar] [CrossRef]
- Panebianco, C.; Trivieri, N.; Villani, A.; Terracciano, F.; Latiano, T.P.; Potenza, A.; Perri, F.; Binda, E.; Pazienza, V. Improving Gemcitabine Sensitivity in Pancreatic Cancer Cells by Restoring miRNA-217 Levels. Biomolecules 2021, 11, 639. [Google Scholar] [CrossRef]
- Wang, F.; Xiang, Z.; Huang, T.; Zhang, M.; Zhou, W.B. ANLN Directly Interacts with RhoA to Promote Doxorubicin Resistance in Breast Cancer Cells. Cancer Manag. Res. 2020, 12, 9725–9734. [Google Scholar] [CrossRef]
- Moyano-Galceran, L.; Pietila, E.A.; Turunen, S.P.; Corvigno, S.; Hjerpe, E.; Bulanova, D.; Joneborg, U.; Alkasalias, T.; Miki, Y.; Yashiro, M.; et al. Adaptive RSK-EphA2-GPRC5A signaling switch triggers chemotherapy resistance in ovarian cancer. EMBO Mol. Med. 2020, 12, e11177. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Lever, J.; Zhao, E.Y.; Grewal, J.; Jones, M.R.; Jones, S.J.M. CancerMine: A literature-mined resource for drivers, oncogenes and tumor suppressors in cancer. Nat. Methods 2019, 16, 505–507. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, A.; Sidiropoulos, K.; Garapati, P.; Gillespie, M.; Hausmann, K.; Haw, R.; Jassal, B.; Jupe, S.; Korninger, F.; McKay, S.; et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016, 44, D481–D487. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Sun, B.F.; Chen, C.Y.; Zhou, J.Y.; Chen, Y.S.; Chen, H.; Liu, L.; Huang, D.; Jiang, J.; Cui, G.S.; et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019, 29, 725–738. [Google Scholar] [CrossRef]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018, 36, 411–420. [Google Scholar] [CrossRef]
- Zhang, X.; Lan, Y.; Xu, J.; Quan, F.; Zhao, E.; Deng, C.; Luo, T.; Xu, L.; Liao, G.; Yan, M.; et al. CellMarker: A manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019, 47, D721–D728. [Google Scholar] [CrossRef] [PubMed]
- Iorio, F.; Knijnenburg, T.A.; Vis, D.J.; Bignell, G.R.; Menden, M.P.; Schubert, M.; Aben, N.; Goncalves, E.; Barthorpe, S.; Lightfoot, H.; et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell 2016, 166, 740–754. [Google Scholar] [CrossRef]
- Finan, C.; Gaulton, A.; Kruger, F.A.; Lumbers, R.T.; Shah, T.; Engmann, J.; Galver, L.; Kelley, R.; Karlsson, A.; Santos, R.; et al. The druggable genome and support for target identification and validation in drug development. Sci. Transl. Med. 2017, 9, eaag1166. [Google Scholar] [CrossRef]
- Ellrott, K.; Bailey, M.H.; Saksena, G.; Covington, K.R.; Kandoth, C.; Stewart, C.; Hess, J.; Ma, S.; Chiotti, K.E.; McLellan, M.; et al. Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell Syst. 2018, 6, 271–281.e7. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]


| Cell Type | Number of Pathways | Consistance | ||
|---|---|---|---|---|
| Normal | Tumor | Common | ||
| Acinar cells | 5 | 6 | 3 | 55.00% |
| Cancer-related ductal cells | - | 29 | - | - |
| Ductal cells | 35 | 30 | 22 | 68.10% |
| Endocrine cells | 27 | 29 | 22 | 78.67% |
| Endothelial cells | 94 | 82 | 64 | 73.07% |
| Fibroblasts | 22 | 29 | 16 | 63.95% |
| Macrophages | 70 | 74 | 64 | 88.96% |
| Stellate cells | 39 | 28 | 23 | 70.56% |
| T cells | 30 | 32 | 28 | 90.42% |
| GEO Accession | Platform | Sample Size | ||
|---|---|---|---|---|
| PDAC | PDAC_Adjacent | Normal | ||
| Unpaired Samples | ||||
| GSE62452 | GPL6244 | 69 | 61 | 0 |
| E-MTAB-1791 | A-MEXP-2271 | 268 | 74 | 41 |
| GSE91035 | GPL22763 | 25 | 0 | 8 |
| GSE62165 | GPL13667 | 118 | 13 | 0 |
| GSE56560 | GPL5175 | 28 | 4 | 3 |
| GSE71989 | GPL570 | 13 | 0 | 8 |
| Total | 521 | 212 | ||
| Paired Samples | - | |||
| GSE60646 | GPL5175 | 10 | 10 | - |
| GSE22780 | GPL570 | 8 | 8 | - |
| GSE28735 | GPL6244 | 45 | 45 | - |
| GSE15471 | GPL570 | 39 | 39 | - |
| Total | 102 | 102 | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, J.; Zheng, L.; Zhang, H.; Fan, Q.; Liu, H.; Wang, O.; Yan, H. Drug Resistance Analysis of Pancreatic Cancer Based on Universally Differentially Expressed Genes. Int. J. Mol. Sci. 2025, 26, 3936. https://doi.org/10.3390/ijms26093936
Xia J, Zheng L, Zhang H, Fan Q, Liu H, Wang O, Yan H. Drug Resistance Analysis of Pancreatic Cancer Based on Universally Differentially Expressed Genes. International Journal of Molecular Sciences. 2025; 26(9):3936. https://doi.org/10.3390/ijms26093936
Chicago/Turabian StyleXia, Jie, Linyong Zheng, Huarong Zhang, Qi Fan, Hui Liu, Ouxi Wang, and Haidan Yan. 2025. "Drug Resistance Analysis of Pancreatic Cancer Based on Universally Differentially Expressed Genes" International Journal of Molecular Sciences 26, no. 9: 3936. https://doi.org/10.3390/ijms26093936
APA StyleXia, J., Zheng, L., Zhang, H., Fan, Q., Liu, H., Wang, O., & Yan, H. (2025). Drug Resistance Analysis of Pancreatic Cancer Based on Universally Differentially Expressed Genes. International Journal of Molecular Sciences, 26(9), 3936. https://doi.org/10.3390/ijms26093936





