Comprehensive Characterization and Functional Analysis of the Lateral Organ Boundaries Domain Gene Family in Rice: Evolution, Expression, and Stress Response
Abstract
1. Introduction
2. Result
2.1. Identification and Physicochemical Property Analysis of the Rice LBD Gene Family Members
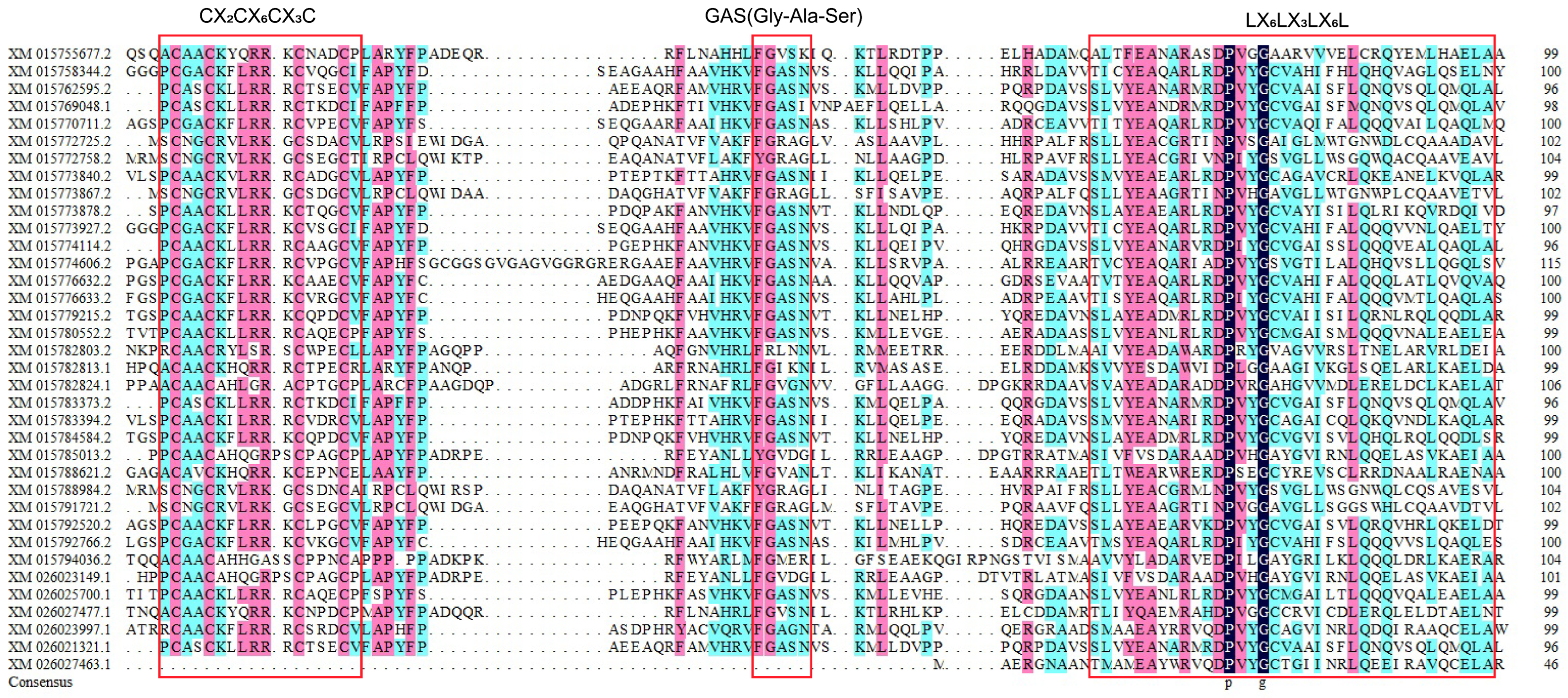
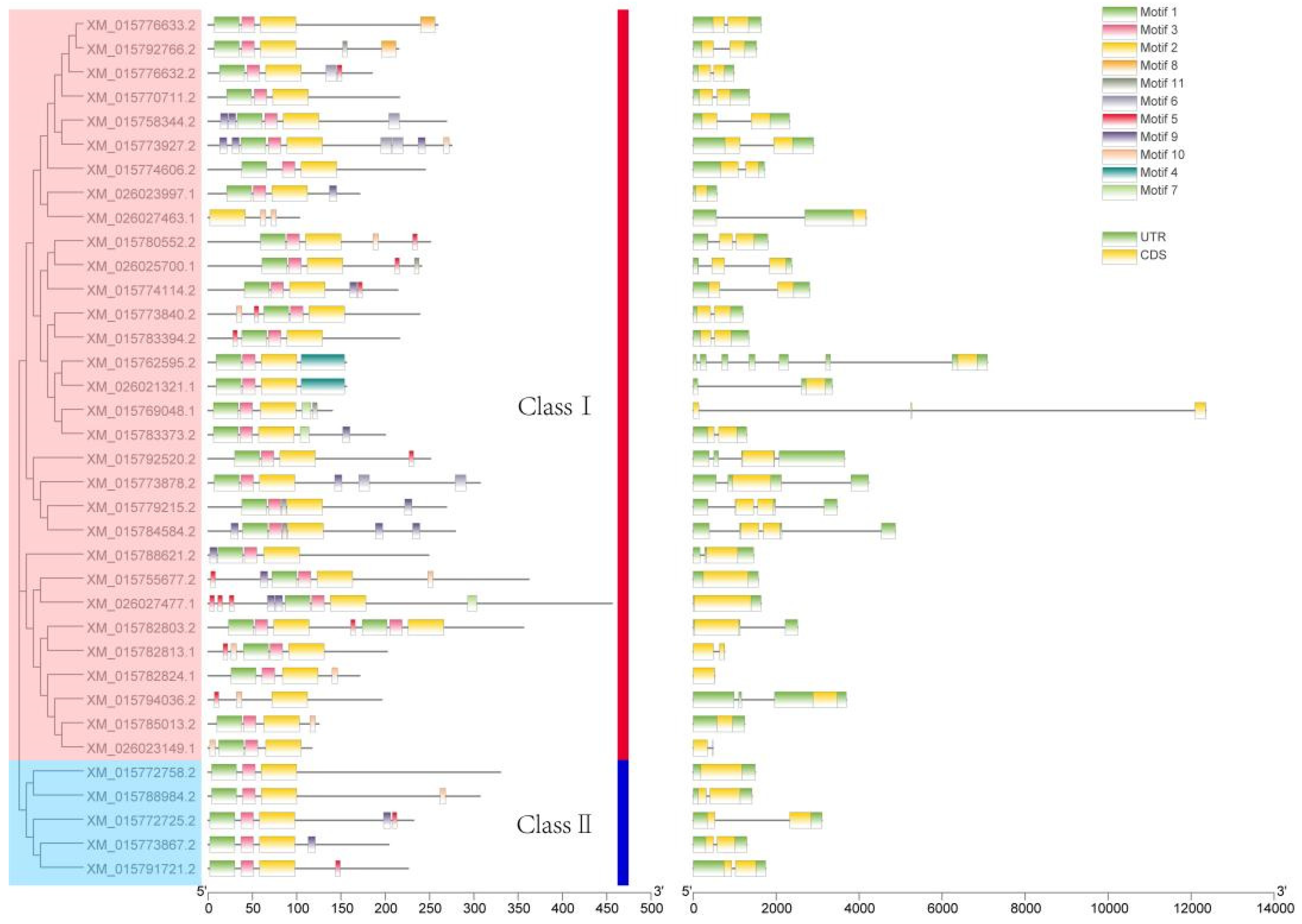
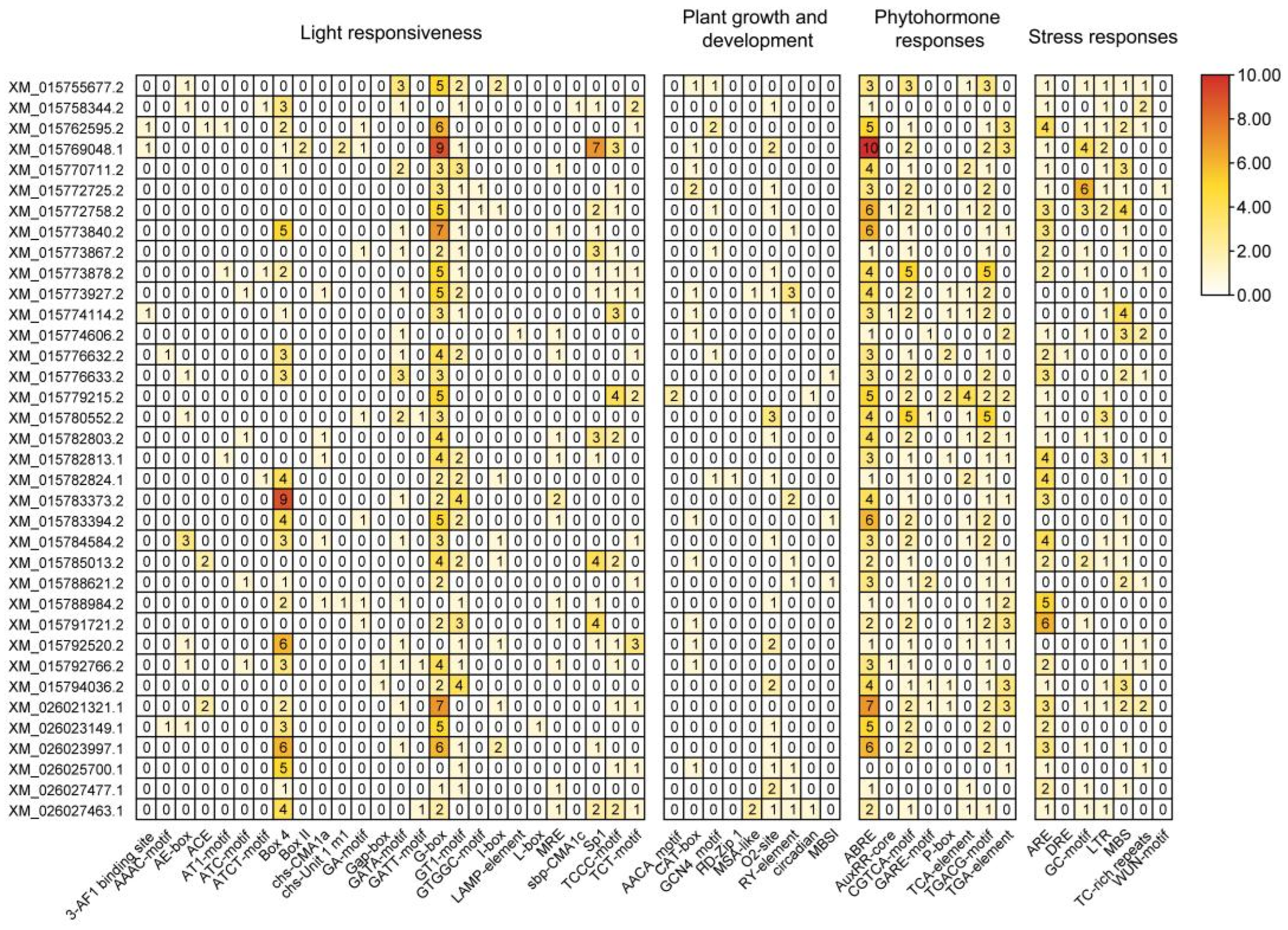
2.2. Phylogenetic, Motif, and cis-Acting Element Analysis of the Rice LBD Gene Family
2.3. Phylogeny Analysis of LBD Gene Family
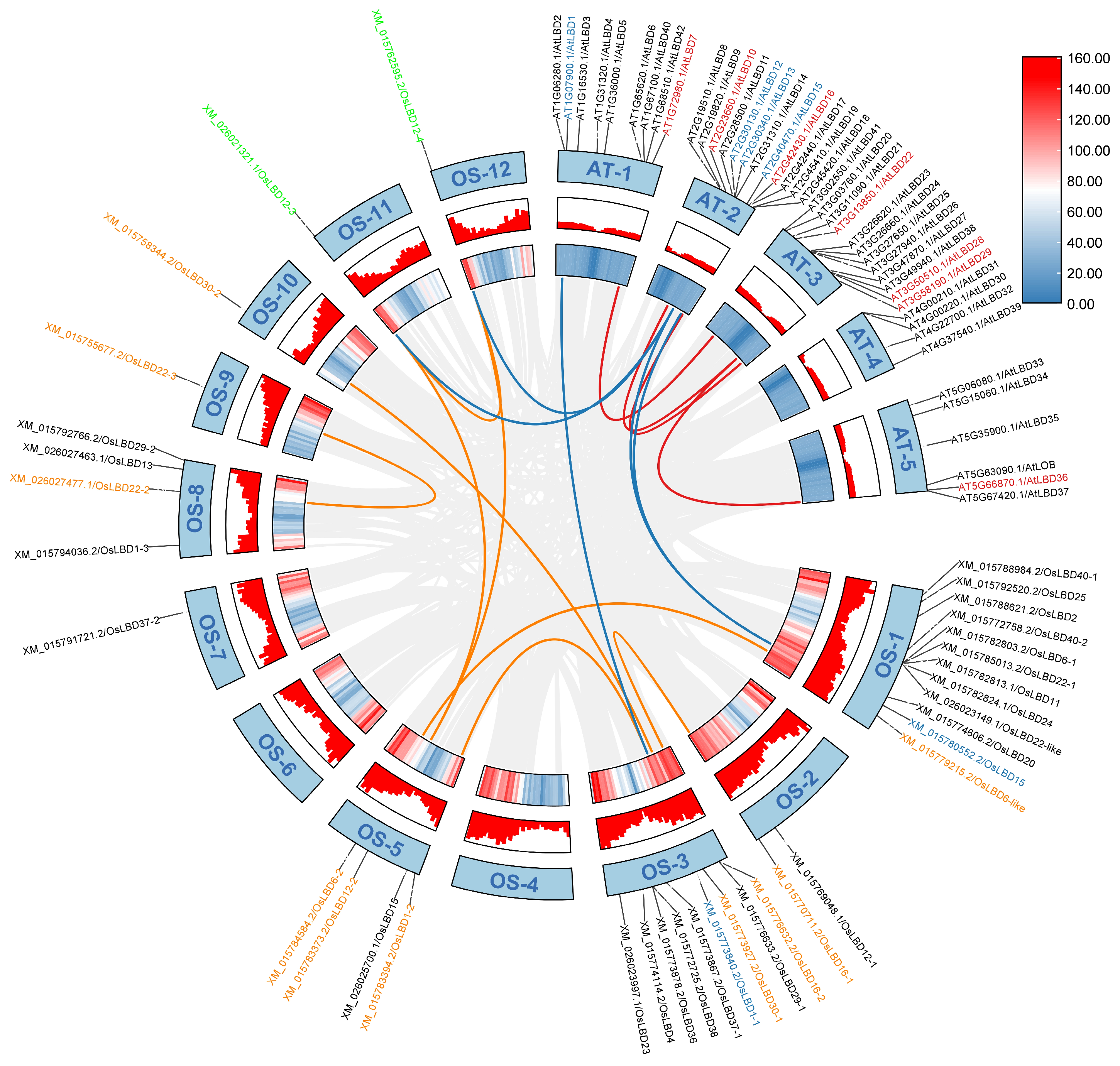
2.4. Collinearity Analysis of LBD Gene Family in Rice
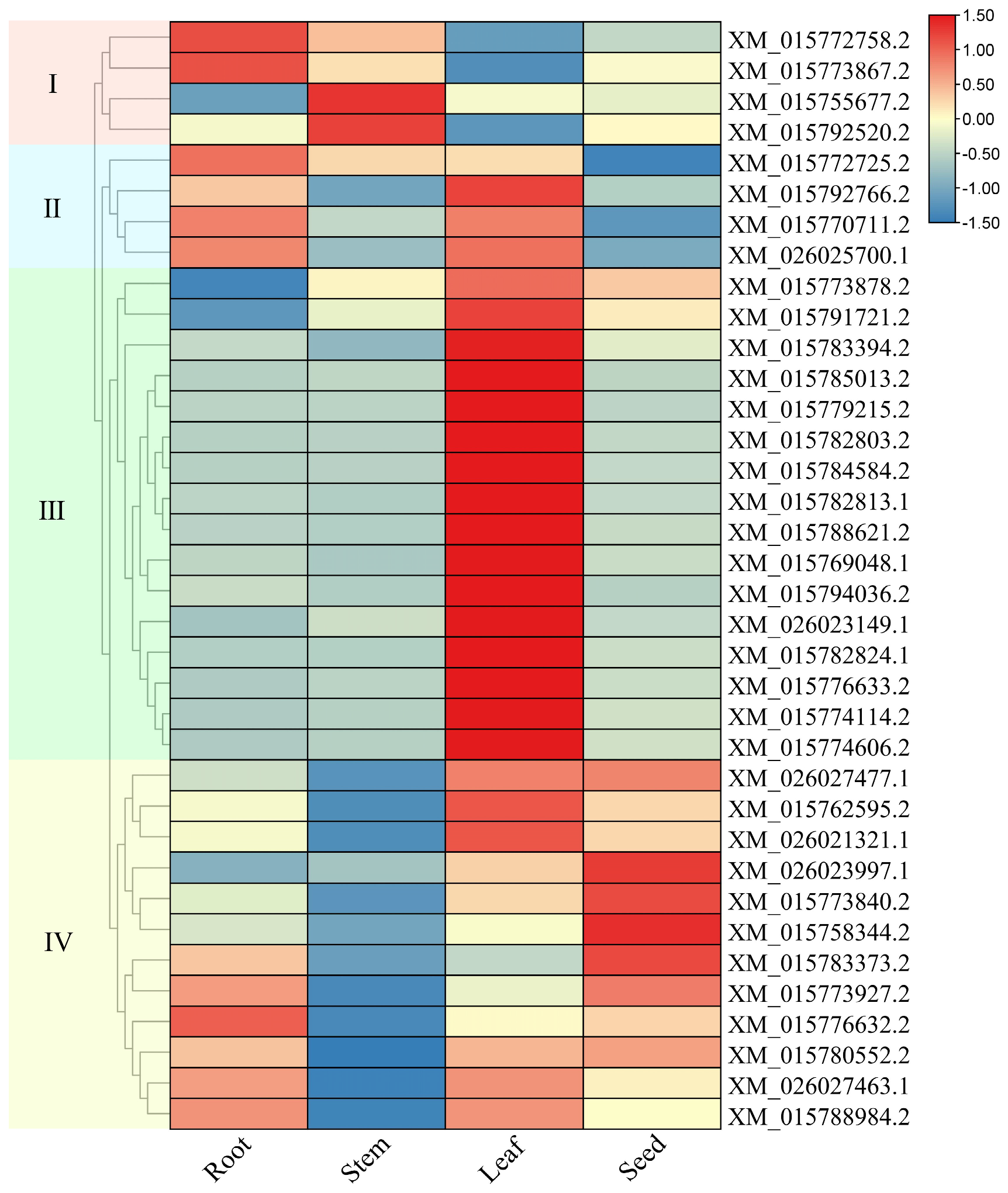
2.5. Analysis of Expression Patterns of Rice LBD Genes
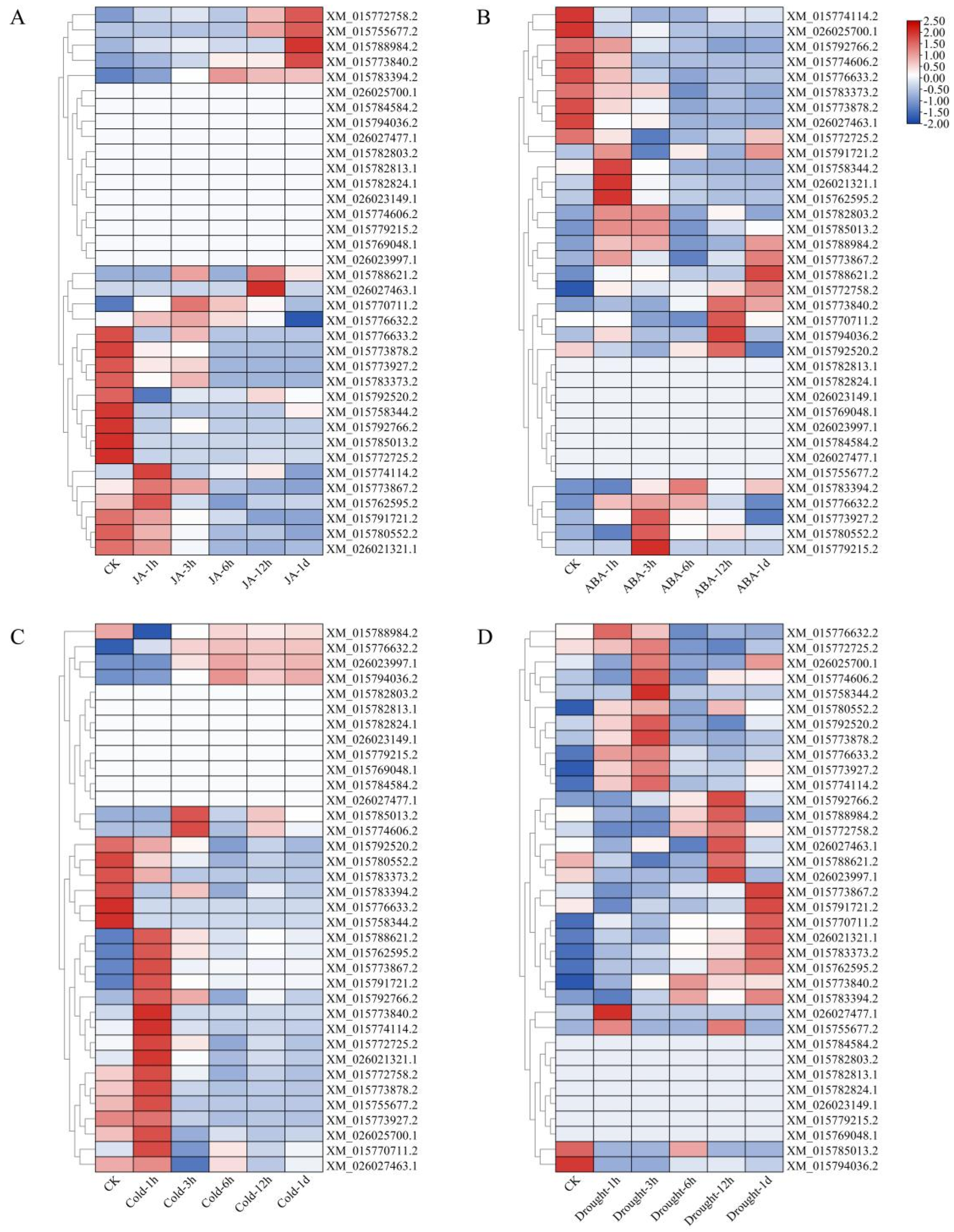
2.6. Transcriptome Data Analysis of Phytohormone and Stress Treatment
2.7. Chromosomal Mapping Analysis of the LBD Gene Family in Rice
3. Discussion
4. Materials and Methods
4.1. Sources of Data
4.2. Identification and Physicochemical Properties Analysis
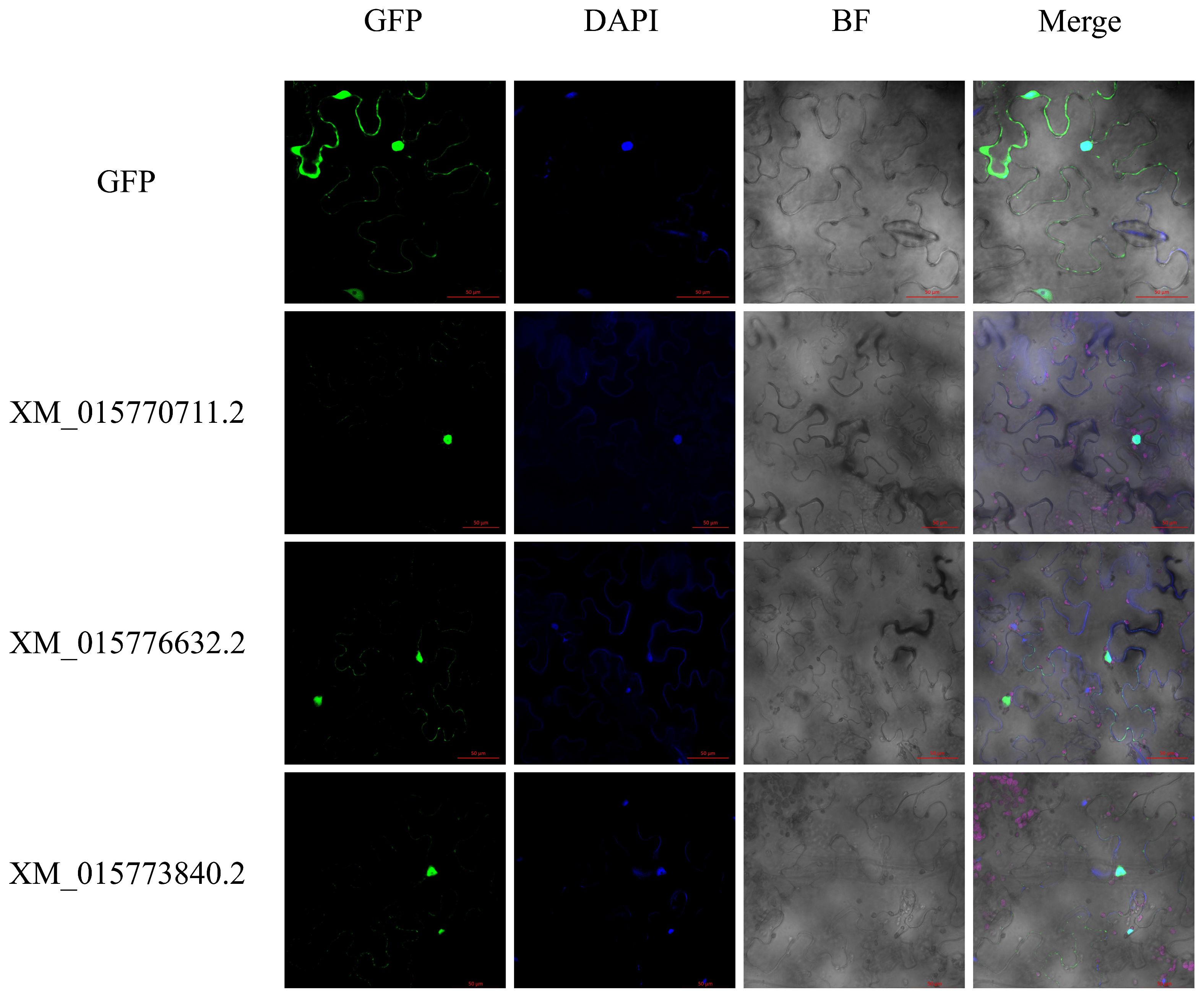
4.3. Subcellular Localization
4.4. Chromosomal Mapping Analysis
4.5. Analysis of Motifs, Gene Structures, and cis-Acting Elements
4.6. Phylogeny Analysis
4.7. Collinearity Analysis
4.8. Expression Patterns Analysis
4.9. Analysis of Transcriptome Data of Phytohormone and Stress Treatment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Iwakawa, H.; Ueno, Y.; Semiarti, E.; Onouchi, H.; Kojima, S.; Tsukaya, H.; Hasebe, M.; Soma, T.; Ikezaki, M.; Machida, C.; et al. The ASYMMETRIC LEAVES2 Gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002, 43, 467–478. [Google Scholar] [CrossRef]
- Matsumura, Y.; Iwakawa, H.; Machida, Y.; Machida, C. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J. 2009, 58, 525–537. [Google Scholar] [CrossRef]
- Shuai, B.; Reynaga-Pena, C.G.; Springer, P.S. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002, 129, 747–761. [Google Scholar] [CrossRef]
- Xie, T.; Zeng, L.; Chen, X.; Rong, H.; Wu, J.; Batley, J.; Jiang, J.; Wang, Y. Genome-Wide analysis of the lateral organ boundaries domain gene family in Brassica Napus. Genes 2020, 11, 280. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Cao, M.; Chen, X.; Ye, M.; Zhao, P.; Nan, Y.; Li, W.; Zhang, C.; Kong, L.; Kong, N.; et al. Genome-Wide analysis of the lateral organ boundaries domain (LBD) gene family in Solanum tuberosum. Int. J. Mol. Sci. 2019, 20, 5360. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Xie, Q.; Li, C.; Dong, Y.; Zhu, S.; Chen, J. Comprehensive characterization and gene expression patterns of LBD gene family in Gossypium. Planta 2020, 251, 81. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, R.; Cheng, Y.; Lei, P.; Song, W.; Zheng, W.; Nie, X. Genome-Wide identification, evolution, and expression analysis of LBD transcription factor family in bread wheat (Triticum aestivum L.). Front. Plant Sci. 2021, 12, 721253. [Google Scholar] [CrossRef]
- Tian, Y.; Han, X.; Qu, Y.; Zhang, Y.; Rong, H.; Wu, K.; Xu, L. Genome-wide identification of the Ginkgo (Ginkgo biloba L.) LBD transcription factor gene and characterization of its expression. Int. J. Mol. Sci. 2022, 23, 5474. [Google Scholar] [CrossRef]
- Li, K.; Wei, Y.; Wang, Y.; Tan, B.; Chen, S.; Li, H. Genome-Wide Identification of LBD Genes in Foxtail Millet (Setaria italica) and Functional Characterization of SiLBD21. Int. J. Mol. Sci. 2023, 24, 7110. [Google Scholar] [CrossRef]
- Jia, R.; Li, C.; Wang, Y.; Qin, X.; Meng, L.; Sun, X. Genome-wide analysis of LBD transcription factor genes in Dendrobiumcatenatum. Int. J. Mol. Sci. 2022, 23, 2089. [Google Scholar] [CrossRef]
- Kim, S.H.; Bahk, S.; An, J.; Hussain, S.; Nguyen, N.T.; Do, H.L.; Kim, J.Y.; Hong, J.C.; Chung, W.S. A Gain-of-function mutant of IAA15 inhibits lateral root development by transcriptional repression of LBD genes in Arabidopsis. Front. Plant Sci. 2020, 11, 1239. [Google Scholar] [CrossRef]
- Rubin, G.; Tohge, T.; Matsuda, F.; Saito, K.; Scheible, W.R. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 2009, 21, 3567–3584. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Kim, M.; Lee, M.R.; Park, S.K.; Kim, J. LATERAL ORGAN BOUNDARIES DOMAIN (LBD)10 interacts with SIDECAR POLLEN/LBD27 to control pollen development in Arabidopsis. Plant J. 2015, 81, 794–809. [Google Scholar] [CrossRef]
- Fisher, K.; Turner, S. PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr. Biol. 2007, 17, 1061–1066. [Google Scholar] [CrossRef] [PubMed]
- Ikeuchi, M.; Sugimoto, K.; Iwase, A. Plant callus: Mechanisms of induction and repression. Plant Cell 2013, 25, 3159–3173. [Google Scholar] [CrossRef] [PubMed]
- Okushima, Y.; Fukaki, H.; Onoda, M.; Theologis, A.; Tasaka, M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 2007, 19, 118–130. [Google Scholar] [CrossRef]
- Xu, C.; Cao, H.; Xu, E.; Zhang, S.; Hu, Y. Genome-Wide Identification of Arabidopsis LBD29 target genes reveals the molecular events behind auxin-induced cell reprogramming during callus formation. Plant Cell Physiol. 2018, 59, 744–755. [Google Scholar] [CrossRef]
- Kim, J.; Lee, H.W. Direct activation of EXPANSIN14 by LBD18 in the gene regulatory network of lateral root formation in Arabidopsis. Plant Signal. Behav. 2013, 8, e22979. [Google Scholar] [CrossRef][Green Version]
- Lee, H.W.; Cho, C.; Kim, J. Lateral organ boundaries domain16 and 18 act downstream of the AUXIN1 and LIKE-AUXIN3 auxin influx carriers to control lateral root development in Arabidopsis. Plant Physiol. 2015, 168, 1792–1806. [Google Scholar] [CrossRef]
- Berckmans, B.; Vassileva, V.; Schmid, S.P.; Maes, S.; Parizot, B.; Naramoto, S.; Magyar, Z.; Alvim, K.C.; Koncz, C.; Bogre, L.; et al. Auxin-dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins. Plant Cell 2011, 23, 3671–3683. [Google Scholar] [CrossRef]
- Soyano, T.; Thitamadee, S.; Machida, Y.; Chua, N.H. ASYMMETRIC LEAVES2-LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell 2008, 20, 3359–3373. [Google Scholar] [CrossRef]
- Yamauchi, T.; Tanaka, A.; Inahashi, H.; Nishizawa, N.K.; Tsutsumi, N.; Inukai, Y.; Nakazono, M. Fine control of aerenchyma and lateral root development through AUX/IAA- and ARF-dependent auxin signaling. Proc. Natl. Acad. Sci. USA 2019, 116, 20770–20775. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, J.; Li, L.; Li, J.; Zhuang, M.; Li, B.; Li, Q.; Huang, J.; Du, Y.; Wang, J.; et al. TaMOR is essential for root initiation and improvement of root system architecture in wheat. Plant Biotechnol. J. 2022, 20, 862–875. [Google Scholar] [CrossRef]
- Zhu, L.; Zheng, C.; Liu, R.; Song, A.; Zhang, Z.; Xin, J.; Jiang, J.; Chen, S.; Zhang, F.; Fang, W.; et al. Chrysanthemum transcription factor CmLBD1 direct lateral root formation in Arabidopsis thaliana. Sci. Rep. 2016, 6, 20009. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.C.; Shuai, B.; Springer, P.S. The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 2003, 15, 2241–2252. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wang, F.; Guo, J.; Zhang, X.S. Rice OsAS2 Gene, a member of LOB domain family, functions in the regulation of shoot differentiation and leaf development. J. Plant Biol. 2009, 52, 374–381. [Google Scholar] [CrossRef]
- Xu, B.; Li, Z.; Zhu, Y.; Wang, H.; Ma, H.; Dong, A.; Huang, H. Arabidopsis genes AS1, AS2, and JAG negatively regulate boundary-specifying genes to promote sepal and petal development. Plant Physiol. 2008, 146, 566–575. [Google Scholar] [CrossRef]
- Kim, M.J.; Kim, M.; Kim, J. Combinatorial interactions between LBD10 and LBD27 are essential for male gametophyte development in Arabidopsis. Plant Signal. Behav. 2015, 10, e1044193. [Google Scholar] [CrossRef]
- Zhang, J.; Tang, W.; Huang, Y.; Niu, X.; Zhao, Y.; Han, Y.; Liu, Y. Down-regulation of a LBD-like gene, OsIG1, leads to occurrence of unusual double ovules and developmental abnormalities of various floral organs and megagametophyte in rice. J. Exp. Bot. 2015, 66, 99–112. [Google Scholar] [CrossRef]
- Ye, L.; Wang, X.; Lyu, M.; Siligato, R.; Eswaran, G.; Vainio, L.; Blomster, T.; Zhang, J.; Mahonen, A.P. Cytokinins initiate secondary growth in the Arabidopsis root through a set of LBD genes. Curr. Biol. 2021, 31, 3365–3373. [Google Scholar] [CrossRef]
- Li, C.; Zhu, S.; Zhang, H.; Chen, L.; Cai, M.; Wang, J.; Chai, J.; Wu, F.; Cheng, Z.; Guo, X.; et al. OsLBD37 and OsLBD38, two class II type LBD proteins, are involved in the regulation of heading date by controlling the expression of Ehd1 in rice. Biochem. Biophys. Res. Commun. 2017, 486, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; He, W.; Wang, L.; Zhang, X.; Wang, K.; Xiang, Y. PheLBD29, an LBD transcription factor from Moso bamboo, causes leaf curvature and enhances tolerance to drought stress in transgenic Arabidopsis. J. Plant Physiol. 2023, 280, 153865. [Google Scholar] [CrossRef] [PubMed]
- Yordanov, Y.S.; Regan, S.; Busov, V. Members of the LATERAL ORGAN BOUNDARIES DOMAIN transcription factor family are involved in the regulation of secondary growth in Populus. Plant Cell 2010, 22, 3662–3677. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Yang, T.; Guo, Y.; Cui, W.H.; Yao, L.J.; Li, G.; Wu, A.M.; Li, J.H.; Liu, L.J. The transcription factor PagLBD3 contributes to the regulation of secondary growth in Populus. J. Exp. Bot. 2021, 72, 7092–7106. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, J.; Xu, J.; Li, Y.; Guo, L.; Wang, Z.; Zhang, X.; Zhao, B.; Guo, Y.D.; Zhang, N. CRISPR/Cas9 targeted mutagenesis of SlLBD40, a lateral organ boundaries domain transcription factor, enhances drought tolerance in tomato. Plant Sci. 2020, 301, 110683. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, J.; Xu, J.; Li, Y.; Lv, H.; Wang, F.; Guo, J.; Lin, T.; Zhao, B.; Li, X.X.; et al. SlMYC2 promotes SlLBD40-mediated cell expansion in tomato fruit development. Plant J. 2024, 118, 1872–1888. [Google Scholar] [CrossRef]
- Thatcher, L.F.; Powell, J.J.; Aitken, E.A.; Kazan, K.; Manners, J.M. The lateral organ boundaries domain transcription factor LBD20 functions in Fusarium wilt Susceptibility and jasmonate signaling in Arabidopsis. Plant Physiol. 2012, 160, 407–418. [Google Scholar] [CrossRef]
- Zhao, D.; Chen, P.; Chen, Z.; Zhang, L.; Wang, Y.; Xu, L. Genome-wide analysis of the LBD family in rice: Gene functions, structure and evolution. Comput. Biol. Med. 2023, 153, 106452. [Google Scholar] [CrossRef]
- Lin, M.; Yan, J.; Ali, M.M.; Wang, S.; Tian, S.; Chen, F.; Lin, Z. Isolation and functional characterization of a green-tissue promoter in Japonica Rice (Oryza sativa subsp. Japonica). Biology 2022, 11, 1092. [Google Scholar] [CrossRef]
- Liu, L.; Xu, W.; Hu, X.; Liu, H.; Lin, Y. W-box and G-box elements play important roles in early senescence of rice flag leaf. Sci. Rep. 2016, 6, 20881. [Google Scholar] [CrossRef]
- Prince, V.E.; Pickett, F.B. Splitting pairs: The diverging fates of duplicated genes. Nat. Rev. Genet. 2002, 3, 827–837. [Google Scholar] [CrossRef]
- Conant, G.C.; Wolfe, K.H. Turning a hobby into a job: How duplicated genes find new functions. Nat. Rev. Genet. 2008, 9, 938–950. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, J.; Diaz-Manzano, F.E.; Sanchez, M.; Rosso, M.N.; Melillo, T.; Goh, T.; Fukaki, H.; Cabello, S.; Hofmann, J.; Fenoll, C.; et al. A role for LATERAL ORGAN BOUNDARIES-DOMAIN 16 during the interaction Arabidopsis-Meloidogyne spp. provides a molecular link between lateral root and root-knot nematode feeding site development. New Phytol. 2014, 203, 632–645. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.Z.; Xue, Y.J.; Zhou, J.J.; Zhang, B.; Chang, H.; Takano, M. Phytochromes regulate SA and JA signaling pathways in rice and are required for developmentally controlled resistance to Magnaporthe grisea. Mol. Plant 2011, 4, 688–696. [Google Scholar] [CrossRef]
- Nahar, K.; Kyndt, T.; De Vleesschauwer, D.; Hofte, M.; Gheysen, G. The jasmonate pathway is a key player in systemically induced defense against root knot nematodes in rice. Plant Physiol. 2011, 157, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Yang, F.; Xu, X.; Peng, Y.; Ji, H. Salicylic Acid, Jasmonate, and Ethylene Contribute to Rice Defense Against White Tip Nematodes Aphelenchoides besseyi. Front. Plant Sci. 2021, 12, 755802. [Google Scholar] [CrossRef]
- Shi, L.; Guo, M.; Ye, N.; Liu, Y.; Liu, R.; Xia, Y.; Cui, S.; Zhang, J. Reduced ABA accumulation in the root system is caused by ABA exudation in upland rice (Oryza sativa L. var. Gaoshan1) and this enhanced drought adaptation. Plant Cell Physiol. 2015, 56, 951–964. [Google Scholar] [CrossRef]
- Schmidt, R.; Schippers, J.H.; Welker, A.; Mieulet, D.; Guiderdoni, E.; Mueller-Roeber, B. Transcription factor OsHsfC1b regulates salt tolerance and development in Oryza sativa ssp. japonica. AoB Plants 2012, 2012, s11. [Google Scholar] [CrossRef]
- Jiao, P.; Wei, X.; Jiang, Z.; Liu, S.; Guan, S.; Ma, Y. ZmLBD2 a maize (Zea mays L.) lateral organ boundaries domain (LBD) transcription factor enhances drought tolerance in transgenic Arabidopsis thaliana. Front. Plant Sci. 2022, 13, 1000149. [Google Scholar] [CrossRef]
- Feng, X.; Xiong, J.; Zhang, W.; Guan, H.; Zheng, D.; Xiong, H.; Jia, L.; Hu, Y.; Zhou, H.; Wen, Y.; et al. ZmLBD5, a class-II LBD gene, negatively regulates drought tolerance by impairing abscisic acid synthesis. Plant J. 2022, 112, 1364–1376. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Feng, Z.; Liu, X.; Bian, L.; Xie, H.; Zhang, C.; Mysore, K.S.; Liang, J. MiR393 and miR390 synergistically regulate lateral root growth in rice under different conditions. BMC Plant Biol. 2018, 18, 261. [Google Scholar] [CrossRef]
- Maruyama, K.; Urano, K.; Yoshiwara, K.; Morishita, Y.; Sakurai, N.; Suzuki, H.; Kojima, M.; Sakakibara, H.; Shibata, D.; Saito, K.; et al. Integrated analysis of the effects of cold and dehydration on rice metabolites, phytohormones, and gene transcripts. Plant Physiol. 2014, 164, 1759–1771. [Google Scholar] [CrossRef]
- Ba, L.J.; Kuang, J.F.; Chen, J.Y.; Lu, W.J. MaJAZ1 attenuates the MaLBD5-Mediated transcriptional activation of jasmonate biosynthesis gene MaAOC2 in regulating cold tolerance of Banana Fruit. J. Agric. Food Chem. 2016, 64, 738–745. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, X.; Wu, P. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol. Phylogenet. Evol. 2006, 39, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Cho, C.; Jeon, E.; Pandey, S.K.; Ha, S.H.; Kim, J. LBD13 positively regulates lateral root formation in Arabidopsis. Planta 2019, 249, 1251–1258. [Google Scholar] [CrossRef]
- Ma, W.; Wu, F.; Sheng, P.; Wang, X.; Zhang, Z.; Zhou, K.; Zhang, H.; Hu, J.; Lin, Q.; Cheng, Z.; et al. The LBD12-1 transcription factor suppresses apical meristem size by repressing argonaute 10 expression. Plant Physiol. 2017, 173, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Inukai, Y.; Sakamoto, T.; Ueguchi-Tanaka, M.; Shibata, Y.; Gomi, K.; Umemura, I.; Hasegawa, Y.; Ashikari, M.; Kitano, H.; Matsuoka, M. Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 2005, 17, 1387–1396. [Google Scholar] [CrossRef] [PubMed]
- To, H.; Pham, D.T.; Le Thi, V.A.; Nguyen, T.T.; Tran, T.A.; Ta, A.S.; Chu, H.H.; Do, P.T. The Germin-like protein OsGER4 is involved in promoting crown root development under exogenous jasmonic acid treatment in rice. Plant J. 2022, 112, 860–874. [Google Scholar] [CrossRef]
- Geng, L.; Tan, M.; Deng, Q.; Wang, Y.; Zhang, T.; Hu, X.; Ye, M.; Lian, X.; Zhou, D.X.; Zhao, Y. Transcription factors WOX11 and LBD16 function with histone demethylase JMJ706 to control crown root development in rice. Plant Cell 2024, 36, 1777–1790. [Google Scholar] [CrossRef]
- Geng, L.; Li, Q.; Jiao, L.; Xiang, Y.; Deng, Q.; Zhou, D.X.; Zhao, Y. WOX11 and CRL1 act synergistically to promote crown root development by maintaining cytokinin homeostasis in rice. New Phytol. 2023, 237, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zou, X.; Zhang, C.; Shao, Q.; Liu, J.; Liu, B.; Li, H.; Zhao, T. OsLBD3-7 Overexpression induced adaxially rolled leaves in Rice. PLoS ONE 2016, 11, e156413. [Google Scholar] [CrossRef]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]

| Accession Number | Gene ID | Gene Name | Protein Size (aa) | MV | PI | GRAVI | Subcellular Localizatio |
|---|---|---|---|---|---|---|---|
| XM_015788984.2 | LOC_Os01g03890.1 | OsLBD40-1 | 307 | 32,379.55 | 6.89 | −0.297 | Nucleus. |
| XM_015792520.2 | LOC_Os01g07480.1 | OsLBD25/OsAS2 | 251 | 25,728.12 | 7.61 | 0.003 | Nucleus. |
| XM_015788621.2 | LOC_Os01g14030.1 | OsLBD2 | 249 | 26,091.37 | 9.72 | −0.321 | Nucleus. |
| XM_015772758.2 | LOC_Os01g32770.1 | OsLBD40-2 | 330 | 35,019.79 | 5.36 | −0.399 | Nucleus. |
| XM_015782803.2 | LOC_Os01g39040.1 | OsLBD6-1 | 356 | 38,510.31 | 4.9 | −0.396 | Nucleus. |
| XM_015785013.2 | LOC_Os01g39070.1 | OsLBD22-1 | 125 | 13,306.08 | 6.26 | −0.425 | Nucleus. |
| XM_015782813.1 | LOC_Os01g39150.1 | OsLBD11 | 202 | 21,537.04 | 7.77 | −0.682 | Nucleus. |
| XM_015782824.1 | LOC_Os01g39160.1 | OsLBD24 | 171 | 17,865.16 | 5.25 | −0.127 | Nucleus. |
| XM_026023149.1 | LOC_Os01g39220.1 | OsLBD22-like | 117 | 12,500.28 | 6.89 | −0.173 | Nucleus. |
| XM_015774606.2 | LOC_Os01g56530.1 | OsLBD20 | 245 | 25,730.04 | 7.72 | −0.147 | Nucleus. |
| XM_015780552.2 | LOC_Os01g60960.1 | OsLBD15-1/OsLBD1-8 | 251 | 26,493 | 6.45 | −0.173 | Nucleus. |
| XM_015779215.2 | LOC_Os01g66590.1 | OsLBD6-like | 269 | 27,621.93 | 8.17 | −0.08 | Nucleus. |
| XM_015769048.1 | LOC_Os02g48270.1 | OsLBD12-1 | 140 | 15,868.16 | 6.81 | −0.481 | Nucleus. |
| XM_015770711.2 | LOC_Os02g57490.1 | OsLBD16-1 | 216 | 22,457.64 | 8.09 | 0.103 | Nucleus. |
| XM_015776632.2 | LOC_Os03g05500.1 | OsLBD16-2 | 185 | 19,631.01 | 6.05 | −0.186 | Nucleus. |
| XM_015776633.2 | LOC_Os03g05510.1 | OsLBD29-1/OsCRL1-like | 259 | 26,736.57 | 5.96 | −0.085 | Nucleus. |
| XM_015773927.2 | LOC_Os03g14270.1 | OsLBD30-1 | 275 | 28,252.75 | 8.27 | −0.321 | Nucleus. |
| XM_015773840.2 | LOC_Os03g17810.1 | OsLBD1-1 | 239 | 25,556.79 | 6.06 | −0.241 | Nucleus. |
| XM_015773867.2 | LOC_Os03g33090.1 | OsLBD37-1 | 204 | 21,037.1 | 8.41 | 0.042 | Nucleus. |
| XM_015772725.2 | LOC_Os03g41330.1 | OsLBD38 | 232 | 23,618.55 | 6.05 | 0.023 | Nucleus. |
| XM_015773878.2 | LOC_Os03g41600.1 | OsLBD36 | 307 | 33,077.8 | 6.65 | −0.577 | Nucleus. |
| XM_015774114.2 | LOC_Os03g45750.1 | OsLBD4 | 214 | 22,012.96 | 9.06 | −0.131 | Nucleus. |
| XM_026023997.1 | LOC_Os03g57670.1 | OsLBD23/OsIAL1-like | 171 | 18,322.4 | 7.63 | −0.458 | Nucleus. |
| XM_015783394.2 | LOC_Os05g03160.1 | OsLBD1-2 | 216 | 22,365.67 | 6.57 | 0.125 | Nucleus. |
| XM_026025700.1 | LOC_Os05g07270.1 | OsLBD15-2 | 241 | 25,300.6 | 8.77 | −0.078 | Nucleus. |
| XM_015783373.2 | LOC_Os05g27980.1 | OsLBD12-2 | 200 | 21,105.84 | 6.28 | −0.145 | Nucleus. |
| XM_015784584.2 | LOC_Os05g34450.1 | OsLBD6-2 | 279 | 28,044.35 | 8.23 | −0.004 | Nucleus. |
| XM_015791721.2 | LOC_Os07g40000.1 | OsLBD37-2 | 226 | 23,216.21 | 7.54 | −0.115 | Nucleus. |
| XM_015794036.2 | LOC_Os08g06659.1 | OsLBD1-3 | 196 | 21,283.44 | 4.62 | −0.88 | Nucleus. |
| XM_026027477.1 | LOC_Os08g31080.1 | OsLBD22-2 | 456 | 48,653.41 | 4.69 | −0.474 | Nucleus. |
| XM_026027463.1 | LOC_Os08g40520.1 | OsLBD13 | 103 | 11,163.64 | 4.34 | −0.565 | Nucleus. |
| XM_015792766.2 | LOC_Os08g44940.1 | OsLBD29-2/OsCRL1-like | 215 | 23,239.17 | 5.85 | −0.16 | Nucleus. |
| XM_015755677.2 | LOC_Os09g19950.1 | OsLBD22-3/OsIAL1 | 362 | 37,637.55 | 5.27 | −0.397 | Nucleus. |
| XM_015758344.2 | LOC_Os10g07510.1 | OsLBD30-2 | 269 | 27,974.12 | 6.41 | −0.336 | Nucleus. |
| XM_026021321.1 | LOC_Os11g01550.1 | OsLBD12-3 | 156 | 17,404.87 | 6.42 | −0.254 | Nucleus. |
| XM_015762595.2 | LOC_Os12g01550.1 | OsLBD12-4 | 156 | 17,404.87 | 6.42 | −0.254 | Nucleus. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, S.; Yi, J.; Gu, P.; Huang, Y.; Huang, X.; Li, H.; Fan, T.; Zhao, J.; Wang, R.; Gaballah, M.M.; et al. Comprehensive Characterization and Functional Analysis of the Lateral Organ Boundaries Domain Gene Family in Rice: Evolution, Expression, and Stress Response. Int. J. Mol. Sci. 2025, 26, 3948. https://doi.org/10.3390/ijms26093948
Sun S, Yi J, Gu P, Huang Y, Huang X, Li H, Fan T, Zhao J, Wang R, Gaballah MM, et al. Comprehensive Characterization and Functional Analysis of the Lateral Organ Boundaries Domain Gene Family in Rice: Evolution, Expression, and Stress Response. International Journal of Molecular Sciences. 2025; 26(9):3948. https://doi.org/10.3390/ijms26093948
Chicago/Turabian StyleSun, Shang, Jingjing Yi, Peiling Gu, Yongtian Huang, Xin Huang, Hanqing Li, Tingting Fan, Jing Zhao, Ruozhong Wang, Mahmoud Mohamed Gaballah, and et al. 2025. "Comprehensive Characterization and Functional Analysis of the Lateral Organ Boundaries Domain Gene Family in Rice: Evolution, Expression, and Stress Response" International Journal of Molecular Sciences 26, no. 9: 3948. https://doi.org/10.3390/ijms26093948
APA StyleSun, S., Yi, J., Gu, P., Huang, Y., Huang, X., Li, H., Fan, T., Zhao, J., Wang, R., Gaballah, M. M., Xiao, L., & Li, H. (2025). Comprehensive Characterization and Functional Analysis of the Lateral Organ Boundaries Domain Gene Family in Rice: Evolution, Expression, and Stress Response. International Journal of Molecular Sciences, 26(9), 3948. https://doi.org/10.3390/ijms26093948







